Abstract
Nuclear Epidermal Growth Factor Receptor (EGFR) has been associated with worse prognosis and treatment resistance for several cancer types. After Epidermal Growth Factor (EGF) binding, the ligand-receptor complex can translocate to the nucleus where it functions in oncological processes. By three-dimensional quantification analysis of super-resolution microscopy images, we verified the translocation kinetics of fluorescent conjugated EGF to the nucleus in two mesenchymal cell types: human adipose tissue-derived stem cells (hASC) and SK-HEP-1 tumor cells. The number of EGF clusters in the nucleus does not change after 10 min of stimulation with EGF in both cells. The total volume occupied by EGF clusters in the nucleus of hASC also does not change after 10 min of stimulation with EGF. However, the total volume of EGF clusters increases only after 20 min in SK-HEP-1 cells nuclei. In these cells the nuclear volume occupied by EGF is 3.2 times higher than in hASC after 20 min of stimulation, indicating that translocation kinetics of EGF differs between these two cell types. After stimulation, EGF clusters assemble in larger clusters in the cell nucleus in both cell types, which suggests specific sub-nuclear localizations of the receptor. Super-resolution microscopy images show that EGF clusters are widespread in the nucleoplasm, and can be localized in nuclear envelope invaginations, and in the nucleoli. The quantitative study of EGF-EGFR complex translocation to the nucleus may help to unravel its roles in health and pathological conditions, such as cancer.
Keywords: EGF, Nucleus, Translocation, Kinetics, Super-resolution microscopy
1. Introduction
Epidermal Growth Factor Receptor (EGFR) is a member of ERBB family of receptor tyrosine kinase, which is expressed in cells of ectodermal and mesodermal lineage (Citri and Yarden, 2006). The interaction of ligands, such as the Epidermal Growth Factor (EGF), with EGFR leads to the formation of asymmetric homodimers or heterodimers with other ERBB receptors and, subsequently, to their activation by autophosphorylation of tyrosine residues in their C-terminal region (Lemmon et al., 2014). Activated EGFR can initiate intracellular signals for cell adhesion, migration, proliferation, and oncogenesis (Yarden and Sliwkowski, 2001).
Increased expression of EGFR and mutations in its gene are associated with neoplasms such as non-small-cell lung cancer, glioblastomas, breast cancer, and head and neck squamous cell carcinoma (reviewed by Yarden and Pines, 2012). EGF family proteins have been documented as potential biomarkers for disease prognosis, and for progression of different tumor types (Briffa et al., 2015; Docea et al., 2013; Loupakis et al., 2014; Miyata et al., 2017; Olsen et al., 2012). Cancer therapies that inhibit EGFR signaling have been developed with beneficial results for patient lifespan; however, tumors ultimately acquire resistance to them, leading to the need for treatment change (Roskoski, 2014). Thus, studies of EGFR biology could unveil mechanisms of therapy resistance, and potential methods to overcome the issue.
It has been observed that after ligand interaction, the EGF-EGFR complex can redistribute in the membrane forming clusters that are endocytosed in coated vesicles to the cytosol (Gorden et al., 1978; Haigler et al., 1979). The internalization is important for signaling control of the receptor, and for its transport to other cell compartments, such as mitochondria, Golgi, endoplasmic reticulum, and nucleus (Wang and Hung, 2012). Inside the nucleus, the EGFR acts as a transcriptional regulator, signal transducer, and protein modulator, leading to cell proliferation, DNA replication and repair, and tumor progression (reviewed in Lee et al., 2015).
Nuclear EGFR is mainly observed in highly-proliferative cells, as in tumor tissues (Lin et al., 2001; Lo et al., 2006b; Pereira et al., 2015; Rodrigues et al., 2016). It is associated with poor prognosis (Hoshino et al., 2007; Li et al., 2012; Lo et al., 2005; Traynor et al., 2013; Xia et al., 2009) and therapy resistance (Huang et al., 2011; Lee et al., 2015) in various types of cancer. Although components participating in the nuclear translocation of EGFR have been unveiled (Campos et al., 2011; Hsu and Hung, 2007; Liao and Carpenter, 2007; Lin et al., 2001; Lo et al., 2006a; Wang et al., 2010a,b), little is known about the translocation kinetics and nuclear compartmentalization after stimulation, and how these profiles vary in cells with different proliferative and oncogenic capacity.
Protein inside cellular compartments can be analyzed and quantified by different approaches, such as cell fractioning following protein extraction, transmission electron microscopy, and light microscopy. Western blot of cell fractions can be performed to verify the EGFR nuclear translocation after EGF stimulation over time (Campos et al., 2011; Faria et al., 2016; Lin et al., 2001). Although this technique enables the analysis of the expression profile, it is not possible to visualize the localization of the protein complexes inside cell compartments. Electron microscopy allows precise protein visualization inside cells, but has limitations regarding volume quantifications of the clusters.
On the other hand, light microscopy is a powerful approach to achieve protein intracellular localization and quantification, with recent advances in both aspects (Hamilton, 2009). Modern microscopes allows acquisition of higher resolution images than conventional confocal microscopy (Weisshart, 2014), and new image analysis methods are being developed to fulfill users requirements for protein quantifications (Coffman and Wu, 2012; Rizk et al., 2014; Szoboszlay et al., 2017). Although some approaches use live cells to track intracellular EGFR (Campos et al., 2011; Du et al., 2013), photobleaching and poor spatial and temporal resolution may affect the localization and quantification of proteins over time (Hamilton, 2009). Thus, new methods for quantification of molecules using super-resolution images of fixed cells could generate more precise data to verify EGFR kinetics and subcellular localization.
Given the importance of studying EGF-EGFR transport for a better understanding of its association with tumor development and therapy resistance, this study aims to use an innovative approach to quantify nuclear EGF clusters after increasing times of stimulation in tumor cells, and to compare them to non-tumor cells of the same embryonic origin. The comparison might unveil aspects of EGF-EGFR biology in normal and abnormal contexts.
2. Material and methods
2.1. Cell culture
Mesenchymal stem cells (hASC) were extracted from human adipose tissue freely donated by patients who underwent liposuction and abdominoplasty, as approved by the “Universidade Federal de Minas Gerais” (UFMG) Research Ethics Committee (COEP: 55698116.2.0000.5149). Cell extraction was performed according to Zuk et al. (2001). Briefly, adipose tissue samples were washed twice in phosphate buffered saline (PBS; pH 7.4, Thermo Fisher Scientific, Waltham, MA, USA), and were digested by 0.1% collagenase solution (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 45 min. The samples were centrifuged at 300×g for 10 min at 25 °C, and the pellet was resuspended in sterile Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin/amphotericin B solution (PSA; Sigma-Aldrich). Cells were transferred to culture flasks and kept in a humid atmosphere at 37 °C and 5% CO2. SK-HEP-1 cells were obtained from American Type Culture Collection. These cells were cultured in sterile DMEM with 10% FBS and 1% PSA and kept in a humid atmosphere at 37 °C and 5% CO2. Culture medium from both cell cultures was changed every 3 days. For all the experiments performed hASC was in passage 3–5.
2.2. Western blot
hASC and SK-HEP-1 plated cells were washed twice with cold PBS and were lysed by NETN buffer (150 mM NaCl; 1 mM EDTA; 20 mM Tris–HCl, pH 8.0; 0.5% Nonidet P-40) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). After cell scraping, cells were collected, homogenized by vortex and sonicated. The samples were incubated on ice for 10 min and centrifuged at 16,000×g for 20 min at 4 °C. Supernatants were collected and proteins were quantified by Bradford method (Bradford, 1976). Immunoblotting was performed as previously described (Campos et al., 2011). Briefly, samples were submitted to polyacrylamide gel electrophoresis (SDS-PAGE) and proteins were transferred to 0.22 μm polyvinylidene fluoride membranes (BioRad, Hercules, CA, USA) using a Trans-Blot® SD semi-dry transfer cell (BioRad). Anti-EGFR (Santa Cruz Biotechnology, Dallas, Texas, USA) and anti-α tubulin (Sigma-Aldrich) were used as primary antibodies. Membranes were incubated with peroxidase-conjugated secondary antibodies and revealed with enhanced chemiluminescence solution ECL (Thermo Fisher Scientific) in BioMax® MR (Carestream/Kodak) films. Quantitative analyses of the blotting were performed using Image J software (https://imagej.nih.gov/ij/).
2.3. Super-resolution microscopy
Cells plated on sterile cover slips were incubated in medium without FBS overnight and were stimulated with 200 ng/mL of EGF labeled with Alexa Fluor® 488 (EGF-488) (Thermo Fisher Scientific) for 0 (control), 5, 10, 20 and 40 min. After removal of the stimulus, cells were washed with PBS, fixed with formaldehyde 3.7% and permeabilized with 0.05% Triton X-100 solution. Cells were blocked with 1% bovine serum albumin (BSA) solution containing 5% goat serum and were incubated at 4 °C overnight with primary antibodies: monoclonal anti-Fibrillarin (Cell Signaling Technology, Danvers, MA, USA), monoclonal anti-lamin B2 (Thermo Fisher Scientific), monoclonal anti-EGFR (Millipore, Temecula, CA, USA). In the other day, cells were labeled with a secondary antibody (Alexa Fluor® 555 and Alexa Fluor® 647) and with Hoechst 33258 (Thermo Fisher Scientific) at room temperature for 1 h. The coverslips were washed with PBS and slides were assembled using Prolong Gold Antifade Reagent (Thermo Fisher Scientific). Cells were analyzed in “Centro de Aquisição e Processamento de Imagens da UFMG” using the LSM 880 with the Airyscan detector (Carl Zeiss, Jena, Germany). For image acquisition, it was used 63 x, 1.4NA objective lens. The lasers used were: Diode 405 nm (excitation of Hoechst), Argonium 488 nm (excitation of Alexa Fluor® 488), HeNe 543 nm (excitation of Alexa Fluor® 555) and HeNe 633 nm (excitation of Alexa Fluor® 647). Labels were detected and processed using the Airyscan system, which enables resolution below the light diffraction limit. Images of serial optical sections were acquired at 1024 × 1024 pixels using 16-bits color depth.
2.4. Three-dimensional quantification of EGF clusters
EGF-488-stimulated cells with labeled nucleus were selected according to their morphology and integrity using bright field images. Super-resolution images of the nucleus of those cells were analyzed using the software Image Pro Plus Premier 3D (Media Cybernetics, Rockville, MD, USA). By means of the “iso surface” function, a three-dimensional rendered surface was created for EGF and nuclei labels and it was adjusted according to the fluorescent structures in the original image. Cells stimulated for 40 min were used to determine a threshold for the minimum pixel intensity considered as a label, and not as background. For each of the images, an inferior threshold of fluorescence intensity to identify a pixel as belonging to a 3D EGF cluster was determined. The average of those thresholds for each cell type was used to generate the three-dimensional EGF clusters of all the other EGF-stimulated cells and control (non-stimulated) cells. The number of EGF clusters, the volume of each cluster and the total volume (given by the number of clusters multiplied by the average volume of each cluster) of EGF clusters per cell nucleus were calculated according to the 3D surface created for both cell types.
2.5. Colocalization analysis between EGF and EGFR
Cells stimulated with EGF-488 for 10 and 40 min and labeled with anti-EGFR antibody were used for a colocalization analysis between EGF and EGFR, which was performed according to Everett, M. using the ZEN Black edition software (Carl Zeiss). Briefly, horizontal and vertical thresholds of the scattered plot were determined using control images, in which only EGF or EGFR were labeled. Threshold values were used to analyze the colocalized pixels of the experimental images in which both ligand and receptor were labeled. Images used in this analysis were originated from serial optical sections reconstruction using the maximum intensity projection mode. Pearson correlation coefficient was calculated for each image using ZEN software.
2.6. Statistical analysis
Cluster data in stimulated cells was normalized to non-stimulated cells (background) when nonspecific staining occurred. Only cells with EGF clusters were considered for the analyses. Kruskal-Wallis with Dunn’s multiple comparisons test was performed to analyze the parameters of the EGF clusters for each cell type separately. Two-way ANOVA with Bonferroni’s multiple comparison tests was used to compare the Pearson correlation coefficient and the total volume occupied by clusters in cell nucleus between the cell types. Student-T test was used for densitometric analysis of the blotting. Data are represented as a mean ± standard error, and all statistical tests were performed using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA).
3. Results and discussion
3.1. EGFR is expressed and colocalizes with EGF-488 in hASC and SK-HEP-1 cells
Translocation of EGFR to the nucleus is associated with a proliferative and tumor phenotype (Han and Lo, 2012; Lee et al., 2015). Based on the significance of this process in different cell types, the present study aimed to quantify the amount of EGFR that can translocate to the nucleus in a non-tumor cell, and in a tumor cell with mesenchymal origin characteristics. A previous study suggested that SK-HEP-1 is a tumor mesenchymal stem cell since they exhibit expression markers and differentiation capacity similar to adipose and bone marrow derived mesenchymal stem cells (Eun et al., 2014). The present work quantitatively verified the translocation kinetics of EGF clusters conjugated with Alexa Fluor® 488 (EGF-488) to the nucleus of cells with a mesenchymal profile, but with different oncogenic capacity. Adipose tissue-derived stem cells used in this work were characterized by expression of mesenchymal surface markers (Supplementary Fig. 1A), and for differentiation into adipogenic and osteogenic lineages (Supplementary Fig. 1B), in accordance with established criteria (Bourin et al., 2013).
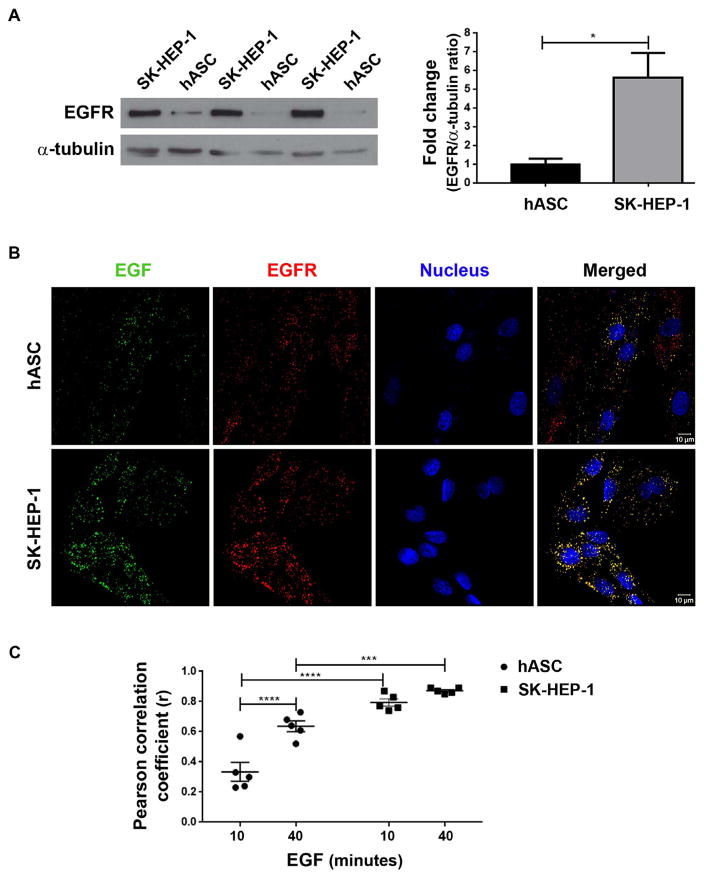
Before quantifying nuclear EGF-EGFR clusters in the nucleus, the expression levels of EGFR and its colocalization with EGF-488 in both cell types were verified. Western blot of total protein of non-stimulated cells showed that SK-HEP-1 cells express a higher quantity of this receptor than hASC (Fig. 1A). Tumor cells are known to show a higher expression of EGFR, which is related to tumorigenic potential (Yarden and Pines, 2012).
Fig. 1.
EGFR colocalizes with labeled EGF in both cell types. A- Western blot of proteins extracted from both cell types cultured for 12–16 h in medium without FBS followed by densitometric analysis of the protein bands related to the expression of EGFR (175 kDa). The expression of α-tubulin (50 kDa) was used as a normalizer. Data were expressed as fold change of hASC bands fluorescence intensity. Statistical test: Student’s T test; N = 3 samples; *: p < .05. B- Super-resolution images of hASC and SK-HEP-1 cells stimulated for 40 min with 200 ng/mL of EGF conjugated to Alexa Fluor® 488 (in green). After stimulation, cells were stained for EGFR (in red) and nucleus (in blue). The merged images show the colocalization between EGF and EGFR in yellow. Scale bar: 10 μm. C- Graph showing a Pearson correlation coefficient (r) of colocalization analysis for cells of both types stimulated by 10 and 40 min with EGF. Statistical test: Two-way ANOVA with Bonferroni’s multiple comparisons test; N = 5 images for each group; ***: p < .001; ****: p < .0001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Colocalization analysis was performed using super-resolution images of cells stimulated for 10 and 40 min with EGF-488, and subsequently fixed and labeled for EGFR by immunofluorescence. In the merged image of both of cell types (Fig. 1B), yellow clusters were observed, indicating overlap between EGF (green) and EGFR (red). The Pearson’s correlation coefficients (r) of the pixel-by-pixel covariance in the signal levels of two images (Dunn et al., 2011) for both of cells were positive, indicating that EGF and EGFR are colocalized. The analysis demonstrated a higher colocalization of EGF with EGFR in SK-HEP-1 cells (r > 0.79), which may be due to higher EGFR expression levels in this cell model (Fig. 1C). Because EGF-488 was shown to interact with EGFR for both cells, the measurements of EGF-EGFR clusters for all the following experiments were performed based on labeled EGF clusters formed after stimulation with EGF-488.
3.2. EGF translocates to the nucleus of hASC and SK-HEP-1 cells
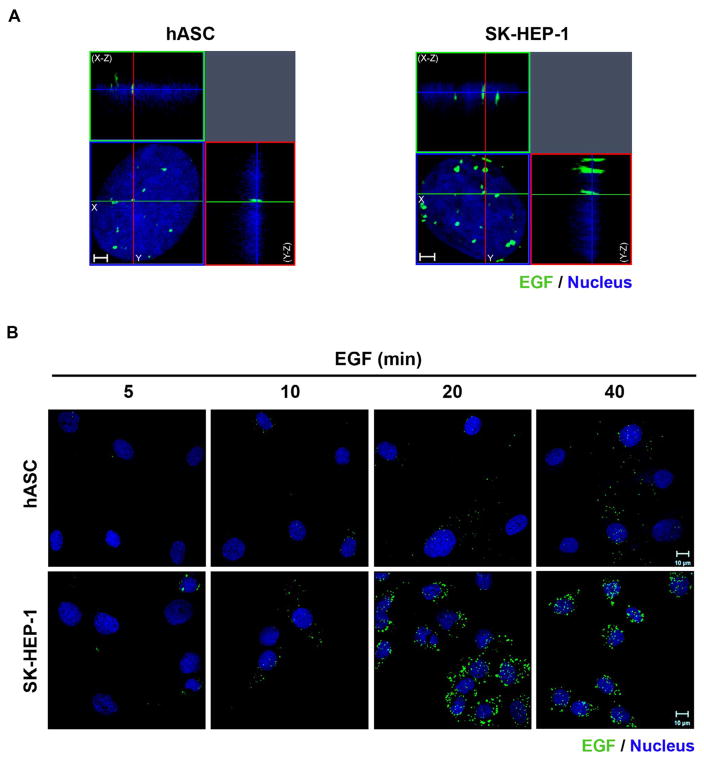
Given that hASC and SK-HEP-1 cells express EGFR that colocalized with EGF, it was possible to verify if labeled EGF translocates to the nucleus after stimulation in both cell types. We have previously shown the translocation kinetics of EGFR for SK-HEP-1 cells by Western Blot analysis (Campos et al., 2011). Here, we show the EGF clusters inside the nucleoplasm of SK-HEP-1 cells and hASC, using the tridimensional reconstruction of optical sections acquired by Airyscan detection of immunofluorescence-labeled cells (Fig. 2A). After 5 min of stimulation with EGF-488, it is possible to verify few EGF clusters in SK-HEP-1 cells and none in hASC cells. With the increase of EGF stimulus time to 10, 20 and 40 min, the clusters visually rise in number or volume in the nuclear region (Fig. 2B).
Fig. 2.
EGF-488 clusters translocate to the nucleus in hASC and SK-HEP-1 cells. A- Three-dimensional reconstruction of optical sections of the hASC and SK-HEP-1 were performed, after cells were stimulated for 40 min with EGF conjugated with Alexa Fluor® 488 (in green). X–Z sections are shown at the top, and Y–Z sections are shown on the right of each image. Images are representative of 41 (hASC) and 37 (SK-HEP-1) cells. Nuclei are in blue. Scale bar: 2 μm. B- Cells were stimulated with EGF conjugated with Alexa Fluor® 488 for 5, 10, 20 and 40 min (in green), had their nuclei labeled with Hoechst (in blue) and were analyzed by super-resolution microscopy. Images are representative of 8 to 10 fields. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Translocation kinetics of EGF to the nucleus differs quantitatively between SK-HEP-1 and hASC
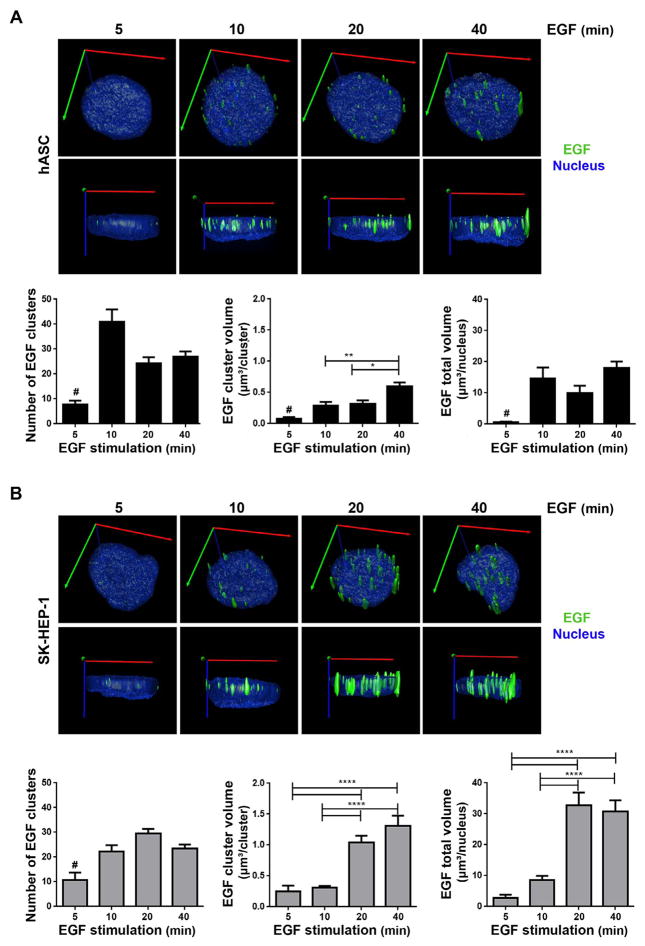
To quantify fluorescent-labeled three-dimensional EGF clusters in images acquired using LSM 880 with the Airyscan detector, the Image Pro Plus Premier 3D software was used. A three-dimensional rendered surface was created for each nucleus and for each EGF cluster of hASC (Fig. 3A) and SK-HEP-1 (Fig. 3B) cells, and clusters inside nucleoplasm were quantified in number and volume for every stimulation time.
Fig. 3.
Three-dimensional quantification analysis of EGF clusters in hASC (A) and SK-HEP-1 (B). A and B: top images - Super-resolution microscopy images of cells stimulated by 5, 10, 20 and 40 min with EGF complexed to Alexa Fluor® 488 were used to create a rendered surface of the nucleus (blue) and EGF clusters (green) in three dimensions. Axes represented in the images: X (red), Y (green) and Z (blue). A and B: bottom images- The number of EGF clusters in each cell, the mean volume of each cluster and the total volume occupied by EGF clusters in each cell nuclei were analyzed. Statistical test: Kruskal-Wallis with Dunn post-test. Bars: mean ± standard error; #: Differs from all other groups with p < .05; *: p < .05; **: p < .01; ****: p < .0001. N = hASC: 28 (5 min), 40 (10 min), 38 (20 min), 41 (40 min); SK-HEP-1: 21 (5 min), 37 (10 min), 34 (20 min), 37 (40 min). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3A, left bottom, shows that the number of EGF clusters for hASC increased and stabilized in 10 min of stimulation with EGF. Similarly, the nuclear total volume occupied by EGF clusters reaches its peak between 5 and 10 min of stimulation (Fig. 3A, right bottom). Based on the results, the EGF translocation kinetic analysis for hASC suggests that these cells achieve translocation equilibrium to the nucleus within 10 min of stimulation. The average volume of each EGF cluster increases in 10 min and is higher for cells stimulated for 40 min (Fig. 3A, middle bottom), indicating an agglomeration of clusters inside the nucleus.
Like hASC, the number of EGF clusters in the nucleus of SK-HEP-1 cells increased in 10 min of stimulation (Fig. 3B, left bottom). However, the total volume of EGF cluster in the nucleus was higher for cells stimulated for 20 and 40 min compared to cells stimulated for 5 and 10 min (Fig. 3B, right bottom). Thus, for SK-HEP-1 cells, the quantitative analysis of total EGF volume suggests that EGF translocation from the plasma membrane to the nucleus reach its peak between 10 and 20 min of stimulation. The average volume of each EGF cluster also indicated an agglomeration of clusters inside the nucleus with longer time of stimulation, once it is higher in cells stimulated for 20 and 40 min (Fig. 3B, middle bottom).
The profile of EGF translocation kinetics differs between the two cell types (Fig. 4). In hASC nuclear EGF clusters were visualized at 10 min of stimulus, and after that, the total volume of EGF clusters did not change. In SK-HEP-1 cells nuclear EGF was visualized at 5 min, and there was an increase in the nuclear volume occupied by them at 20 min of stimulus. The difference in translocation kinetics profile between hASC and SK-HEP-1 cells might be related to their differential expression levels of EGFR. The higher expression of EGFR by SK-HEP-1 cells (Fig. 1A) might be responsible for bigger EGF clusters in the nucleus. Therefore, the total volume occupied by EGF clusters in the nucleus of those cells may become larger after longer periods of stimulus. On the other hand, hASC expresses less EGFR, which might be associated with lower EGF internalization and achievement of translocation equilibrium within 10 min of stimulus. Additionally, translocation of EGF-EGFR complexes might reflect the different phenotypes of the cells analyzed. Investigations into specific and differential functions of nuclear EGF-EGFR in each cell type are necessary to explain the pattern of EGF translocation kinetics found in this study.
Fig. 4.
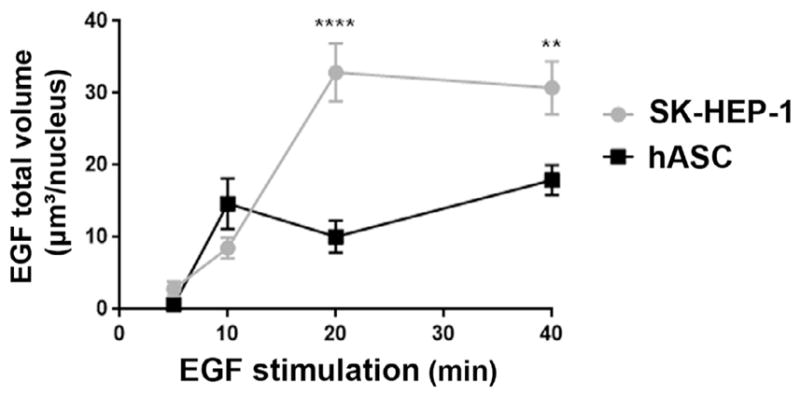
Profile of EGF translocation kinetics to the nucleus differs between hASC and SK-HEP-1. Cells of both cell types were stimulated by 5, 10, 20 and 40 min with EGF complexed to Alexa Fluor® 488. The graphic represents the mean ± standard error of volume occupied in the nucleus by the EGF clusters in each cell. **: p < .01; ****: p < .0001. Statistical test: Two-way ANOVA with Bonferroni’s multiple comparisons test; N = hASC: 28 (5 min), 40 (10 min), 38 (20 min), 41 (40 min); SK-HEP-1: 21 (5 min), 37 (10 min), 34 (20 min), 37 (40 min).
3.4. EGF has a diffuse distribution in the nucleoplasm, and may be found in the nucleoplasmic reticulum and nucleoli of hASC and SK-HEP-1 cells
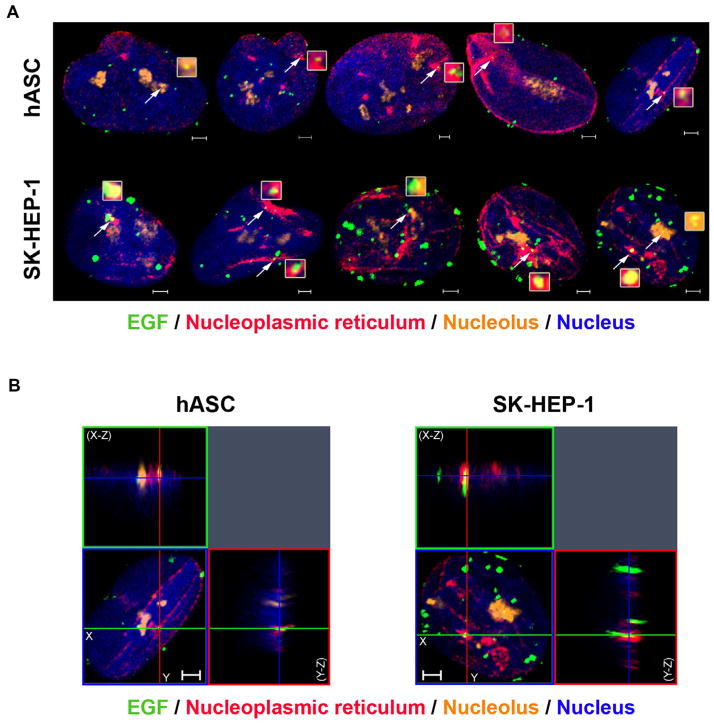
The agglomeration of EGF clusters into larger clusters suggested by the quantitative three-dimensional analysis for both cell types (Fig. 3) might indicate that EGF-EGFR complexes have preferential localization in nuclear organelles that could be related to their transport to the nucleus, or their function in specific nuclear compartments. A model of EGFR translocation to the nucleus suggests that the process occurs via integral trafficking from the endoplasmic reticulum to the nuclear envelope transport (INTERNET), in which the EGFR passes through an endomembrane pathway until reaching the inner nuclear membrane (INM), where it associates with Sec61β, which participates in the translocation of EGFR to the nucleoplasm (Wang et al., 2010b, 2012). It is possible that the nucleoplasmic reticulum participates in the translocation process because it is composed of invaginations of nuclear envelope membranes inside the nucleoplasm (reviewed by Malhas et al., 2011).
To verify if EGF clusters are localized in nucleoplasmic reticulum and in the nucleolus, which might be associated with the nucleoplasmic reticulum (Bourgeois et al., 1979; Malhas et al., 2011), we stimulated cells with EGF-488 and labeled the nuclear envelope and the nucleolus using immunofluorescence. Super-resolution images of nuclei of both hASC and SK-HEP-1 cells show the presence of EGF clusters in nuclear envelope invaginations and in the nucleolus (Fig. 5A). Three-dimensional reconstruction of serial optical sections of the cells confirms the localization of EGF clusters in those organelles, which reside inside the nucleoplasm (Fig. 5B). This result suggests that the nucleoplasmic reticulum might be involved in the translocation process of EGF-EGFR to the nucleus. EGF-EGFR complexes might have specific functions in the nucleolus, similar to what was suggested for ERBB2 (Li et al., 2011). We observed EGFR either in the nucleoplasm or nucleoplasmic reticulum of tumor samples. For example, EGFR was identified in the nucleoplasm of unicystic and multicyst ameloblastomas, and in the inner nuclear membrane in spontaneous canine model of invasive micropapillary carcinoma of the mammary gland (Pereira et al., 2015; Rodrigues et al., 2016). In general, EGF clusters are distributed around the nucleoplasm, not only in the organelles analyzed. Other nuclear compartments should be labeled to verify if EGF-EGFR complexes are present in different nuclear organelles and further studies are required to investigate the functional relevance of the differential nuclear localization of the complexes.
Fig. 5.
Nuclear EGF localizes in nuclear envelope invaginations and in nucleoli of both cell types. A- hASC and SK-HEP-1 cells were stimulated with EGF conjugated to Alexa Fluor® 488 for 40 min and had the nuclear envelope stained with an anti-lamin B2 antibody, the nucleolus stained with an anti-brillarin antibody and the nucleus labeled with Hoechst (in blue). The white arrows point to EGF clusters (in green) located in invaginations of the nuclear envelope (in red) and in the nucleolus (in orange). These clusters are in light yellow and in higher magnitude in the inserts next to the arrows. Scale bar: 2 μm. B- Three-dimensional reconstruction of the serial optical sections of 40 min-stimulated hASC and SK-HEP-1 cells. X–Z sections are shown at the top, and Y–Z sections are shown on the right of each image. Images are representative of 41 (hASC) and 37 (SK-HEP-1) cells. Scale bar: 2 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
The Airyscan detector allows acquisition of three-dimensional images with higher lateral and axial resolution compared to conventional confocal microscopy (Weisshart, 2014), making it a powerful technique to quantitatively analyze molecule distribution inside cells. Here, we showed that the quantification of EGF-488 clusters using a three-dimensional analysis of super-resolution images was effective for analyzing their translocation kinetics to the nucleus in hASC and SK-HEP-1 cells.
EGF-EGFR translocation was previously observed in tumor cell lines, and its function in the cell nucleus is associated with transcriptional regulation of genes such as cyclin D1 (Lin et al., 2001), Aurora-A (Hung et al., 2008), and STAT1 (Han et al., 2013). Furthermore, nuclear EGFR is associated with phosphorylation signaling (Wang et al., 2006), and can interact with DNA repair proteins (Dittmann et al., 2005). In this study, we evaluated and compared the kinetics profile of EGF-488 translocation to the nucleus in hASC and SK-HEP-1 cells, both of mesenchymal origin. We verified that the translocation equilibrium might occur earlier for hASC than for SK-HEP-1 cells, which show an increase of nuclear EGF cluster volume at later times of stimulation. Differences in the translocation kinetics profile of the two cell types might be associated with the higher EGFR expression shown by SK-HEP-1 or to unknown specific functions of the EGF-EGFR in the nucleus of this tumor cell type.
The agglomeration of EGF clusters in the nucleus over time suggests that they have preferable sub-nuclear localization that might be related to other functions. We observed that EGF clusters are diffusely widespread in the nucleoplasm and are also localized in nucleoplasmic reticulum of both cell types. The finding indicates possible involvement of the nucleoplasmic reticulum in EGF-EGFR translocation to the nucleus (Malhas et al., 2011), which is also involved in EGF-EGFR trafficking (Wang et al., 2012). EGF-EGFR clusters were also found in nucleoli of both cell types, which suggest new roles of the complex in this nuclear compartment. Further investigations into the functions of the nucleoplasmic reticulum, nucleoli, and other nuclear organelles in EGF-EGFR translocation are required to better understand their roles.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Elio Anthony Cino for advice and comments. The microscopic data shown in this work was obtained using the microscopes of the “Centro de Aquisição e Processamento de Imagens” (CAPI- ICB/UFMG). This work was supported by the NIH grant 1R03TW008709 and by grants from the “INCT-Regenera,” “Rede Mineira de Engenharia de Tecidos e Terapia Celular (REMETTEC, RED-00570-16),” FAPEMIG, CAPES, and CNPq.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2017.12.007.
References
- Bourgeois CA, Hemon D, Bouteille M. Structural relationship between the nucleolus and the nuclear envelope. J Ultrastruct Res. 1979;68:328–340. doi: 10.1016/s0022-5320(79)90165-5. [DOI] [PubMed] [Google Scholar]
- Bourin P, Bunnell BA, Casteilla L, Dominici M, Katz AJ, March KL, Redl H, Rubin JP, Yoshimura K, Gimble JM. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT) Cytotherapy. 2013;15:641–648. doi: 10.1016/j.jcyt.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briffa R, Um I, Faratian D, Zhou Y, Turnbull AK, Langdon SP, Harrison DJ. Multi-scale genomic, transcriptomic and proteomic analysis of colorectal cancer cell lines to identify novel biomarkers. PloS one. 2015;10:e0144708. doi: 10.1371/journal.pone.0144708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ACDA, Rodrigues MA, de Andrade C, de Goes AM, Nathanson MH, Gomes DA. Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochem Biophys Res Commun. 2011;412:341–346. doi: 10.1016/j.bbrc.2011.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Wu J-Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- Docea AAO, Mitruţ P, Cernea D, Georgescu C, Olimid D, Margaritescu C. Immunohistochemical expression of EGF, c-erbB-2 and EGFR in intestinal variant of gastric adenocarcinomas. Rom J Morphol Embryol. 2013;54:545–554. [PubMed] [Google Scholar]
- Du Y, Shen J, Hsu J, Han Z, Hsu M, Yang C, Kuo H, Wang Y, Yamaguchi H, Miller S, et al. Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene. 2013;33:756–770. doi: 10.1038/onc.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating co-localization in biological microscopy. Am J Physiol-Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun JR, Jung YJ, Zhang Y, Zhang Y, Tschudy-Seney B, Ramsamooj R, Wan YJY, Theise ND, Zern MA, Duan Y. Hepatoma SK Hep-1 cells exhibit characteristics of oncogenic mesenchymal stem cells with highly metastatic capacity. PloS one. 2014;9:e110744. doi: 10.1371/journal.pone.0110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett M. Acquiring and analyzing data for colocalization experiments in AIM or ZEN Software. Carl Zeiss MicroImaging n.d [Google Scholar]
- Faria JA, de Andrade C, Goes AM, Rodrigues MA, Gomes DA. Effects of different ligands on epidermal growth factor receptor (EGFR) nuclear translocation. Biochem Biophys Res Commun. 2016;478:39–45. doi: 10.1016/j.bbrc.2016.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden P, Carpentier J-L, Cohen S, Orci L. Epidermal growth factor: morphological demonstration of binding, internalization, and lysosomal association in human fibroblasts. Proc Natl Acad Sci. 1978;75:5025–5029. doi: 10.1073/pnas.75.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler HT, McKANNA JA, Cohen S. Direct visualization of the binding and internalization of a ferritin conjugate of epidermal growth factor in human carcinoma cells A-431. J Cell Biol. 1979;81:382–395. doi: 10.1083/jcb.81.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton N. Quantification and its applications in fluorescent microscopy imaging. Traffic. 2009;10:951–961. doi: 10.1111/j.1600-0854.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- Han W, Carpenter RL, Cao X, Lo H-W. STAT1 gene expression is enhanced by nuclear EGFR and HER2 via cooperation with STAT3. Mol Carcinog. 2013;52:959–969. doi: 10.1002/mc.21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Lo H-W. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Fukui H, Ono Y, Sekikawa A, Ichikawa K, Tomita S, Imai Y, Imura J, Hiraishi H, Fujimori T. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- Hsu S-C, Hung M-C. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- Huang W-C, Chen Y-J, Li L-Y, Wei Y-L, Hsu S-C, Tsai S-L, Chiu P-C, Huang W-P, Wang Y-N, Chen C-H, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung L-Y, Tseng JT, Lee Y-C, Xia W, Wang Y-N, Wu M-L, Chuang Y-H, Lai C-H, Chang W-C. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucl Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-H, Wang Y-N, Hung M-C. Non-canonical signaling mode of the epidermal growth factor receptor family. Am J Cancer Res. 2015;5:2944. [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harbor Persp Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-F, Fang F-M, Wang J-M, Tzeng C-C, Tai H-C, Wei Y-C, Li S-H, Lee Y-T, Wang Y-H, Yu S-C, et al. EGFR nuclear import in gallbladder carcinoma: nuclear phosphorylated EGFR upregulates iNOS expression and confers independent prognostic impact. Ann Surg Oncol. 2012;19:443–454. doi: 10.1245/s10434-011-1942-6. [DOI] [PubMed] [Google Scholar]
- Li L-Y, Chen H, Hsieh Y-H, Wang Y-N, Chu H-J, Chen Y-H, Chen H-Y, Chien P-J, Ma H-T, Tsai H-C, Lai C-C, Sher Y-P, Lien H-C, Tsai C-H, Hung M-C. Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 2011;71:4269–4279. doi: 10.1158/0008-5472.CAN-10-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung M-C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Lo H-W, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu S-C, Hung M-C. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006a;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- Lo H-W, Hsu S-C, Hung M-C. EGFR signaling pathway in breast cancers: from traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006b;95:211–218. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- Lo H-W, Xia W, Wei Y, Ali-Seyed M, Huang S-F, Hung M-C. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- Loupakis F, Cremolini C, Fioravanti A, Orlandi P, Salvatore L, Masi G, Schirripa M, Di Desidero T, Antoniotti C, Canu B, et al. EGFR ligands as pharmacodynamic biomarkers in metastatic colorectal cancer patients treated with cetuximab and irinotecan. Targeted Oncol. 2014;9:205–214. doi: 10.1007/s11523-013-0284-7. [DOI] [PubMed] [Google Scholar]
- Malhas A, Goulbourne C, Vaux DJ. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Miyata K, Yotsumoto F, Fukagawa S, Kiyoshima C, Ouk NS, Urushiyama D, Ito T, Katsuda T, Kurakazu M, Araki R, et al. Serum heparin-binding epidermal growth factor-like growth factor (HB-EGF) as a biomarker for primary ovarian cancer. Anticancer Res. 2017;37:3955–3960. doi: 10.21873/anticanres.11779. [DOI] [PubMed] [Google Scholar]
- Olsen DA, Bechmann T, Østergaard B, Wamberg PA, Jakobsen EH, Brandslund I. Increased concentrations of growth factors and activation of the EGFR system in breast cancer. Clin Chem Lab Med. 2012;50:1809–1818. doi: 10.1515/cclm-2011-0823. [DOI] [PubMed] [Google Scholar]
- Pereira NB, Do-Carmo AC, Diniz MG, Gomez RS, Gomes DA, Gomes CC. Nuclear localization of epidermal growth factor receptor (EGFR) in ameloblastomas. Oncotarget. 2015;6:9679–9685. doi: 10.18632/oncotarget.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk A, Paul G, Incardona P, Bugarski M, Mansouri M, Niemann A, Ziegler U, Berger P, Sbalzarini IF. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat Protoc. 2014;9:586–596. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- Rodrigues MA, Gamba CO, Faria JAQA, Ferreira Ê, Goes AM, Gomes DA, Cassali GD. Inner nuclear membrane localization of epidermal growth factor receptor (EGFR) in spontaneous canine model of invasive micropapillary carcinoma of the mammary gland. Pathol-Res Pract. 2016;212:340–344. doi: 10.1016/j.prp.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Szoboszlay M, Kirizs T, Nusser Z. Objective quantification of nanoscale protein distributions. Sci Rep. 2017;7:15240. doi: 10.1038/s41598-017-15695-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor AM, Weigel TL, Oettel KR, Yang DT, Zhang C, Kim K, Salgia R, Iida M, Brand TM, Hoang T, Campbell TC, Hernan HR, Wheeler DL. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer. 2013;81:138–141. doi: 10.1016/j.lungcan.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-C, Nakajima Y, Yu Y-L, Xia W, Chen C-T, Yang C-C, McIntush EW, Li L-Y, Hawke DH, Kobayashi R, Hung M-C. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Wang Y-N, Hung M-C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:1. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-N, Lee H-H, Lee H-J, Du Y, Yamaguchi H, Hung M-C. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J Biol Chem. 2012;287:16869–16879. doi: 10.1074/jbc.M111.314799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-N, Wang H, Yamaguchi H, Lee H-J, Lee H-H, Hung M-C. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem Biophys Res Commun. 2010a;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-N, Yamaguchi H, Huo L, Du Y, Lee H-J, Lee H-H, Wang H, Hsu J-M, Hung M-C. The translocon Sec61$\beta$ localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010b;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K. The Basic Principle of Airyscanning. Zeiss Technology; 2014. Note 22. [Google Scholar]
- Xia W, Wei Y, Du Y, Liu J, Chang B, Yu Y-L, Huo L-F, Miller S, Hung M-C. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.