Abstract
Myceliophthora thermophila (ATCC 42464) is a thermophilic fungus that produces cellulolytic enzymes with high thermal stability. Unlike its mesophile counterparts, study on gene expression regulation of cellulolytic enzymes in M. thermophila is inadequate. This work identified the function of MHR1, a putative transcription regulator of cellulolytic enzymes in M. thermophila that was found through RNA-Seq based gene expression profile analysis. RNA interference was used to study the role of MHR1. A recombinant plasmid, pUC19–Ppdc–mhr1–Tpdc, which contained the RNAi sequence for mhr1 was constructed and transformed into M. thermophila. One of the transformants, MtR5, in which the RNA interference efficiency was the highest, was used for the following studies. In the mhr1-silenced strain MtR5, the filter paper hydrolyzing activity was 1.33-fold; β-1, 4-endoglucanase activity was 1.65-fold; and xylanase activity was 1.48-fold higher than those of the parental strain after induction, respectively, by wheat straw powder. qRT-PCR showed that gene expression of cbh1, cbh2, egl3 and xyr1 were 9.56-, 37.36-, 56.14- and 28.30-fold higher in MtR5 than in wild type, respectively. Our findings suggest that the transcription factor MHR1 of M. thermophila can repress cellulase and xylanase activities. Silenced mhr1 results in increased expression not only of the main cellulase genes, but also of the positive regulatory gene xyr1. This work is relevant to the development of M. thermophila as an industrial production host for cellulolytic and hemicellulolytic enzymes, which could be used to degrade a wide range of different biomass, converting lignocellulosic feedstock into sugar precursors for biofuels.
Keywords: Myceliophthora thermophila, Cellulase, Xylanase, RNAi, Transcription factor
Introduction
Myceliophthora thermophila is a thermophilic fungus that grows optimally at 45 °C. Its extracellular enzymes have high thermal stability, can degrade cellulose effectively and be used for the production of biofuels (Maheshwari et al. 2000). The genome of M. thermophila strain ATCC 42464 was sequenced in 2011 (Berka et al. 2011). This strain secretes a large number of thermostable cellulolytic and hemicellulolytic enzymes that can degrade a wide range of biomass, including cellulose and hemicellulose. These enzymes have potential industrial applications, turning different kinds of lignocellulosic feedstock into sugar precursors for biofuels and chemicals (Kolbusz et al. 2014). M. thermophila could be developed as an industrial production host for cellulolytic and hemicellulolytic enzymes, on the basis of genomic analysis (Karnaouri et al. 2014).
Numerous recent studies have focused on M. thermophila ATCC 42464. Xu et al. developed a high efficiency Agrobacterium tumefaciens-mediated transformation system for M. thermophila, including targeted gene deletion using green fluorescent protein (GFP) as a selectable marker, and a ku70 deletion mutant was constructed in the ATCC 42464 background population (Xu et al. 2015). Overexpression of xyr1 under the control of the MtPpdc (pyruvate decarboxylase) promoter results in enhanced xylanase activity in M. thermophila (Wang et al. 2015). CRE1 negatively regulates cellulase gene expression in M. thermophila, as shown by an increase in cellulase production using RNA interference of cre1 (Yang et al. 2015). This previous work suggested that other transcription factors could regulate cellulase and hemicellulase activity in M. thermophila.
M. thermophila ATCC42464 was cultured using glucose and Avicel as the sole carbon source, respectively. The gene expression profilings were analyzed based on Solexa sequencing technology of RNA-Seq (Lockhart and Winzeler 2000; Wang et al. 2009). NOIseq methods (Tarazona et al. 2015) were used for screening differentially expressed genes in two groups. Thirty-five potential transcription factors were found (unpublished data). MHR1 is one of them.
MHR1 is a hypothetical protein, 733 amino acids long. The fungal_TF_MHR (fungal transcription factor regulatory middle homology region) runs from residue positions 135 to 558. It has 63% amino acid sequence homology with a putative transcription factor from Chaetomium thermophilum var. thermophilum DSM 1495, 48% homology with a fungal-specific transcription factor from Colletotrichum higginsianum and 44% homology with a hypothetical protein M419DRAFT_95750 from Trichoderma reesei RUT C-30. However, the function of MHR1 and its relationship with regulation of cellulase genes are still unknown. In the present study, we designed an RNAi sequence for mhr1 and constructed a recombinant plasmid. Cellulase activities and gene expression of the main cellulase genes were studied in both the recombinant and wild-type strains cultured in the inducing media. Results indicated that RNA interference of mhr1 resulted in enhanced cellulase production in M. thermophila.
Materials and methods
Strains, plasmids and cultivation conditions
Escherichia coli JM107 (Thermo Fisher Scientific Fermentas, Canada) was used as the host strain for recombinant plasmid construction. M. thermophila (ATCC 42464) was purchased from the American Type Culture Collection center and used as the parent strain throughout this study. The E. coli strain was cultivated in LB medium supplemented with 100 µg/mL ampicillin (Applied Biosystems, USA) at 37 °C. M. thermophila was maintained on potato dextrose agar (PDA) at 45 °C. Transformants (MtR(n)) were selected on PDA with 50 μg/mL hygromycin B (AMRESCO, USA). Spores were washed and suspended in a 0.1% Tween-80 solution. For liquid culture, M. thermophila was grown in Mandel’s medium with 2% glucose at 45 °C, in 250-ml Erlenmeyer flasks, shaking at 250 rpm. M. thermophila produces cellulase when 3% wheat straw powder (Solarbio, China) is used as inducer (Van den Brink et al. 2013). The pUC19-Mtpdc plasmid contains the promoter and terminator of the pyruvate decarboxylase gene of M. thermophila (Ppdc and Tpdc, respectively) and was previously constructed in our laboratory (Wang et al. 2015). Plasmid pAN7-1, which contains the hygromycin B-resistance cassette, was used as an assisting plasmid for the transformation of M. thermophila (Punt et al. 1987).
Construction of the RNAi plasmid
The mhr1 gene sequence was obtained from GenBank (accession AEO57591.1). Small interfering RNA (siRNA) sequences of the mhr1 gene were designed using a siRNA design program (http://rnaidesigner.lifetechnologies.com/rnaiexpress/) (Table 1). siRNA sequences were synthesized by Sangon (Shanghai, China). A stable stem-loop structure 5′–TTCAAGAGA–3′ connected complementary oligonucleotides (underlined in Table 1; Brummelkamp et al. 2002). The restriction site at the 5′ end of the sense strand is Not1, while Xbal1 is at the 5′ end of the antisense strand. Double-stranded siRNA-mhr1 was obtained using PCR, with the following reaction conditions: 40% nuclease-free water, 20% annealing buffer for DNA oligos (5×), 20% DNA oligo S (50 µM), and 20% DNA oligo A (50 µM) (Sangon, China); initial denaturation at 95 °C for 2 min, falling 0.1 °C per second to 25 °C (Yang et al. 2015). The resulting double-stranded siRNA-mhr1 with a cohesive terminus was inserted into the pUC19–pdc plasmid containing the MPpdc–mhr1–MTpdc cassette (Wang et al. 2015). Sequence analysis was performed (Sangon, China) to verify the insertion of siRNA-mhrl in the recombinant plasmid.
Table 1.
The siRNA oligonucleotide sequences of target gene mhr1
| Name | Sequence (5′–3′) |
|---|---|
| mhr1 sense | GGCC GCAAGGCTTCTTGCGAAAT TTCAAGAGA ATTTCGCAAGAAGCCTTGC |
| mhr1 anti | CTAG GCAAGGCTTCTTGCGAAAT TCTCTTGAA ATTTCGCAAGAAGCCTTGC |
The 5′ end of the sense and antisense siRNA sequences are Not I and Xba I restriction sites, respectively (shown in italics). Underlined and double underlined sequences are complementary oligonucleotides. The bold sequences are stem-loop structures
Protoplast preparation and transformation of M. thermophila
Protoplast preparation and transformation was modified from our previous study (Wang et al. 2015). For a better rate of germination, spores were cultured at 45 °C and 160 rpm for 9 h. A few spores of single colonies of candidate transformations were maintained on PDA plates at 2 days, for picking of mycelium. The picked mycelium was mixed with 15 μL lysis buffer for direct PCR (Takara, Japan) and lysed at 80 °C for 15 min. The resultant genomic DNA was used as the template for PCR to verify whether the MPpdc–mhr1–MTpdc cassette had integrated into the genome of transformants. Primers used are listed in Table 2. Sequence analysis was performed at Sangon (Shanghai, China).
Table 2.
Primer sequences used for verifying the recombinant vector, and for qRT-PCR
| Primer name | Sequence (5′–3′) |
|---|---|
| Mt-Ppdc-F | CCCAAGCTTCCGAGTGTACTCCGTAAGGA |
| Mt-Tpdc-R | CCGGAATTCGGATTACAGCGCAGTGCACG |
| RTq-tub-F | AGGGTATGGATGAGATGGAG |
| RTq-tub-R | AGAAGCAAGCCCTGGAAC |
| RTq-mhr1-R | CGCAGTACGCTACGATGTCT |
| RTq-cbh1-F | GATGGTCCTTCTTGCGCCTCC |
| RTq-cbh1-R | TTACCGCTCGTGGTGATGC |
| RTq-cbh2-F | TTCTCGGGCGTCCGGCTCTTC |
| RTq-cbh2-R | GGAATGGCGAGATTGTGGA |
| RTq-egl1-F | TCGGGCAACGGTTACGAGG |
| RTq-egl1-R | AGCTCTTGACCTGGTTCTGG |
| RTq-egl3-F | TCCGCCACCAGCACCGCCCCTC |
| RTq -egl3-R | TTGCTGCCGAGCCACTTC |
| RTq-xyr1-F | CCAGATCTTCGGGGGCAGC |
| RTq-xyr1-R | GTACTCGCAGCCCAGTTGGAAC |
RNA extraction
Mycelia were collected by centrifugation at 5000 rpm and 4 °C for 10 min. Total RNA of each of the samples was isolated from frozen mycelia, using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA quantity and quality were measured with a NanoDrop ND-2000 spectrometer (Thermo, USA), and RNA integrity was assessed using standard denaturing agarose gel electrophoresis.
Quantitative real-time reverse transcription PCR (qRT-PCR)
The PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Japan) was used to remove genomic DNA from the total RNA samples. Complementary DNA (cDNA) was synthesized for each sample from 1.0 µg prepared total RNA. The qRT-PCR protocol has been described previously (Li et al. 2012) and was performed on a StepOnePlus™ qPCR machine (ABI, USA). Gene expression levels were normalized with the β-tubulin gene (GenBank AEO58945.1) as an endogenous control. The primers used for qRT-PCR were designed using the software Prime 5.0 and are listed in Table 2.
Enzyme activity assays and total extracellular protein concentration
Supernatants from liquid cultures were collected by centrifuging at 12,000 rpm for 10 min to remove mycelia and residual medium. For cellulase activity assays, each supernatant was diluted fivefold with 0.05 M sodium phosphate (Na2HPO4) citric acid buffer (pH 4.8). Filter paper activity (FPA) and endoglucanase (EGL) activities were assayed using filter paper (Whatman, UK) with a 10 mm × 30 mm standard and 1% sodium carboxymethyl cellulose (CMC-Na) solution as substrates, respectively (Ghose 1987). Enzyme reactions were performed in 0.05 M sodium pyrophosphate (Na2HPO4) citric acid buffer (pH 4.8) at 50 °C for 60 and 30 min, respectively; then the released reducing sugar was quantified using the DNS method (Sun et al. 2008). Xylanase activity was assayed using 1% xylan from birchwood (Sigma, USA) as the substrate, according to the method of Bailey et al. (1992).
The concentration of total extracellular protein was quantified using Modified Bradford Protein Assay Kit (Sangon), following the manufacturer’s instructions.
Results
Construction of mhr1-silenced strains in M. thermophila
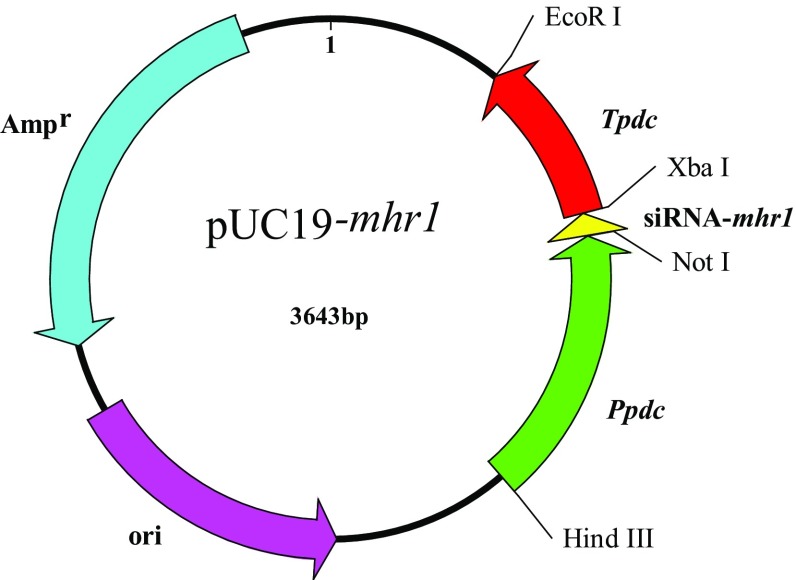
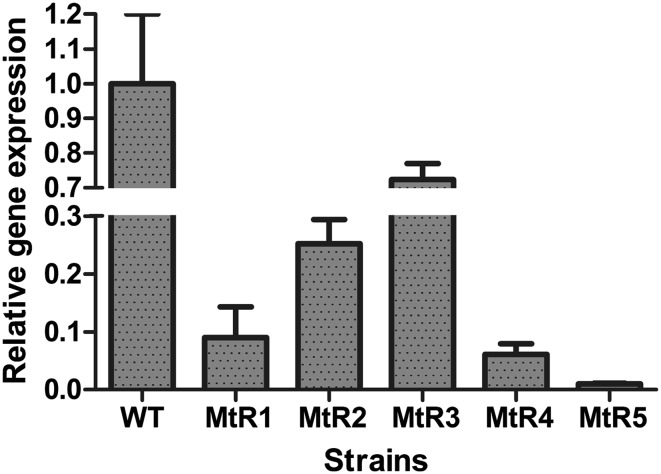
To silence the expression of the mhr1 gene in M. thermophila, the pUC19–Ppdc–mhr1–Tpdc plasmid (Fig. 1) was transformed into M. thermophila with the aid of plasmid pAN7.1, through co-transformation. Six single mycelial colonies were screened from the PDA plates. Subsequently, five colonies named MtR(n) (MtR1, MtR2, MtR3, MtR4, MtR5) were verified as containing the mhr1-interference sequence by PCR and DNA sequencing. From these five transformants, MtR5 was found to have the best RNAi efficiency by qRT-PCR. The transcript level of the target gene mhr1 in transformant MtR(n) was reduced to 72.4, 25.2, 9, 6.1 and 1%, respectively, in MtR1–5, as compared to wild-type (WT) transcript levels (Fig. 2). As MtR5 showed the highest RNAi efficiency, it was used for further study.
Fig. 1.
Schematic representation of the RNA interference vector. Ppdc, pdc promoter of M. thermophila; siRNA-mhr1, small interfering RNA sequences including the mhr1 sense sequence, stem-loop, and mhr1 antisense sequence; Tpdc, pdc terminator of M. thermophila
Fig. 2.
qRT-PCR analysis of mhr1 mRNA levels in M. thermophila transformants under inducing conditions
The morphology of MtR5
The growth and development of WT and MtR5 strains were almost synchronous on PDA plates. When spores matured after 7 days, the spore layer of the transformant MtR5 was thinner, and the spores of MtR5 were more mucilaginous, compared with those of the WT strain. From the same surface area of the medium, the quantity of MtR5 spores obtained was about half that of the WT spores (Fig. 3). Obvious hyphae were present on the surface of the MtR5 medium, making it difficult to scrape clean.
Fig. 3.

Conidiation of wild-type and transformant (MtR5) growth at 45 °C for 7 days on PDA plates without a selective reagent
Enhancing cellulase and xylanase production in the mhr1-silenced strain MtR5
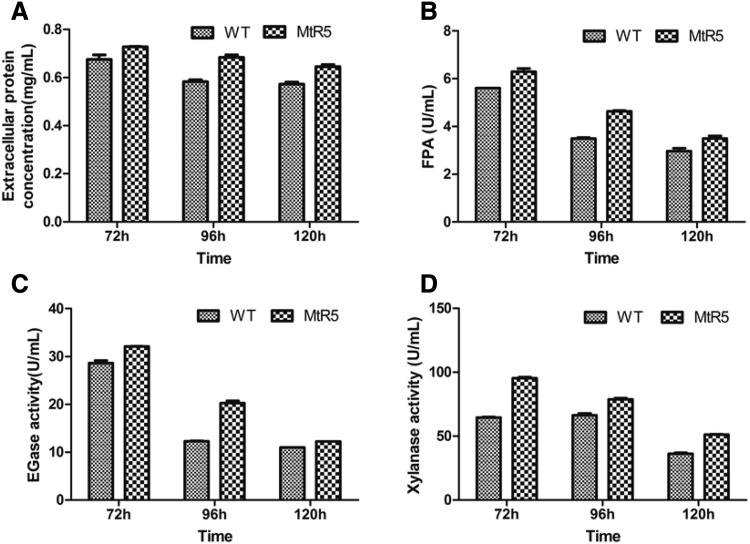
WT and MtR5 strains were cultured in Mandel’s medium with 0.2% glucose and 3% wheat straw powder. Extracellular protein concentrations, filter paper activity (FPA), endoglucanase (EG) and xylanase activities were determined for the supernatants of WT and MtR5 strains, at 72, 96 and 120 h. The concentration of extracellular protein in strain MtR5 was 1.17-fold higher than that of WT when cultured for 96 h (Fig. 4a). Cellulase activity was elevated in MtR5 at each time point, compared with WT. FPA (Fig. 4b) and EGase activities of MtR5 (Fig. 4c) were 1.33- and 1.65-fold higher, respectively, at 96 h, and xylanase activity was 1.48-fold higher at 72 h, compared with WT (Fig. 4d).
Fig. 4.
Extracellular protein concentration and enzyme activities of the original strain and the transformant. a–d Extracellular protein concentration, the filter paper (FP), β-1, 4-endoglucanase (EG) and xylanase activities, respectively
Expression of the main cellulase genes in the mhr1-silenced transformant under inducing conditions
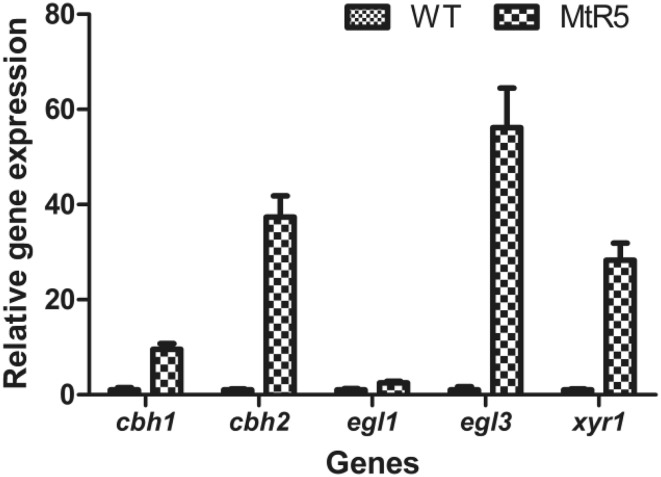
To illustrate the relationship between MHR1 and cellulase, the expressions of the main cellulase genes (cbh1, cbh2, egl1 and egl3) and a positive regulator, XYR1, were measured by qRT-PCR. Compared with WT, expressions of cbh1, cbh2, egl3 and xyr1 were increased in MtR5, being 9.56-, 37.36-, 56.14- and 28.30-fold higher, respectively (Fig. 5). The expression of egl1 was slightly increased in MtR5, being 2.52-fold higher than that of WT (Fig. 5). These results were concordant with the enzyme activity results.
Fig. 5.
Expression of the main cellulase genes and regulator xyr1 in WT and MtR5 under inducing conditions
Discussion
MHR1 includes the fungal_TF_MHR region, making it a transcription factor of fungi in the broadest sense. The present results showed that RNA interference of mhr1 can increase cellulase activities, when the strains are cultured in inducing medium. Fluorescence quantitative results showed that the expression of various cellulase-related genes increased in the recombinant strain MtR5, which indicates that MHR1 is associated with carbon metabolic repression, and this transcription factor functions as a repressor. Thus, silencing of mhr1 facilitates the production of cellulase in the recombinant strain. However, the total extracellular protein of the recombinant strain was also slightly higher than that of wild type.
This study demonstrated that the interference of mhr1 can also increase xylanase activity. The results of qRT-PCR showed that the expression of xyr1 increased in the recombinant strain MtR5. The transcriptional activator XYR1 is the central regulator that governs cellulolytic and xylanolytic gene expression in Trichoderma reesei (Furukawa et al. 2009; Stricker et al. 2008; Mach-Aigner et al. 2008), Aspergillus niger (van Peij et al. 1998; Omony et al. 2016), and Neurospora crassa (Sun et al. 2012). However, the function of (hemi-) cellulolytic regulator Xyr1 (XlnR/Xlr1) is different in filamentous fungi (Klaubauf et al. 2014; Amore et al. 2013; Brunner et al. 2007). XYR1 is an activator of xylanase in Myceliophthora thermophila ATCC 42464 (Wang et al. 2015). Therefore, interference with mhr1 expression either resulted in a direct increase of xylanase activity in strain MtR5, or the increase in xyr1 expression resulted in an indirect increase of xylanase activity.
In the present study, the silence of MHR1 resulted in the dramatic increase of transcription levels of several genes (cbh1, cbh2, egl3 and xyr1). However, the enzyme activity measured was not increased too much. We also observed the same phenomena, and the possible reason has been explained in our previous study (Yang et al. 2015).
There is no published report describing the function of the MHR1 protein. Here, we have described the relationship between MHR1 and the regulation of cellulase. However, it remains unclear whether this transcription factor has any connection with known cellulase repressors CRE1 and ACE1; and where MHR1 binds to cellulase genes.
Conclusion
MHR1 is a regulatory gene, with the generic fungal transcription factor middle homology region. RNA interference of mhr1 resulted in enhanced cellulase and xylanase productions in M. thermophila. The expression of the regulatory gene xyr1 was also enhanced. This is the first description of the relationship between MHR1, cellulase and xylanase.
Acknowledgements
This research was supported by the Shenzhen Municipal Science and Technology key projects of the Basic Research Program (JYJC20140418091413587).
Compliance with ethical standards
Conflict of interest
The authors have declared that no conflicts of interest exist.
Contributor Information
Juan Wang, Phone: 86-755-26534152, Email: wangjuan@szu.edu.cn.
Gang Liu, Phone: 86-755-26534966, Email: zjuliug@szu.edu.cn.
References
- Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Genom. 2013;4:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23:257–270. doi: 10.1016/0168-1656(92)90074-J. [DOI] [Google Scholar]
- Berka RM, Grigoriev IV, Otillar R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29:922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Brunner K, Lichtenauer AM, Kratochwill K, Delic M, Mach RL. Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr Genet. 2007;6:213–220. doi: 10.1007/s00294-007-0154-x. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Shida Y, Kitagami N, Mori K, Kato M, Kobayashi T, Okada H, Ogasawara W, Morikawa Y. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet Biol. 2009;8:564–574. doi: 10.1016/j.fgb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Ghose T. Measurement of cellulase activities. Pure Appl Chem. 1987;59:257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- Karnaouri A, Topakas E, Antonopoulou I, Christakopoulos P. Genomic insights into the fungal lignocellulolytic system of Myceliophthora thermophila. Front Microbiol. 2014;5:281–302. doi: 10.3389/fmicb.2014.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S, Narang HM, Post H, Zhou M, Brunner K, Mach-Aigner AR, Mach RL, Heck AJ, Altelaar AF, de Vries RP. Similar is not the same: differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet Biol. 2014;72:73–81. doi: 10.1016/j.fgb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Kolbusz MA, Di Falco M, Ishmael N, Marqueteau S, Moisan MC, Baptista Cda S, Powlowski J, Tsang A. Transcriptome and exoproteome analysis of utilization of plant-derived biomass by Myceliophthora thermophila. Fungal Genet Biol. 2014;72:10–20. doi: 10.1016/j.fgb.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Li JX, Wang J, Wang SW, Xing M, Yu SW, Liu G. Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters. Microb Cell Fact. 2012;11:84–93. doi: 10.1186/1475-2859-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405(6788):827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl Environ Microbiol. 2008;21:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari R, Bharadwaj G, Bhat MK. Thermophilic fungi: their physiology and enzymes. Microbiol Mol Biol Rev. 2000;3:461–488. doi: 10.1128/MMBR.64.3.461-488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omony J, Mach-Aigner AR, van Straten G, van Boxtel AJ. Quantitative modeling and analytic assessment of the transcription dynamics of the XlnR regulon in Aspergillus niger. BMC Syst Biol. 2016;10:13–22. doi: 10.1186/s12918-016-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;1:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- Stricker AR, Mach RL, de Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;2:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- Sun X, Liu Z, Qu Y, Li X. The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Appl Biochem Biotechnol. 2008;1–3:119–128. doi: 10.1007/s12010-007-8049-3. [DOI] [PubMed] [Google Scholar]
- Sun J, Tian C, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012;4:482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, Conesa A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucl Acids Res. 2015;43(21):e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Brink J, van Muiswinkel GC, Theelen B, Hinz SW, de Vries RP. Efficient plant biomass degradation by thermophilic fungus Myceliophthora heterothallica. Appl Environ Microbiol. 2013;4:1316–1324. doi: 10.1128/AEM.02865-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;10:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu Y, Gong Y, Yu S, Liu G. Enhancing xylanase production in the thermophilic fungus Myceliophthora thermophila by homologous overexpression of Mtxyr1. J Ind Microbiol Biotechnol. 2015;9:1233–1241. doi: 10.1007/s10295-015-1628-3. [DOI] [PubMed] [Google Scholar]
- Xu J, Li J, Lin L, Liu Q, Sun W, Huang B, Tian C. Development of genetic tools for Myceliophthora thermophila. BMC Biotechnol. 2015;15:35–44. doi: 10.1186/s12896-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gong YF, Liu G, Zhao SM, Wang J. Enhancing cellulase production in thermophilic fungus Myceliophthora thermophila ATCC42464 by RNA interference of cre1 gene expression. J Microbiol Biotechnol. 2015;7:1101–1107. doi: 10.4014/jmb.1501.01049. [DOI] [PubMed] [Google Scholar]