Abstract
Microglia, the resident macrophages of the brain, play a key role in the pathogenesis of Human Immunodeficiency Virus (HIV)-associated neurocognitive disorders (HAND) due to their productive infection by HIV. This results in the release of neurotoxic viral proteins and pro-inflammatory compounds which negatively affect the functionality of surrounding neurons. Because models of HIV infection within the brain are limited, we aimed to create a novel microglia cell line with an integrated HIV-provirus capable of recreating several hallmarks of HIV infection. We utilized CRISPR/Cas9 gene editing technology and integrated a modified HIV provirus into CHME-5 immortalized microglia to create HIV-NanoLuc CHME-5. In the modified provirus, the Gag-Pol region is replaced with the coding region for Nanoluciferase (NanoLuc), which allows for the rapid assay of HIV LTR activity using a luminescent substrate, while still containing the necessary genetic material to produce established neurotoxic viral proteins (e.g. tat, nef, gp120). We confirmed that HIV-NanoLuc CHME-5 microglia express NanoLuc, along with the HIV viral protein Nef. We subsequently exposed these cells to a battery of experiments to modulate the activity of the provirus. Proviral activity was enhanced by treating the cells with pro-inflammatory factors LPS and TNF-α and by overexpressing the viral regulatory protein Tat. Conversely, genetic modification of the TLR-4 gene by CRISPR/Cas9 reduced LPS-mediated proviral activation, and pharmacological application of NF-κB inhibitor sulfasalazine similarly diminished proviral activity. Overall these data suggest that HIV-NanoLuc CHME-5 may be a useful tool in the study of HIV-mediated neuropathology and proviral regulation.
Keywords: HIV, CRISPR/Cas9, Microglia, NanoLuciferase, NeuroAIDS
Graphical Abstract
In the brain, microglia are the resident macrophage and the prominent cell type infected by the Human Immunodeficiency Virus (HIV). We have developed a renewable microglia cell line harboring a non-replicative HIV provirus with a luciferase based reporter termed HIV-NanoLuc CHME-5 E9. This cell line is amenable to pharmacological, genetic, and viral manipulations to study HIV proviral activity.
1. Introduction
The presence of Human Immunodeficiency Virus-1 (HIV) infected cells within the central nervous system (CNS) remains an important facet of the disease. Brain-resident glial cell populations including microglia and astrocytes are infected by the virus [1, 2], negatively impacting the functionality and viability of surrounding neurons through mechanisms involving pro-inflammatory cytokine release [3-5], neurotrophic dysregulation [6, 7], aberrant glutamate homeostasis [8, 9], and direct damage from viral protein production [10, 11]. These mechanisms are thought to contribute to the development of HIV-associated neurocognitive disorders (HAND), a spectrum disorder which adversely affects cognitive, motor, and behavioral processes [12, 13]. Furthermore, it has been established that microglia harbor latent provirus [1, 14], which can be activated by certain pro-inflammatory stimuli (e.g. interleukin-1β, tumor necrosis factor-α) and leads to productive viral replication [15]. Thus, the microglia within the CNS may serve as “viral reservoirs”, allowing for viral compartmentalization, evolution, and escape [16, 17]. Understanding the pathobiology of HIV-infected microglia is paramount to the development of novel therapeutic approaches that delay or cure HIV-mediated pathogenesis in the CNS.
To date, the models used to study HIV-infected microglia are limited. For in vitro studies, a common method involves the introduction of one or more recombinant HIV viral proteins (e.g. tat, nef, gp120) into microglia/neuronal cultures and assessing the pro-inflammatory/cytotoxic effects [18-20]. While this method is useful to identify the individual contributions of viral proteins in microglial activation, the relevance to neuroinflammation caused by HIV-infected microglia releasing these factors in combination is unclear. For example, understanding how the state of microglia influences transcriptional regulation and viral protein production from a harbored provirus is not possible from exposure of single viral proteins. One approach to address this issue, is the use of primary cultures of human microglia infected with strains of HIV [21, 22] leading to productive infection and the release of infectious virus and viral proteins. However, the limited availability and the rigorous culture conditions necessary to utilize primary human microglia is a disadvantage when compared to microglial cell lines. Therefore, a reusable microglia model which harbors a stably integrated HIV provirus and allows consistent high throughput experimentation would be of value.
The microglia cell line CHME-5 was originally created through SV40 transfection of human embryonic brain derived macrophages [23]. Recent studies have used this cell line to examine microglia processes including toll-like receptor expression and signaling [24], arginase activity [25], and drug abuse/HIV mediated impact on microglial energetic metabolism [26]. However, recent data acquired from PCR analysis of human vs. rat CYCT1 shows that CHME-5 cells are rat in origin [27]. Given their established properties of a microglial cell, we chose to use this cell line to express our modified HIV provirus to allow for comparisons with previously published data on CHME-5/HIV cell lines [28].
Here, our lab utilized the CRISPR/Cas9 system and integrated a modified HIV-provirus. This modified provirus has the Gag-Pol region replaced by Nanoluciferase (NanoLuc), which has the dual purpose of rendering the provirus replication-defective and creating a rapid and robust assay for monitoring the transcriptional activity associated with the long terminal repeat (LTR). Furthermore, coding regions for the neurotoxic viral proteins such as tat, nef and gp120 remain intact. In this study, we generated stable cell lines by integrating a modified HIV provirus and compared the LTR activity and pro-inflammatory phenotype of four selected clones. Using pharmacological and genetic manipulation, we demonstrated that the modified provirus expresses a Nanoluciferase reporter and viral proteins in a manner consistent with activity of a wild-type HIV provirus. Collectively, we developed an in vitro model of HIV-infected microglia to study the pathobiology and evaluate therapeutics for HIV-associated neuroinflammation and neurodegeneration.
2. Results
2.1. Generation of a stable microglial cell line containing a modified HIV provirus targeted to the ROSA26 locus
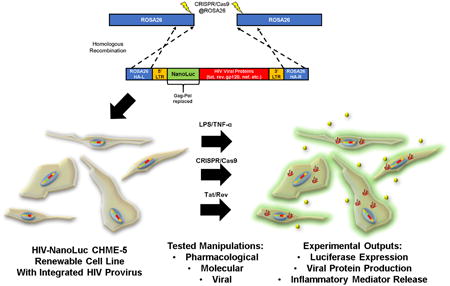
We designed a targeting construct to integrate a replication defective HIV (YU-2, M-tropic) provirus in the ROSA26 locus (Fig. 1A), a “safe harbor” locus for expressing transgenes based on their cis-containing elements [29]. A safe harbor locus is an area of the genome that is resistant to epigenetic silencing, ubiquitously expressed in all tissues, and the transcription of surrounding genes remains intact after foreign transgene insertion. In our modified HIV provirus, the gag-pol genes were replaced with a Nanoluciferase (NanoLuc) coding sequence to enable monitoring of HIV LTR activation. The resulting construct, “HIV-NanoLuc” (Fig. 1A) was co-transfected with plasmids expressing Cas9 nickase (Cas9n) and gRNAs targeting the ROSA26 locus of CHME-5 cells, a microglia cell line. The nickase form of Cas9 system utilizes two independent gRNAs to create proximal nicks in target DNA and greatly decreases off-target mutation events in the genome [30].
Figure 1. Using CRISPR/Cas9 to stably integrate the HIV-NanoLuc provirus at the ROSA26 locus.
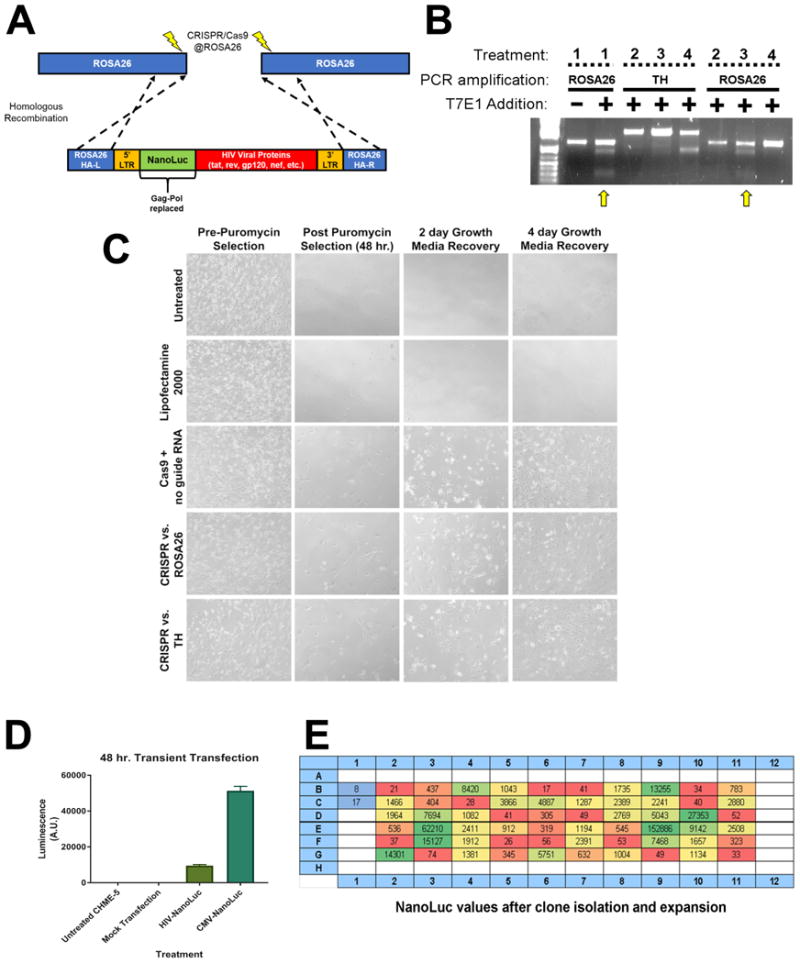
(A) Model of modified HIV provirus targeting to ROSA26 locus. Cas9 nickase is used to create mutagenic events at ROSA26. The HIV-NanoLuc provirus is flanked by ROSA26 homologous arms to facilitate integration. (B) CHME5 cells were transfected with CRISPR/Cas9 components and gRNAs targeting the ROSA26 locus, or tyrosine hydroxylase (TH) as a control. Puromycin selection was performed to select cell populations that were successfully transfected. PCR was then performed on genomic DNA to amplify the ROSA26 or TH genomic sequences. T7 endonuclease 1 enzyme was added to identify mutagenic events, and shows specific targeting of the ROSA26 locus in CHME-5 microglia by CRISPR/Cas9. Treatments are as follows 1: PC12 cells + ROSA26 gRNA (positive control), 2: CHME-5 cells + no gRNA, 3: CHME-5 cells + ROSA26 gRNA, 4: CHME-5 cells + TH gRNA. (C) Photomicrographs of CHME-5 cells undergoing puromycin selection. (D) Transient transfection of the HIV-NanoLuc provirus in CHME-5 microglia results in luciferase activity. Mock transfection of pBSII or a CMV driven NanoLuc were used as a negative and positive control respectively. (E) CRISPR/Cas9 at the ROSA26 locus was performed with the addition of the donor HIV-nanoLuc provirus. After puromycin selection, cells were clonally isolated in a 96 well plate and assayed for luciferase activity. Wells B1, B2: Parental CHME-5, Red wells: low NanoLuc expression, Yellow wells: medium NanoLuc expression, Green wells: High NanoLuc expression.
We first confirmed CRISPR/Cas9n targeting of the ROSA26 locus. ROSA26-specific CRISPR/Cas9n components were transfected into CHME-5 cells followed by puromycin selection and regrowth (Fig. 1C). A combination of Cas9n and ROSA26 gRNAs caused mutations and genetic diversity at the ROSA26 locus in CHME-5 cells as verified using the T7E1 assay [31]. Cas9n + gRNAs to ROSA26 showed evidence of nuclease activity at the ROSA26 locus but not at the control gRNA locus TH (Fig. 1B). Conversely, Cas9n plus control gRNAs targeting TH did not show evidence of nuclease activity when ROSA26 was amplified (Fig. 1B). These data suggest that the ROSA26 locus can be successfully targeted with CRISPR/Cas9n in CHME-5 microglia. Therefore, we proceeded with co-transfection of the CRISPR/Cas9n components and the HIV-NanoLuc targeting DNA.
After transfection, puromycin selection, and regrowth, cells were plated at one cell per well in a 96 well plate and allowed to grow to confluency. We have previously observed detectable luciferase activity using transient transfections of our targeting construct (Fig. 1D). Therefore, the luciferase assay was used to evaluate our stable clones. Following expansion, cells were replica-plated and assayed for luciferase which identified a wide range of NanoLuc values (Fig. 1E). We chose four clones with relatively high levels of NanoLuc expression within the clonal populations to further characterize; E3, F3, D10 and E9. Two negative controls were included, parental CHME-5 cells and clone B2 that was subjected to transfection, puromycin selection and clonal expansion, but expressed background levels of NanoLuc (Fig 1D). To confirm ROSA26 targeted integration of the HIV provirus, we PCR amplified the 5′ junction, 3′ junction, and the Env region of the HIV-NanoLuc provirus (Fig. 2A). The 5′ junction primer pairs spanned from the 5′ genomic region of ROSA26 through the NanoLuc gene within the provirus (purple arrows; expected size 2071 bp). The 3′ junction primers spanned the Nef gene to the 3′ end of the ROSA26 genomic region (green arrows; expected side 1786 bp). To determine if truncation of the provirus occurred, primer pairs spanning a 2285 bp region (orange arrows) of the Env gene were used. After PCR amplification, we verified that clones E3, D10 and E9 have integration of the provirus at the ROSA26 locus (Fig. 2B). Furthermore, our negative controls showed no bands, suggesting specificity of our primer pairs. However, while clone F3 amplified the Env gene, improper products were observed for the 5′ and 3′ integration junctions. This may indicate off-target integration, or that recombination events occurred which altered the PCR primer sites.
Figure 2. Molecular Characterization of HIV-NanoLuc clonal cell lines.
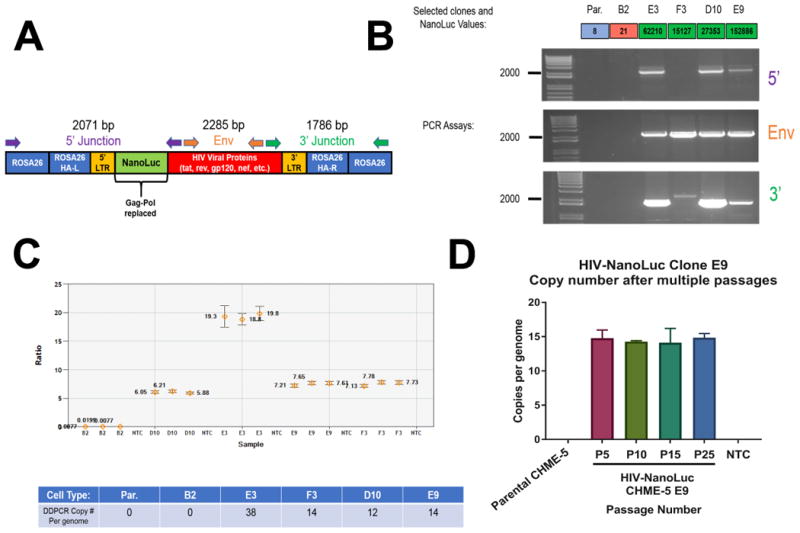
(A) Schematic of integrated provirus at the ROSA26 locus with PCR assays indicated. Junction PCR was performed to confirm proper integration. (B) PCR assays were performed on four clones with high NanoLuc expression; E3, F3, D10, E9 with parental CHME-5 and clone B2 serving as controls. Clones E3, D10 and E9 show expected bands for proper insertion. DDCPR was performed on the genomic DNA of HIV-NanoLuc clones E3, F3, D10, and E9 assaying for NanoLuc vs. autosomal gene GGT1. (C) Ratio of NanoLuc and GGT1 copy number. Table: Copy number of the provirus is ((copies/μL NanoLuc)/(copies/μL GGT1))*(2) = copy number per genome. (D) Stability of provirus integration was determined in clone E9. There is no change in copy number of the HIV provirus after multiple passages.
We used Droplet-Digital PCR (ddPCR) to assay the number of HIV-NanoLuc provirus copies within the selected clones. We compared the number of copies of NanoLuc transgene and the autosomal reference gene Ggt1 (Fig. 2C). Our results show multiple copies of the transgene sequence in clones E3, F3, D10 and E9. No amplification of NanoLuc was observed with parental CHME-5 or clone B2, our negative controls. To determine if provirus copy number is stable through multiple passages, we assayed the copy number of clone E9 at passage numbers 5, 10, 15, and 25. The copy number of the HIV provirus does not change (Fig. 2D) indicating stable integration. We proceeded with the phenotypic characterization of clones E3, D10 and E9.
2.2. Phenotypic characterization of HIV-NanoLuc clones
For the following set of validation experiments, we evaluated clones with confirmed insertion of the provirus at the ROSA26 locus- E3, D10 and E9 (pictured in Fig. 3A). We first examined each clonal line for response to a known activator of the HIV LTR promoter. We used lipopolysaccharide (100 ng/ml) as an inducer of the NF-κB transcription factor, which has been shown to bind to the HIV-LTR and enhance proviral activity [32-34]. Parental CHME-5 cells and clones D10, E3 and E9 were stimulated with LPS for 24 hrs. and lysates were prepared. Clones E3 and E9 showed a significant increase in NanoLuc activity after LPS stimulation (Fig. 3B). The lysates from these cells were also assayed for expression of the HIV protein, Nef. For clones E3 and E9 we observed a LPS-induction of a 35 kDa Nef band (Fig. 3C). Nef expression was further validated by immunohistochemistry using a Nef specific antibody (Fig. 3D). Clones E3 and E9 displayed Nef immunoreactivity. Further confirmation of antibody specificity was performed by mixing Parental CHME-5 cells with E9 cells, where Nef immunoreactivity was observed in only a portion of the population.
Figure 3. Phenotypic characterization of HIV-NanoLuc clonal cell lines.
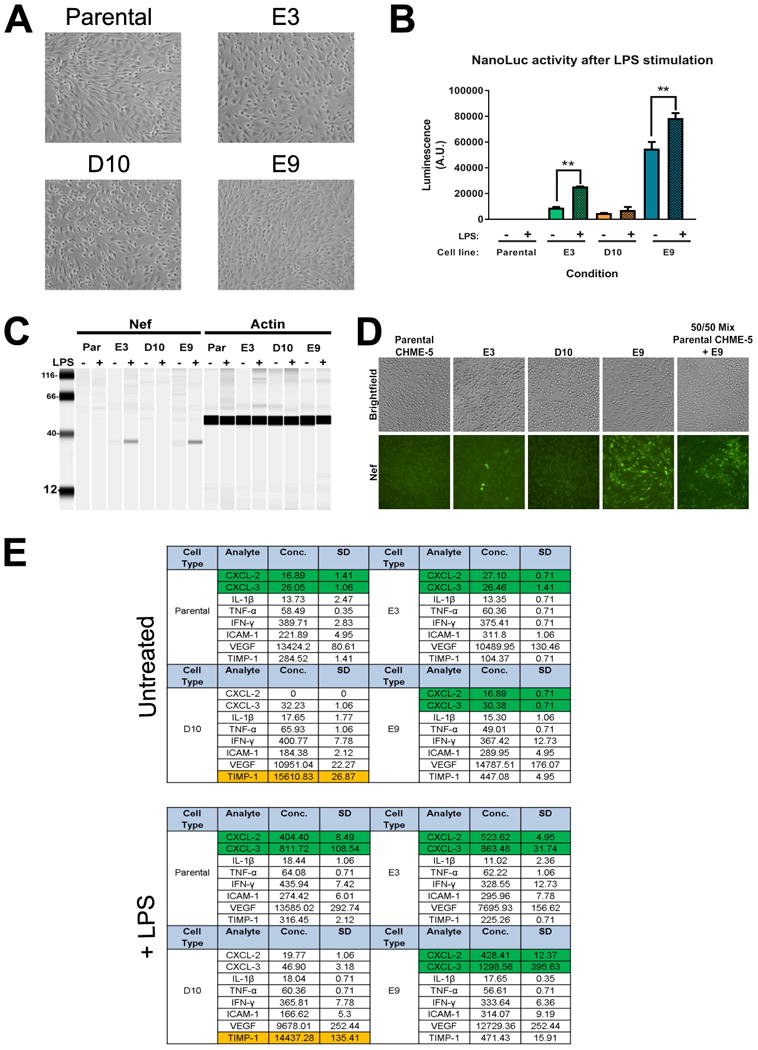
(A) Images of clones with confirmed ROSA26 proviral insertion (E3, D10, E9) and parental CHME-5 cells. (B) Clones with proper integration were stimulated with LPS (100 ng/ml) to determine if the HIV-NanoLuc provirus expression could be enhanced. Significant increases in NanoLuc activity is seen in clones E3 and E9. **p<0.01 (C) Viral protein expression of the integrated HIV-NanoLuc provirus was examined by Wes analysis for Nef. A 35 kDa protein band is visible for clones E3 and E9 before and after LPS stimulation. (D) Immunocytochemistry was performed to confirm Nef expression in clones after TNF-α (50 ng/ml) stimulation. A few E3 cells show Nef immunoreactivity while a majority of E9 cells are positive for Nef. Antibody specificity is further confirmed by mixing parental CHME-5 and E9 cells. (E) Luminex Multiplex was performed on the media of cells to determine the inflammatory profile of clones after LPS (100 ng/ml) stimulation. We observe time increases in CXCL-2 and CXCL-3 by parental CHME-3, E3 and E9 cells (Green cells, p<0.01). D10 cells failed to produce these effects. Additionally, D10 cells constitutively express TIMP-1 at a concentration much higher than other cell lines (Orange cells). Data are expressed as the mean ± SEM for (B). N=3 independent cell passages. Data are expressed at the mean + SD for (E). N=2 independent cell passages.
It is important to note that even though we confirmed stable provirus integration in clone D10, this clone failed to be responsive to LPS stimulation by both NanoLuc and Nef expression. To further understand the lack of proviral activation of clone D10, we analyzed the pro-inflammatory profile of our clones using a Luminex multiplex assay (Fig. 3E). After LPS stimulation, Parental CHME-5 cells, clone E3 and E9 display a significant induction of chemokines CXCL2 and CXCL3 while D10 does not show this effect. Furthermore, clone D10 constitutively expresses TIMP-1 several fold above the other cell lines. These data suggest that clone D10 may have a different cellular phenotype, thus altering its ability to activate the provirus. Overall, these data demonstrate the methodology and experimental caveats in using the CRISPR/Cas9 system to facilitate the integration of a modified HIV provirus to the genome of CHME-5 cells. Clone E9 was chosen for further characterization due to its robust NanoLuc expression, inducible Nef production, and pro-inflammatory phenotype. Now referred to as HIV-NanoLuc E9, we performed a series of experiments to determine the potential of this cell line for evaluating therapeutic strategies to modulate proviral activity in microglia.
2.3. Enhancing proviral activity in the HIV-NanoLuc E9 cell line
The presence of a LTR-driven NanoLuc reporter enables us to easily monitor proviral activity. Therefore, we ran a battery of different experiments in a 96 well plate format to either enhance or inhibit proviral activity in HIV-NanoLuc E9 cells. We first compared our previously shown LPS response (Fig. 3B) to pro-inflammatory mediator Tumor Necrosis Factor-α (TNF-α), a proinflammatory cytokine that promotes NF-κB signaling and activates latent HIV provirus [15]. Cells were treated with LPS (100 ng/ml) or TNF-α (50 ng/ml) by media exchange in a reverse time course for 72, 48, or 24 hours (Fig. 4A1). The cells were lysed and luciferase activity was measured for each timepoint. LPS enhanced NanoLuc activity at 24 hrs of stimulation, which decreased with longer treatment times of 48 and 72 hrs (Fig. 4A2). At the concentrations tested, TNF-α showed a stronger stimulatory effect in enhancing NanoLuc activity compared to LPS (Fig. 4A2 and 4A3). We next wanted to establish if multiple treatments with pro-inflammatory factors would further drive proviral activity. HIV-NanoLuc clone E9 cells were stimulated 1x, 2x, or 3x times with LPS or TNF-α by full media exchange and NanoLuc levels were assayed (Fig. 4B1). A single (1x) treatment of LPS increased luminescence over untreated control. A 3x treatment of LPS showed a significant enhancement of proviral activity over 1x treatment (Fig. 4B2). TNF-α also increased NanoLuc activity after single (1x) treatment, and further increased activity at 2x and 3x treatments (Fig. 4B3). Therefore, multiple pro-inflammatory stimulations can promote proviral activity.
Figure 4. Enhancing proviral activity in HIV-NanoLuc clone E9.
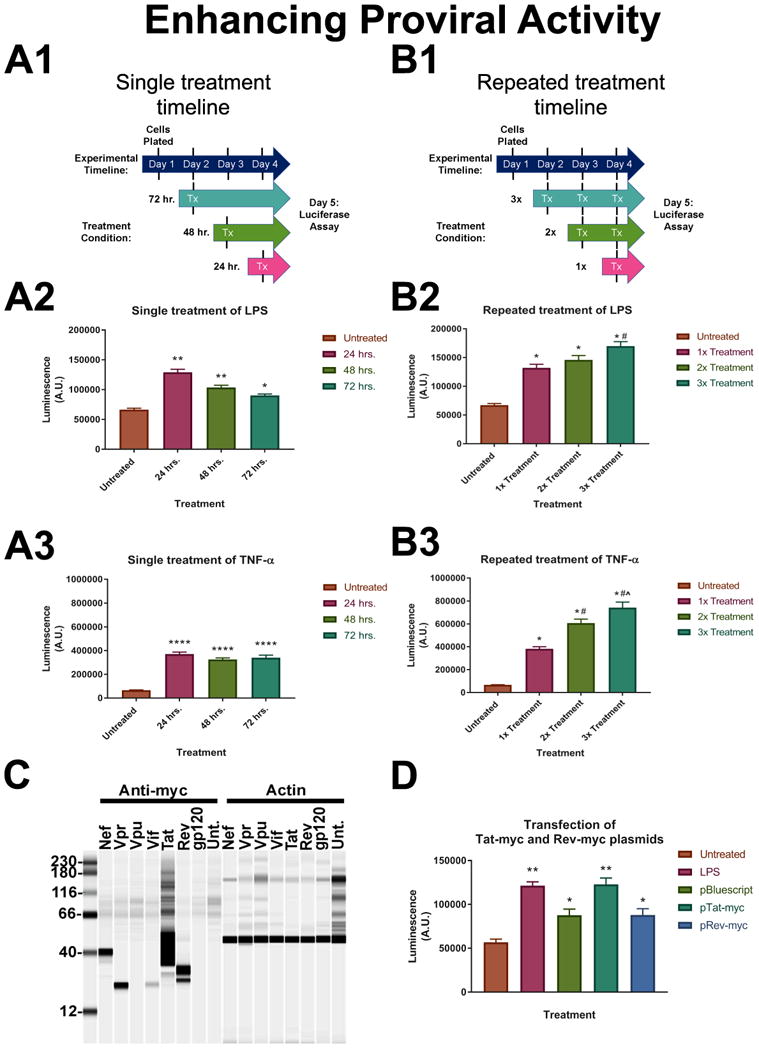
A series of experiments were conducted to test the activity of the integrated provirus in clone E9. (A) Cells were plated in a 96 well plate and stimulated one time with either LPS (100ng/ml) or TNF-α (50ng/ml) for 24, 48, or 72 hrs (timeline in A.1). We observed significant increases in luciferase activity after simulation with both LPS (A.2) and TNF-α (A.3). Significant induction persists up to the 72 hr. time point. LPS: **p<0.0001, *p<0.0002; TNF-α: ****p<0.0001. (B) Cells were treated 1x, 2x or 3x with LPS or TNF-α to determine if proviral activity could be increased by multiple treatments (timeline in B.1). There is a significant effect of proviral induction with multiple treatments of pro-inflammatory factors; LPS (B.2), TNF-α (B.3). LPS: *p<0.0001 vs Untreated, #p<0.001 vs 1x treatment; TNF-α: *p<0.0001 vs Untreated, #p<0.0001 vs 1x treatment, (x0005E)p<0.0004 vs 2x treatment. (C) Myc-tagged HIV protein expression plasmids were developed from their corresponding coding sequence in the provirus. Plasmids were transfected into HEK293 cells for expression testing and Wes analysis was performed to confirm protein expression. (D) E9 cells were transfected with pTat-myc and pRev-myc to determine the effect of HIV regulator proteins on proviral activity. LPS was used as a positive control for induction and pBSII plasmid was used as a transfection control. pRev-myc transfection produced the same relative NanoLuc levels as pBSII transfection. pTat-myc significantly increased NanoLuc activity similar to the levels of LPS. *p<0.05 vs. untreated, **p<0.0001 vs. untreated. Data are expressed as the mean ± SEM. N=3 independent cell passages.
Because the previous experiments examined pro-inflammatory factors that work on a cell surface receptor (e.g. TLR-4 and TNFR), we wanted to determine if intracellular components directly involved with viral replication would also drive proviral activity. Tat is a regulatory protein that enhances HIV proviral transcription and Rev facilitates transport of full length provirus from the nucleus [35]. We developed two plasmids that express myc-tagged viral proteins Tat-myc and Rev-myc that are under control of the CMV promoter (Fig. 4C). These two plasmids were transfected into HIV-NanoLuc clone E9 cells for 48 hrs, and proviral activity was determined by luciferase. LPS was used as a positive control and pBluescript II SK (+), or pBSII, plasmid was used as a transfection control (Fig. 4D). We observed that transfection with pBSII increased NanoLuc activity which may be due to contamination of the plasmid with bacterial endotoxin or the process of transfection activating cellular processes that have downstream effects on the provirus. Transfection of pRev-myc had similar effect as pBSII. However, transfection with pTat-myc significantly enhanced proviral activity to levels similar to LPS. These data suggest that overexpression of the HIV viral protein Tat can drive expression of the proviral construct.
2.4. Inhibiting proviral activity in the HIV-NanoLuc (E9) cell line
Our next set of experiments aimed to inhibit proviral activity by genetic and pharmacological means. We tested the ability of CRISPR/Cas9 to alter the LPS response observed in HIV-NanoLuc E9. Specifically, we utilized gRNAs in combination with wild-type Cas9 to mutate the Toll-like receptor 4 (TLR-4) receptor, a primary receptor for LPS, in order to decrease LPS mediated proviral activation. We performed TLR-4 targeted CRISPR/Cas9 mutagenesis and puromycin selection (as previously performed in Fig. 1) on cell populations of HIV-NanoLuc E9 cells. Confirmation of mutagenesis in the TLR-4 gene was achieved by using the T7E1 assay (Fig. 5A). After stimulating mixed populations of TLR-4 mutant cells and non-TLR-4 mutant cells with LPS for 24 hrs, we observed that TLR-4 mutants showed less induction by LPS (CRISPR(x00040)TLR4; 1.17 fold) compared to the control cells (HIV-NanoLuc E9 or CRISPR(x00040)TH; 1.43 fold and 1.52 fold respectively; Fig. 5B). To further confirm the observed effects were due to TLR4 mutagenesis and not an overall decrease in inflammatory response, the same cell types were stimulated with TNF-α which does not signal via TLR-4. Each cell type exhibited similar changes in response to TNF-α stimulation (HIV-NanoLuc E9; 3.9 fold, CRISPR(x00040)TH; 4.1 fold, and CRISPR(x00040)TLR4; 4.2 fold; Fig. 5C).
Figure 5. Inhibiting proviral activity in HIV-NanoLuc clone E9.
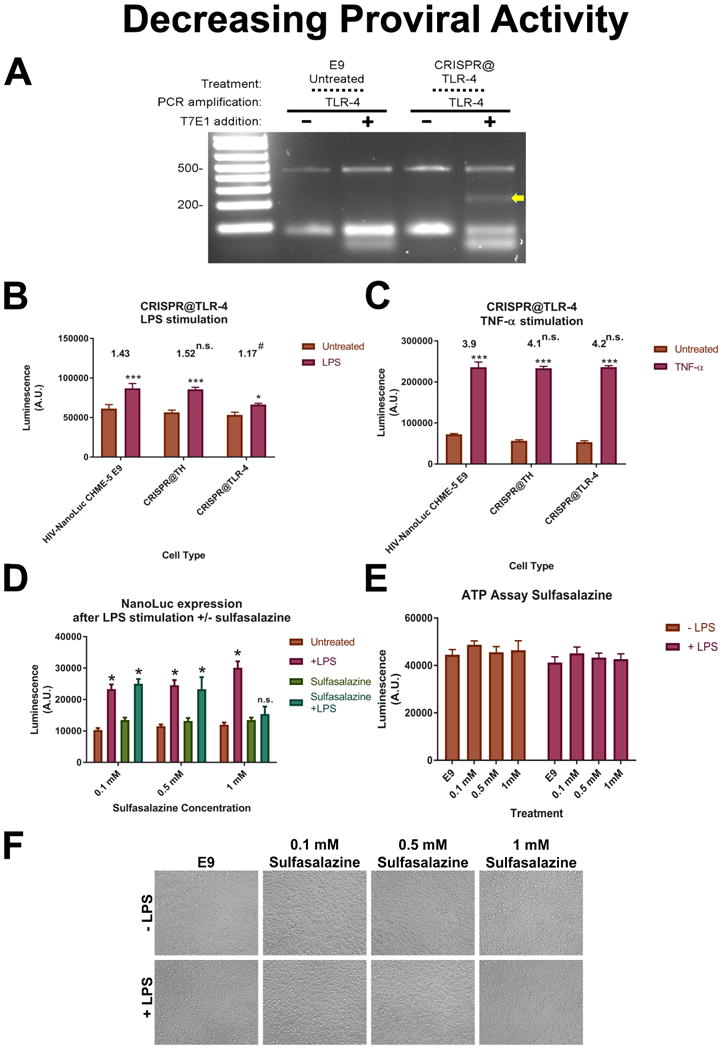
A gRNA designed to mutate the TLR-4 receptor using wildtype Cas9 were developed and transfected into clone E9. (A) T7E1 assay confirms mutagenesis at the TLR-4 genomic sequence in E9 cell populations. (B) TLR-4 mutated cell populations were stimulated with LPS (100 ng/ml) with untreated E9 cells and E9 cells receiving TH gRNAs were used as a comparison. ***p<0.0001 vs untreated, *p<0.05 vs untreated. Average fold changed listed above graph. #p<0.05 vs fold change of HIV-NanoLuc CHME-5 E9. (C) Cells were treated with TNF-α (50 ng/ml) to show that the LPS effect is specific to TLR-4 mutation. ***p<0.0001 vs untreated. Average fold changes listed above graphs. No significant difference vs. fold change of HIV-NanoLuc CHME-5. (D) Inhibition of proviral activity using NF-κB inhibitor sulfasalazine. Sulfasalazine (0.1mM, 0.5 mM or 1 mM) was added to cells one hour before LPS (100 ng/ml) stimulation. Luciferase was measured after 24 hours. 1mM sulfasalazine prevented LPS mediated proviral activty. p<0.0001 vs untreated, n.s. vs untreated. (E) Cell viability assay shows no significant difference among treatment groups. (F) Photomicrographs of cells density. Data are expressed as the mean ± SEM. N=3 independent cell passages.
NF-κB is an established transcription factor that regulates HIV proviral activity. We tested whether the NF-kB inhibitor sulfasalazine could block LPS-mediated proviral activation. Sulfasalazine (0.1, 0.5, or 1 mM) was prepared in serum free media and added to HIV-NanoLuc E9 cells one hour before LPS (100 ng/ml) stimulation. We observed that 1 mM sulfasalazine inhibited LPS-mediated proviral activity (Fig. 5D). We observed no difference in ATP activity among sulfasalazine concentrations or LPS stimulation indicating the loss of NanoLuc activity is not due to decreased cell viability (Fig. 5E). Photomicrographs of the wells also indicated that no cell loss occurred as a result of the treatments (Fig. 5F). Overall, the data from characterization of HIV-NanoLuc E9 cells show that the modified HIV provirus can be manipulated in pharmacological, viral, and molecular means consistent with the known biology of the HIV provirus.
2.5. Targeting the HIV-NanoLuc provirus with CRISPR/Cas9
The CRISPR/Cas9 gene editing system has also been utilized to mutate and excise a provirus which lead to decreased proviral activity, replication and subsequent re-infectivity [28]. In addition, other investigators have observed proviral escape and establishment of “CRISPR/Cas9 resistant strains of HIV [36, 37]. Our next aim was to determine if CRISPR/Cas9 could be directly used on the provirus within the HIV-NanoLuc CHME-5 E9 line and determine if proviral activity can be altered. We developed two gRNA sequences (LTR gRNA A and LTR gRNA B) to target the long terminal repeat (LTR) of our provirus (Fig. 6A). LTR gRNA A targets a region similar to previously published studies, while LTR gRNA B targets within an untested region of a NF-κB binding site. We tested these gRNAs on the HIV-NanoLuc CHME-5 E9 cell line in combination with Cas9. Transfection with Cas9 alone was used as a control. These treated cells were then plated onto a 96 well plate at equal cell density, stimulated with LPS (100ng/ml) or TNF-α (50ng/ml), and luciferase was assayed 24 hours post stimulation. We observe that LTR gRNA 2 resulted in a significantly decreased luciferase value under basal conditions, and after stimulation with pro-inflammatory factors (Fig. 6B). Furthermore, we determined the mutagenic effects of the gRNA's on the HIV LTR and assessed proviral excision/mutation. We developed primers within the HIV LTR region (Table 1) and amplified a 620 bp product from the genomic DNA of mutated cell lines. The T7E1 assay was the performed and produced the expected splice products for LTR gRNA 2 (358 bp and 262 bp), suggesting CRISPR/Cas9 mutated the target site (Fig. 6C). Finally, DDPCR was performed using NanoLuc primer and probe sets as previously described in Fig 2 C-D. LTR gRNA 2 treatment decreased ratio the of NanoLuc to GGT1 indicating a significant reduction of proviral genome copies per cellular genome (Fig. 6D). Overall, these experiments demonstrate that the HIV-NanoLuc CHME-5 E9 cell line can also be a useful model to test gRNAs targeting the HIV provirus. Furthermore, the presence of NanoLuc allows for a rapid assay of proviral activity after gRNA targeting in a 96 well plate format, allowing you to combine gRNA treatment pharmacological stimulation in one plate. Finally, the stability of the provirus allows for the assay of proviral excision efficiency, a factor that is not easily achieved in productive viral infection.
Figure 6. CRISPR/Cas9 Targeting of HIV-NanoLuc Provirus.
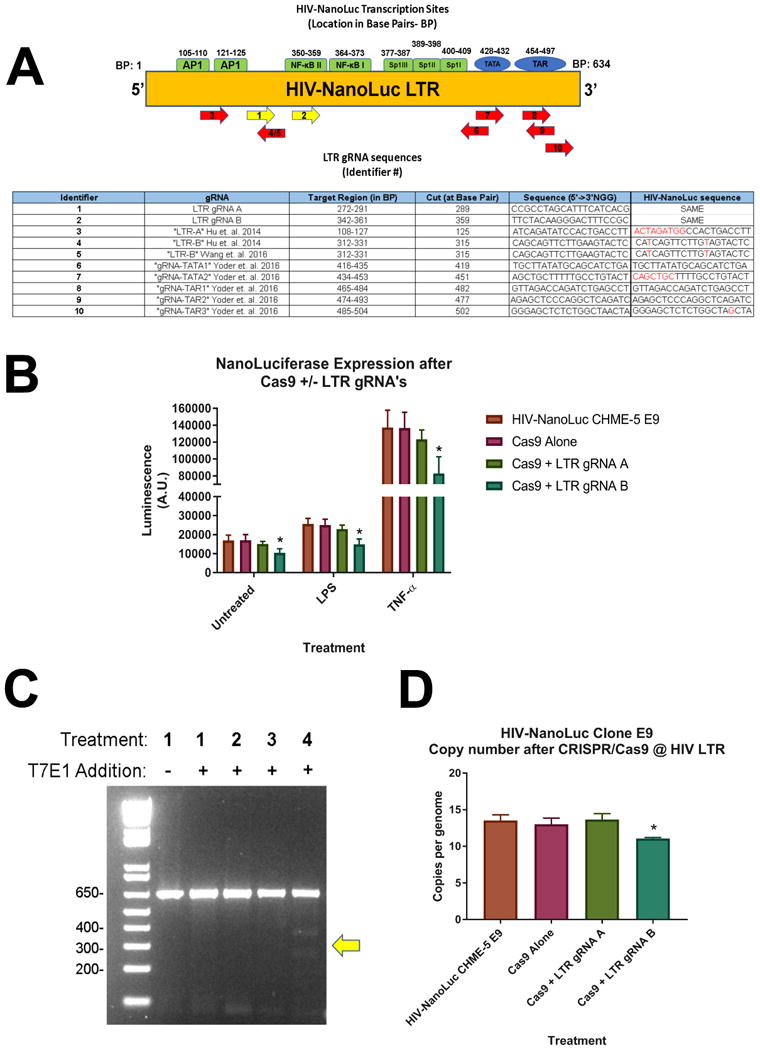
Two gRNAs (LTR gRNA A and LTR gRNA B) were developed to perform CRISPR/Cas9 directed mutagenesis on the HIV-NanoLuc provirus in HIV-NanoLuc CHME-5 E9 cell line. (A) Schematic of the HIV-NanoLuc LTR region denoting transcription sites and gRNA targeted regions. Yellow arrows denote gRNAs used for the subsequent experiments. Table: Overview of gRNA binding sites, target region, cut site, and sequence comparison with published studies. (B) HIV-NanoLuc CHME-5 E9 cells were transfected with Cas9 ± LTR gRNA A or LTR gRNA B. After passaging, cells were plated on at 96 well plate and stimulated with LPS (100ng/ml) or TNF-α (50ng/ml) for 24 hrs. Luciferase assay shows a significant reduction of proviral activity using gRNA B *p<0.05 vs HIV-NanoLuc CHME-5 E9. (C) T7E1 mutagenesis assay was performed using LTR specific primers and confirms mutagenesis by Cas9 + LTR gRNA B. (D) DDPCR was performed to determine changed in copy number of integrated HIV-NanoLuc provirus. Cas9 + LTR gRNA B significantly lowered the copy number of integrated provirus p<0.05 vs HIV-NanoLuc CHME-5 E9. Data are expressed as the mean ± SEM. N=3 independent cell passages.
Table 1. List of oligos used.
| Name | Sequence 5′ to 3′ | Use |
|---|---|---|
| Rosa26 gRNA1 | GCAGATCACGAGGGAAGAAG | Cas9 nickase based gRNA pair #1 for ROSA 26 |
| Rosa26 gRNA2 | GAGTCTTTCTGGAAGATAGG | Cas9 nickase based gRNA pair #2 for ROSA 26 |
| TH gRNA1 | ACGGCCCTTCTGAAGCCCTT | Cas9 nickase based gRNA pair #1 for TH |
| TH gRNA 2 | AGGCCGAGGCTGTCACGGTG | Cas9 nickase based gRNA pair #2 for TH |
| ROSA26 T7E1 Fwd | GGGATTCCTCCTTGAGTTGTGGC | Used to amplify ROSA26 genomic region for T7E1 assay |
| ROSA26 T7E1 Rev | GGAGGAGATATTCATCTGTAAACCATTAACAGG | Used to amplify ROSA26 genomic region for T7E1 assay |
| TH T7E1 Fwd | GAGATGGCTACCACTAGCTCGAG | Used to amplify TH genomic region for T7E1 assay |
| TH T7E1 Rev | GAGCCTGAGACAGGGTGATCC | Used to amplify TH genomic region for T7E1 assay |
| TLR-4 gRNA | TTCCTTTCCTGCCTGAGACC | Cas9 wild-type based gRNA for TLR-4 |
| TLR-4 T7E1 Fwd | TCTTGCTCTCTAGCCAGTATACTTTG | Used to amplify TLR4 genomic region for T7E1 assay |
| TLR-4 T7E1 Rev | TGATCCTGCCCGGTGTTAACAC | Used to amplify TLR 4 genomic region for T7E1 assay |
| GGT1 DDPCR Fwd | CCACCCCTTCCCTACTCCTAC | Used to amplify GGT1 genomic region for DDPCR |
| GGT1 DDPCR Rev | GGCCACAGAGCTGGTTGTC | Used to amplify GGT1 genomic region for DDPCR |
| GGT1 Probe | HEX-CCGAGAAGCAGCCACAGCCATACCT-Iowa Black | DDPCR GGT1 Probe |
| NanoLuc DDPCR Fwd | ATTGTCCTGAGCGGTGAAA | Used to amplify NanoLuc genomic region for DDPCR |
| NanoLuc DDPCR Rev | CACAGGGTACACCACCTTAAA | Used to amplify NanoLuc genomic region for DDPCR |
| NanoLuc probe | FAM- TGGGCTGAAGATCGACATCCATGT-lowaBlack | DDPCR NanoLuc Probe |
| HIV LTR Fwd | TGGAAGGGCTAATTCACTCCCAAC | Used to amplify HIV LTR for T7E1 assay |
| HIV LTR Rev | CACACTGACTAAAAGGGTCTGAGGG | Used to amplify HIV LTR for T7E1 assay |
3. Discussion
This study describes the development and characterization of a reusable microglia cell line for the study of HIV pathobiology in microglia. We use CRISPR/Cas9 technology to facilitate targeted integration of a modified HIV provirus to the safe harbor locus ROSA26. We show that the HIV provirus was successfully integrated at the targeted locus in three of our chosen clonal lines. The provirus is also active in clones E3 and E9, producing the NanoLuc reporter and the HIV viral protein Nef after activation by LPS, while still maintaining the same inflammatory profile as parental CHME-5 cells. Furthermore, a battery of experiments using HIV-NanoLuc E9 reveals this cell line is a useful model for identifying changes in proviral activity after pharmacological, viral, and molecular manipulations. Overall, HIV-NanoLuc E9 may be used as a first step in proviral experimentation to determine optimal conditions before transitioning to primary cells, live virus, and in vivo models. However, throughout the experimental procedures of this manuscript there are several important aspects to review.
CRISPR/Cas9 technology has been widely regarded for its efficient and targeted genome modification. In this study, we used this technique to facilitate homologous recombination and insertion of our modified provirus. Interestingly, while we do observe proper insertion of the provirus at ROSA26 (validated by cross junction PCR Fig. 1F), there is evidence of off-target insertion. Clone F3 failed to show junction PCR products, despite confirming genomic presence of the provirus by successfully amplifying the Env gene. Using droplet digital PCR, we identified multiple copies of NanoLuc in all the clones that were picked by screening for greater than 10000 luciferase activity, possibly indicating additional proviral insertions. Although we used Cas9 nickase to minimize off-target effects (Shen et al 2014), we do not know whether the additional integrations are due to off target effects of CRISPR or the random insertion of donor DNA into the genome. Despite this caveat, we were able to obtain stable microglial cell lines with a modified HIV provirus targeted to the ROSA26 locus.
The effect that HIV infection and integration has on the inflammatory profile in microglia is varied. Typically infection enhances secretion of pro-inflammatory factors such as IL-1β and TNF-α [38], and results in a dysregulation of glutamate homeostasis and release of detrimental metabolites [39] with alterations in metabolic processes and caspase activation [5, 40]. However, our Magpix analysis reveals that clones E3 and E9 continue to produce inflammatory factors that are comparable to the profile of parental CHME-5 microglia (Fig. 2E). The largest different observed was with clone D10, which not only appears to be unable to respond to LPS stimulation, but displays a different inflammatory profile (e.g. no/low induction of CXCL-2/3, constitutive expression of TIMP-1). This may be due to random integration events or Cas9 nickase events that change the overall phenotype away from a M1-like microglia (pro-inflammatory) to one that resembles M2 (anti-inflammatory). There is evidence of HIV-infected microglia from human subjects that have a decreased expression of pro-inflammatory factors after LPS stimulation [41]. Furthermore, newer evidence supports the role of TIMP-1 expression as a neuroprotective factor against HIV mediated neurotoxicity [42]. Therefore, a subset of infected microglia may change to this anti-inflammatory phenotype which can contribute to evasion and identification by host immune responses, leading to establishment of viral reservoirs within the CNS. Further study is needed to explore this hypothesis.
Using clone HIV-NanoLuc E9, we demonstrate its potential as a model to rapidly assess proviral activity by a variety of methods. Previous HIV-proviral reporter cell lines and constructs have utilized GFP, and subsequently flow cytometry, as a readout of activity [28, 43]. Because our proviral construct contains the luciferase NanoLuc, we can easily assess proviral activity using a luminescence assay in high throughput formats (i.e. 96 well plates), bypassing the need for time-consuming flow cytometry measures. However, if populations studies are still desired by FACS sorting or flow cytometry, the Nef protein is highly expressed in this cell line (Fig. 2D) and could be used as a target protein for immunolabeling with a fluorophore.
In our studies enhancing the proviral activity in HIV-NanoLuc E9 cells, we show longitudinal activation, repeated activation, and comparative activation between two pro-inflammatory factors, LPS and TNF-α (Fig. 3). We observed that proviral activity decreased over time after a single treatment with LPS but not with TNF-α treatment, suggesting a prolonged state of proviral activity. Additional studies, such as dose response curves, may be performed to elucidate the difference in magnitude in luminescence observed after stimulation between LPS and TNF-α. Indeed, proviral “activators” may not be equal in their potency. Some factors that may play a role in this effect may be the difference in expression between the TLR4 and TNF-α receptor, respectively. One study has shown that reactivation of latent HIV provirus by TNF- α differs by cell types (both primary and immortalized) and this was contingent on TNFR expression [44]. With this in mind, HIV-NanoLuc E9 cells may be utilized to quantitatively measure and compare compounds (e.g. other cytokines/chemokines, drugs of abuse, co-infection with other viruses, etc.) with their ability to activate proviral activity.
The use of CRISPR/Cas9 has been applied to the study of HIV. For example, recreation of the CCR5Δ32 mutation that renders cells immune to HIV infection has been applied in human cell studies [45], and Cas9 + gRNAs targeted to regions of the provirus (e.g. LTR, Gag-Pol, Env) can excise the provirus and knock down viral activity [28, 43]. In this study, we target the cellular receptor TLR4 by CRISPR/Cas9 and show a reduction in LPS-stimulated proviral gene expression. Bacterial infection and subsequent LPS circulation during HIV infection continues to be a co-morbidity concern due to constitutive activation of the provirus by LPS [46]. Our TLR4 mutation supports the use of therapies that target toll-like receptors to modify proviral activity. As a model, the HIV-NanoLuc E9 cells can be used for further studies involving pharmacological and CRISPR-mediated manipulations of proviral activity. Using this cell line, we were able to perform CRISPR/Cas9 targeting of the HIV LTR region. We show not only proviral mutation and excision, but also decreased proviral activity after stimulation with pro-inflammatory factors, further supporting the use of this cell line for CRISPR mediated experimentation. Future studies may include a compatible CRISPR library screen [47] which could be used to identify genes that alter HIV proviral activation/reactivation in microglial cells.
We have previously used the CHME-5 microglial cell line for HIV LTR expression studies [32] and chose it as our surrogate microglia for integrating a modified HIV provirus. However, proviral activity is subject to the phenotype of host cells. For CHME-5 cells, we did not observe increased secretion of pro-inflammatory cytokines after LPS stimulation (Fig. 2E). This is not surprising as it has been characterized that CHME-5 cells require a cytomix formula (e.g. interferon-γ, TNF-α, IL1-β) to produce pro-inflammatory substances such as nitric oxide [25]. Furthermore, by using rat-specific gRNAs and PCR assays in our experiments, we confirmed that CHME-5 cells are derived from rat. Overall, new microglia models would be beneficial in conjunction with the CRISPR mediated integration technique. At least two investigators have developed techniques to produce reusable human microglia either through immortalization [27] or cell preparation [48], and as these cell line become more readily available, integrating our HIV-NanoLuc provirus would be of value to compare the inflammatory profile and proviral effects between the cell lines.
Collectively, we describe the creation of an in vitro model for studying the pathobiology of HIV in microglia. Having a renewable microglia cell line with a stably integrated HIV provirus may provide a foundational starting point for many NIH-AIDS priority investigations. For example, this model is amenable to pharmacological manipulation to alter HIV proviral gene expression, which may facilitate experimentation into next generation compounds that ether suppress, or activate latent HIV proviral activity (i.e. the “kick and kill” hypothesis). As precedence, the CHME-5 cell line has previously been used to test antiretroviral compounds [25]. The addition of the HIV-NanoLuc provirus into CHME-5 cells will allow for rapid assay of proviral activity by luciferase, in addition to a pro-inflammatory screen (Fig. 3E) and viability assay (Fig. 5E) all in one cell line. This will allow for identification of compounds that have a combination of high therapeutic effects, but with minimal inflammatory and toxicity profiles. Furthermore, because the original cell line is microglia derived, experiments can be performed to understand provirus regulation in a cell type that is part of the HIV “viral reservoir”. Microglia are dynamic cells, and it would be of interest to uncover how proviral activity is regulated under classical microglia functions including phagocytosis, migration, scavenging, etc. Co-morbidity studies, such as HIV and drug abuse, are also of high priority in AIDS-related research. Our lab has previously shown that methamphetamine enhances LTR activity in CHME-5 cells [32]. Future experiments could include treatments of methamphetamine and other drugs of abuse (e.g. morphine, heroin, cocaine) on the HIV-NanoLuc CHME-5 E9 cell line to assay effects of proviral activation/deactivation by these compounds. Overall, this cell line provides an additional model for the study of HIV proviral activity and regulation in microglia.
4. Materials and Methods
4.1. Cell culture and Transfections
CHME-5 microglia were grown in microglia growth media: High Glucose DMEM (Gibco/Thermo-Fisher, Waltham, MA, USA) supplemented with 5% fetal bovine serum (Hyclone/GE, Logan, UT) and 1% penicillin/streptomycin (Gibco). Cells were passaged every 4 days using 0.01% trypsin (Gibco). Transfections of CHME-5 cells were carried out using Lipofectamine ® 2000 (Thermo-Fisher) according to the manufacturer's instructions.
4.2. CRISPR/Cas9
4.2.1. ROSA26 targeting by Cas9 nickase system
A Cas9-nickase based system was used for targeted integration of the modified HIV provirus into the ROSA26 locus. A Cas9 nickase [30] -compatible pair of gRNAs were designed using the crispr.mit.edu website and cloned into the BbsI restriction sites of the pX462-pSpCas9n(BB)-2A-Puro construct [49]; Addgene #48141;see Table 1). pspCas9n(BB)-2A-Puro (PX462 was a gift from Feng Zhang (Addgene plasmid #48141). A similar workflow was used to create a pair of gRNAs for tyrosine hydroxylase (TH) as a control. The gRNA/Cas9 nickase constructs were tested by co-transfection into CHME-5 microglia cells and assaying for heterogeneity at the ROSA26 locus by the T7E1 assay.
4.2.2. DNA constructs
A replication-defective HIV provirus was constructed by replacing the final 1500 nucleotides of gag region and the leading 2306 nucleotides of the pol region of pYU2 [50, 51];GenBank #M93258) with a loxP site and the coding region for NanoLucifierase (NanoLuc). The pYU2 plasmid was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH and donated there by B. Hahn and G. Shaw. The entire proviral sequence (bounded by and including the flanking long terminal repeats) were transferred into a backbone containing sequences homologous to 1021 nucleotides upstream and 998 nucleotides downstream of the Rosa26-targeted CRISPR-induced breakpoint (pOTTC1085, Addgene 89306). The homology-flanked modified provirus was then modified further by replacement of the LTR with the intermediate-early promoter from human cytomegalovirus (CMV-IE) using ligation-independent cloning (pOTTC1166, Addgene 89307).
The epitope-tagged HIV protein expression constructs were created by amplifying the appropriate ORF using pYU2 as a template, or by synthesizing a “spliced” form of a discontiguous ORF using the sequence of pYU2 as a guide, and recombining these inserts with pCMV6-Entry vector (Origene). See Table 2 for Addgene #s.
Table 2. List of constructs used.
| Plasmid # | Plasmid Name | Addgene |
|---|---|---|
| pOTTC1085 | prRosa26 HIV(NLuc) | 89306 |
| pOTTC1166 | prRosa26 CMV-IE NLuc Env | 89307 |
| pOTTC1156 | pCMV6 Nef-Myc-DDK | 89301 |
| pOTTC1157 | pCMV6 Vpr-Myc-DDK | 89302 |
| pOTTC1158 | pCMV6 Vif-Myc-DDK | 89303 |
| pOTTC1162 | pCMV6 Vpu-Myc-DDK | 89309 |
| pOTTC1302 | pCMV6 Tat-Myc-DDK | 89304 |
| pOTTC1340 | pCMV6 Rev-Myc-DDK | 89305 |
4.2.3. Provirus integration by CRISPR/Cas9
Provirus integration through homologous recombination was performed on CHME-5 microglia in a 6 well plate by lipofectamine transfection of Cas9-nickase + gRNA constructs (1 μg of each gRNA per well) plus the HIV provirus (2 μg per well). Forty-eight hours' post transfection cells were split into a new dish and puromycin (Thermo/Fisher) selection (2 μg/ml) was performed for another 48 hrs. Dead cells were washed off with 1x PBS and microglia growth media was added. Surviving cells were allowed to recover for 4 days, after which they were split and plated at 1 cell per well in a 96 well dish. Cells were incubated and single cell clonal populations were confirmed by visualization under a microscope. Non-single cell clonal populations were excluded when passaging the plate. After cells were grown to confluence replica plating was performed to assay for proviral integration by the luciferase assay.
4.2.4. TLR-4 knock out by CRISPR/Cas9
A guide-RNA sequence (see Table 1) was developed to target exon 1 of the Toll-like receptor 4 (TLR-4) gene. The gRNA was cloned into the phU6-gRNA backbone (Addgene #53188) for expression. phU6-gRNA was a gift from Charles Gersbach [52]. This construct was co-transfected with pX459-pSpCas9(B)-2A-Puro [49]; wild-type Cas9, (Addgene #48139) for mutation of the TLR4 gene. Forward and Reverse primer sets (Table 1) were used to amplify the targeted region of TLR4 and introduction of mutations were assayed by the T7E1 assay.
4.2.4. HIV NanoLuc LTR Targeting by CRISPR/Cas9
Two guide-RNA sequences (LTR gRNA A and LTR gRNA B; see Fig. 6A) were developed to target the HIV-NanoLuc long terminal repeat (LTR) region. The gDNA was cloned into the p-Guide-it backbone (Takara, Mountain View, CA, USA). HIV-NanoLuc CHME-5 E9 cells were transfected twice with Cas9 + LTR gRNA A/B and a stable population of cells were established for experimentation. HIV-NanoLuc LTR specific primer sets (Table 1) were utilized to determine mutagenesis by the T7E1 assay.
4.3. Genomic DNA preparation and PCR analysis
The genomic DNA was isolated using the NucleoSpin® Tissue Column (Macherey-Nagel Bethlehem, PA, USA) according to the manufacturer's instructions. DNA concentrations were measured using a Nanodrop 2000 (Thermo-Fisher). PCR reactions were prepared using Phusion High Fidelity GC Buffer (New England Biolabs, Ipswich, Ma, USA) with 5 μM forward and reverse primers. Primers are listed in Table 1. PCR was carried out in a Bio-Rad C1000TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the following cycles. For HIV proviral integration by PCR of the 5′ junction, 3′ junction and Env region of the modified HIV provirus: 98°C for 30 sec., 40x cycle of 98°C 10 sec. → 68°C 30 sec. → 72°C 90 sec, 72°C 5 min, 12°C hold. For amplification of ROSA26 and TLR4: 98°C for 30 sec., 40x cycle of 98°C → 72°C, 72°C 5min, 12°C hold. PCR products were run on a 1% agarose gel made with 1xTAE (VWR, Radnor, PA, USA) with SYBR Safe DNA gel stain (Thermo-Fisher) for 45 min.
4.4. Droplet Digital PCR
The relative copy number per genome of the modified HIV provirus was determined using Droplet Digital PCR (Bio-Rad-QX200) to quantify the number of provirus templates per microliter for the NanoLuc transgene relative to the autosomal reference gene GGT1. Primers and probes used are listed on Table 1. Reaction conditions consisted of a master mix containing: 1x ddPCRTM Supermix for Probes no dUTP (Bio-Rad), 450 nM forward and reverse primers, 50 nM probe, 0.1U MseI restriction enzyme with 50 ng genomic DNA. Reactions were run analyzing NanoLuc and GGT1 simultaneously as a duplex reaction. Equation for calculating copy number is ((copies/μL NanoLuc)/(copies/μL GGT1))*(2) = copy number per genome.
4.5. T7E1 assay for CRISPR-mediated mutagenesis
The T7 Endonuclease 1 enzyme- “T7E1” (New England Biolabs) was used to detect mutations in the ROSA26 and TLR4 PCR amplified region. Amplified products were denatured and slowly annealed in NEBuffer 2 (New England Biolabs) using the following cycle: 95°C 5 min., 85°C 30 sec. → ramp 2°C per 5 sec., 25°C 30 sec. → ramp 0.1°C per sec., 25°C hold. T7E1 was then added to the reannealed products and incubated at 37°C for 1 hour. DNA was run on a 1% agarose gel and presence of T7E1 digestion was used to verify CRISPR/Cas9 mediated mutation.
4.6. Cell culture treatments
CHME-5 or HIV-NanoLuc CHME-5 were stimulated with lipopolysaccharide or tumor necrosis factor-α. Lipopolysaccharide - “LPS” (Cat#L3012 Sigma-Aldrich Allentown, PA, USA) was used at a 100 ng/ml concentration in microglia growth media [53]. Tumor necrosis factor-α “TNF-α” (Cat#T5944 Sigma-Aldrich) was used at a 50 ng/ml concentration in microglia growth media [54]. For repeated treatments of LPS and TNF-α, full media exchange with diluted compounds were performed. Sulfasalazine experiments were carried out in serum free media with full media exchange. Sulfasalazine (Cat# 4935, Tocris Biosciences, Bristol, UK) was pre-incubated at a 0.1 mM, 0.5 mM and 1 mM on cells for 1 hr. After the incubation time, LPS was added by spiking the media to a 100 ng/ml final concentration.
4.7. Western blot analysis (Wes)
Cell lysates for Wes (ProteinSimple, San Jose, CA) analysis were produced using RIPA buffer (50 mM Tris HCl pH 7.5, 0.25% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA) with 1% NP40 detergent (Thermo-Fisher) and protease inhibitor (Sigma-Aldrich). Cells were lysed for 20 min on a rotating plate at 4°C, after which lysates were collected, and spun down for 10 min. at 10000 RPM in a microcentrifuge. The supernatant was extracted and protein levels were read using the BCA assay (Thermo-Fisher). The Wes was run using 400 μg/ml of protein per sample using buffers and antibodies provided by the Master Kit with Split Buffer (Cat# PS-MK15) according to the manufacturer's instructions. Proteins for Nef (The following reagent was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: Nef Antibody Cat# 3689 from Dr. James Hoxie), Myc (Cat# 06-340, Millipore), and β-actin (Sigma-Aldrich) were detected by Wes using specific antibodies.
4.8. Luciferase Assay
Cells that were prepared for luciferase assay were lysed directly in an opaque 96 well using RIPA lysis buffer/1% NP 40 with protease inhibitor as previously described [55]. Luminescence was measured in the opaque plate using the substrate coelenterazine (Regis Technologies Morton Grove, IL, USA) in a Bio-Tek Synergy 2 plate reader (Winooski, VT, USA).
4.9. Cell viability
Cell viability was assessed using CellTiter-Glo ATP assay (Promega Fitchburg, WI, USA) according to the manufacturer's instructions.
4.10. Immunohistochemistry
CHME-5 microglia and clones were washed with 1X PBS and fixed with 4% paraformaldehyde solution for 30 minutes. Cells were then permeabilized using blocking buffer with triton x-100 for 30 minutes, after which the primary antibody (Nef, 1:500 dilution) was added. The following reagent was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: Nef Antibody Cat# 3689 from Dr. James Hoxie. Secondary antibody Alexa Fluor ® 488 (Cat# Z25002, Thermo-Fisher and 1:1000) was added for 1 hour to determine fluorescence.
4.11. Magpix Multiplex Analysis
Media taken from cell lines ± LPS stimulation (100ng/ml for 24 hrs.) were run on a Luminex Multiplex Cytokine Array (Cat#LXSAR-M, R&D, Minneapolis, MN, USA) and analyzed on a Magpix ® System (Millipore, Billerica, MA, USA) according to the manufactures instructions.
4.12. Statistical Analysis
All analyses were evaluated by Graphpad Prism, GraphPad Software (Inc., La Jolla, CA, USA). Data were analyzed with one-way ANOVA with Tukey's multiple comparisons test. Data that compared 2 factors (e.g. sulfasalazine experiments) utilized 2-way ANOVA with Tukey's multiple comparisons test. Data are expressed as the mean ± SEM. Data from Magpix analysis expressed as mean ± SD. Statistically significant differences were considered as p<0.05.
Acknowledgments
The authors would like to thank Dr. K. Whitaker for support with MAGPIX Multiplexing assays.
The authors would like to thank F. Zhang for pspCas9n(BB)-2A-Puro (PX462) from (Addgene plasmid #48141) and pSpCas9(BB)-2A-Puro (PX459) (Addgene plasmid # 48139)
The authors would like to thank C. Gersbach for phU6-gRNA (addgene plasmid # 53188)
This project was supported by the Intramural Research Program at the National Institute on Drug Abuse.
Abbreviations used in text
- CRISPR
Clustered regularly interspaced short palindromic repeats
- HIV
Human Immunodeficiency Virus
- LPS
lipopolysaccharide
- TNF-α
Tumor necrosis factor alpha
- TLR-4
Toll-like receptor 4
- TNFR
TNF-α receptor
- gRNA
guide RNA
- TH
tyrosine hydroxylase
Footnotes
Author Contributions: LAC: Designed and performed the majority of the experiments, developed the HIV-NanoLuc CHME-5 cell line, wrote the manuscript.
CTR: Developed the HIV-NanoLuc provirus, ROSA26 based CRISPR experiments, revised manuscript.
YZ: Developed the DDPCR assay, revised the manuscript.
EJH: Developed the myc-tagged constructs, revised the manuscript.
LMC: Performed DDPCR for excision experiments, revised the manuscript.
EYP: Performed the CRISPR/Cas9 (x00040) TLR-4, revised the manuscript.
BKH: Corresponding author, designed experiments, detailed revision of manuscript.
References
- 1.Thompson KA, Cherry CL, Bell JE, McLean CA. Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol. 2011;179:1623–9. doi: 10.1016/j.ajpath.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conant K, Tornatore C, Atwood W, Meyers K, Traub R, Major EO. In vivo and in vitro infection of the astrocyte by HIV-1. Adv Neuroimmunol. 1994;4:287–9. doi: 10.1016/s0960-5428(06)80269-x. [DOI] [PubMed] [Google Scholar]
- 3.González-Scarano F, Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 4.Mangino G, Famiglietti M, Capone C, Veroni C, Percario ZA, Leone S, Fiorucci G, Lülf S, Romeo G, Agresti C, Persichini T, Geyer M, Affabris E. HIV-1 Myristoylated Nef Treatment of Murine Microglial Cells Activates Inducible Nitric Oxide Synthase, NO2 Production and Neurotoxic Activity. PLoS One. 2015;10:e0130189. doi: 10.1371/journal.pone.0130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh JG, Reinke SN, Mamik MK, McKenzie BA, Maingat F, Branton WG, Broadhurst DI, Power C. Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology. 2014;11:35. doi: 10.1186/1742-4690-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nosheny RL, Bachis A, Acquas E, Mocchetti I. Human immunodeficiency virus type 1 glycoprotein gp120 reduces the levels of brain-derived neurotrophic factor in vivo: potential implication for neuronal cell death. Eur J Neurosci. 2004;20:2857–64. doi: 10.1111/j.1460-9568.2004.03764.x. [DOI] [PubMed] [Google Scholar]
- 7.Bachis A, Wenzel E, Boelk A, Becker J, Mocchetti I. The neurotrophin receptor p75 mediates gp120-induced loss of synaptic spines in aging mice. Neurobiol Aging. 2016;46:160–8. doi: 10.1016/j.neurobiolaging.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing HQ, Hayakawa H, Gelpi E, Kubota R, Budka H, Izumo S. Reduced expression of excitatory amino acid transporter 2 and diffuse microglial activation in the cerebral cortex in AIDS cases with or without HIV encephalitis. J Neuropathol Exp Neurol. 2009;68:199–209. doi: 10.1097/NEN.0b013e31819715df. [DOI] [PubMed] [Google Scholar]
- 9.Xing HQ, Zhang Y, Izumo K, Arishima S, Kubota R, Ye X, Xu Q, Mori K, Izumo S. Decrease of aquaporin-4 and excitatory amino acid transporter-2 indicate astrocyte dysfunction for pathogenesis of cortical degeneration in HIV-associated neurocognitive disorders. Neuropathology. 2016 doi: 10.1111/neup.12321. [DOI] [PubMed] [Google Scholar]
- 10.Bansal AK, Mactutus CF, Nath A, Maragos W, Hauser KF, Booze RM. Neurotoxicity of HIV-1 proteins gp120 and Tat in the rat striatum. Brain Res. 2000;879:42–9. doi: 10.1016/s0006-8993(00)02725-6. [DOI] [PubMed] [Google Scholar]
- 11.Khan MB, Lang MJ, Huang MB, Raymond A, Bond VC, Shiramizu B, Powell MD. Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Aβ(1-42) secretion in SH-SY5Y neural cells. J Neurovirol. 2016;22:179–90. doi: 10.1007/s13365-015-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArthur JC, Haughey N, Gartner S, Conant K, Pardo C, Nath A, Sacktor N. Human immunodeficiency virus-associated dementia: an evolving disease. J Neurovirol. 2003;9:205–21. doi: 10.1080/13550280390194109. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Saksena NK. HIV Associated Neurocognitive Disorders. Infect Dis Rep. 2013;5:e8. doi: 10.4081/idr.2013.s1.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray LR, Roche M, Flynn JK, Wesselingh SL, Gorry PR, Churchill MJ. Is the central nervous system a reservoir of HIV-1? Curr Opin HIV AIDS. 2014;9:552–8. doi: 10.1097/COH.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atwood WJ, Tornatore CS, Traub R, Conant K, Drew PD, Major EO. Stimulation of HIV type 1 gene expression and induction of NF-kappa B (p50/p65)-binding activity in tumor necrosis factor alpha-treated human fetal glial cells. AIDS Res Hum Retroviruses. 1994;10:1207–11. doi: 10.1089/aid.1994.10.1207. [DOI] [PubMed] [Google Scholar]
- 16.Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, Leblanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, Collier AC, Marra CM, Morgello S, Mindt MR, Taylor MJ, Marcotte TD, Atkinson JH, Wolfson T, Gelman BB, McArthur JC, Simpson DM, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol. 2011;17:3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol. 2010;84:2395–407. doi: 10.1128/JVI.01863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podhaizer E, Zou S, Fitting S, Samano K, El-Hage N, Knapp PE, Hauser KF. Morphine and gp120 toxic interactions in striatal neurons are dependent on HIV-1 strain. J Neuroimmune Pharmacol. 2012;7:877–91. doi: 10.1007/s11481-011-9326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Hage N, Wu G, Wang J, Ambati J, Knapp PE, Reed JL, Bruce-Keller AJ, Hauser KF. HIV-1 Tat and opiate-induced changes in astrocytes promote chemotaxis of microglia through the expression of MCP-1 and alternative chemokines. Glia. 2006;53:132–46. doi: 10.1002/glia.20262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaul M, Ma Q, Medders KE, Desai MK, Lipton SA. HIV-1 coreceptors CCR5 and CXCR4 both mediate neuronal cell death but CCR5 paradoxically can also contribute to protection. Cell Death Differ. 2007;14:296–305. doi: 10.1038/sj.cdd.4402006. [DOI] [PubMed] [Google Scholar]
- 21.Zenón F, Cantres-Rosario Y, Adiga R, Gonzalez M, Rodriguez-Franco E, Langford D, Melendez LM. HIV-infected microglia mediate cathepsin B-induced neurotoxicity. J Neurovirol. 2015;21:544–58. doi: 10.1007/s13365-015-0358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Hage N, Rodriguez M, Dever SM, Masvekar RR, Gewirtz DA, Shacka JJ. HIV-1 and morphine regulation of autophagy in microglia: limited interactions in the context of HIV-1 infection and opioid abuse. J Virol. 2015;89:1024–35. doi: 10.1128/JVI.02022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janabi N, Peudenier S, Héron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–8. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 24.Dutta R, Krishnan A, Meng J, Das S, Ma J, Banerjee S, Wang J, Charboneau R, Prakash O, Barke RA, Roy S. Morphine modulation of toll-like receptors in microglial cells potentiates neuropathogenesis in a HIV-1 model of coinfection with pneumococcal pneumoniae. J Neurosci. 2012;32:9917–30. doi: 10.1523/JNEUROSCI.0870-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisi L, Laudati E, Miscioscia TF, Dello Russo C, Topai A, Navarra P. Antiretrovirals inhibit arginase in human microglia. J Neurochem. 2016;136:363–72. doi: 10.1111/jnc.13393. [DOI] [PubMed] [Google Scholar]
- 26.Samikkannu T, Atluri VS, Nair MP. HIV and Cocaine Impact Glial Metabolism: Energy Sensor AMP-activated protein kinase Role in Mitochondrial Biogenesis and Epigenetic Remodeling. Sci Rep. 2016;6:31784. doi: 10.1038/srep31784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Mesa Y, Jay TR, Checkley MA, Luttge B, Dobrowolski C, Valadkhan S, Landreth GE, Karn J, Alvarez-Carbonell D. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol. 2016 doi: 10.1007/s13365-016-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111:11461–6. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A. 1997;94:3789–94. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–56. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 32.Wires ES, Alvarez D, Dobrowolski C, Wang Y, Morales M, Karn J, Harvey BK. Methamphetamine activates nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and induces human immunodeficiency virus (HIV) transcription in human microglial cells. J Neurovirol. 2012;18:400–10. doi: 10.1007/s13365-012-0103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–3. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 34.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6:347–63. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Rep. 2016;15:481–9. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 37.Yoder KE, Bundschuh R. Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9. Sci Rep. 2016;6:29530. doi: 10.1038/srep29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brabers NA, Nottet HS. Role of the pro-inflammatory cytokines TNF-alpha and IL-1beta in HIV-associated dementia. Eur J Clin Invest. 2006;36:447–58. doi: 10.1111/j.1365-2362.2006.01657.x. [DOI] [PubMed] [Google Scholar]
- 39.Cassol E, Misra V, Dutta A, Morgello S, Gabuzda D. Cerebrospinal fluid metabolomics reveals altered waste clearance and accelerated aging in HIV patients with neurocognitive impairment. AIDS. 2014;28:1579–91. doi: 10.1097/QAD.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosati A, Khalili K, Deshmane SL, Radhakrishnan S, Pascale M, Turco MC, Marzullo L. BAG3 protein regulates caspase-3 activation in HIV-1-infected human primary microglial cells. J Cell Physiol. 2009;218:264–7. doi: 10.1002/jcp.21604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghorpade A, Persidsky Y, Swindells S, Borgmann K, Persidsky R, Holter S, Cotter R, Gendelman HE. Neuroinflammatory responses from microglia recovered from HIV-1-infected and seronegative subjects. J Neuroimmunol. 2005;163:145–56. doi: 10.1016/j.jneuroim.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 42.Ashutosh, Chao C, Borgmann K, Brew K, Ghorpade A. Tissue inhibitor of metalloproteinases-1 protects human neurons from staurosporine and HIV-1-induced apoptosis: mechanisms and relevance to HIV-1-associated dementia. Cell Death Dis. 2012;3:e332. doi: 10.1038/cddis.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 47.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MeC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 48.Rawat P, Spector SA. Development and characterization of a human microglia cell model of HIV-1 infection. J Neurovirol. 2016 doi: 10.1007/s13365-016-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–85. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Hui H, Burgess CJ, Price RW, Sharp PM, Hahn BH, Shaw GM. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang PK, Khatchadourian A, McKinney RA, Maysinger D. Docosahexaenoic acid (DHA): a modulator of microglia activity and dendritic spine morphology. J Neuroinflammation. 2015;12:34. doi: 10.1186/s12974-015-0244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang J, Han D, Hong J, Tan Q, Tian Y. The chemokine, macrophage inflammatory protein-2γ, reduces the expression of glutamate transporter-1 on astrocytes and increases neuronal sensitivity to glutamate excitotoxicity. J Neuroinflammation. 2012;9:267. doi: 10.1186/1742-2094-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henderson MJ, Wires ES, Trychta KA, Richie CT, Harvey BK. SERCaMP: a carboxy-terminal protein modification that enables monitoring of ER calcium homeostasis. Mol Biol Cell. 2014;25:2828–39. doi: 10.1091/mbc.E14-06-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]


