Abstract
The advent of mobile health (mHealth) technologies challenges the capabilities of current visualizations, interactive tools, and algorithms. We present Chronodes, an interactive system that unifies data mining and human-centric visualization techniques to support explorative analysis of longitudinal mHealth data. Chronodes extracts and visualizes frequent event sequences that reveal chronological patterns across multiple participant timelines of mHealth data. It then combines novel interaction and visualization techniques to enable multifocus event sequence analysis, which allows health researchers to interactively define, explore, and compare groups of participant behaviors using event sequence combinations. Through summarizing insights gained from a pilot study with 20 behavioral and biomedical health experts, we discuss Chronodes’s efficacy and potential impact in the mHealth domain. Ultimately, we outline important open challenges in mHealth, and offer recommendations and design guidelines for future research.
Additional Key Words and Phrases: Mobile health sensor data, mHealth, sequence mining, cohort discovery, event alignment
CCS Concepts: Human-centered computing → Visual analytics, Applied computing → Health care information systems
1 INTRODUCTION
As revolutionary developments in mobile sensor technology extend to more applications in healthcare, the potential for understanding patterns and relationships among physiological factors is burgeoning. With these sensors, health researchers are afforded new capabilities to collect and analyze health data from study participants with more accuracy and precision than ever before. We have commenced a pioneering initiative to develop innovative methods that turn the wealth of mobile health (mHealth) sensor data unlocked by these emerging and evolving sensors into reliable and actionable health information. As smoking is the leading yet preventable cause of death in the United States, and as smoking lapse contributes to a high failure rate in quit attempts, smoking cessation is one of the most challenging research problems in this area. For this reason, we have selected smoking cessation and lapse as our primary topic of focused analysis and are currently developing tools to assist health researchers in discovering temporal, physiological, and behavioral patterns and factors that cause abstinent smokers to lapse.
To assess the behaviors surrounding smoking abstinence and relapse, health researchers need to to understand lifestyle choices and activities that are related to smoking relapse, and are particularly interested in the sequences of events leading up to lapse [5, 25]. Furthermore, whereas analysts are interested in determining universal indicators and causes of smoking relapse, behavioral variation between participant groups is equally relevant [2, 10]. Should younger individuals’ daily routines be distinguished from those of older? Will early morning and late night smokers exhibit different presmoking behaviors [26]?
To begin answering these questions, researchers need tools to discover which subsets (cohorts) of abstinent smokers exhibit similar or dissimilar behaviors, and to compare the groups as such. Ultimately, these methods must also represent longitudinal, high-resolution mHealth datasets. Figure 1 shows a snapshot of this complexity for only a few evenings’ worth of mHealth data—visualizing the data as-is is complex and difficult to make sense of. Exploring how chronological patterns relate to common behaviors is nontrivial, as it is a challenge not only to define groups of behaviors from mHealth data but also to examine and compare these groups interactively. In contrast to traditional electronic health records (EHRs) where temporal events are already explicitly defined, largely nonoverlapping, and do not repeat frequently throughout the day, mHealth analysis requires that event chronology is derived from overlapping sensor data streams that span daytime continuously.
Fig. 1.
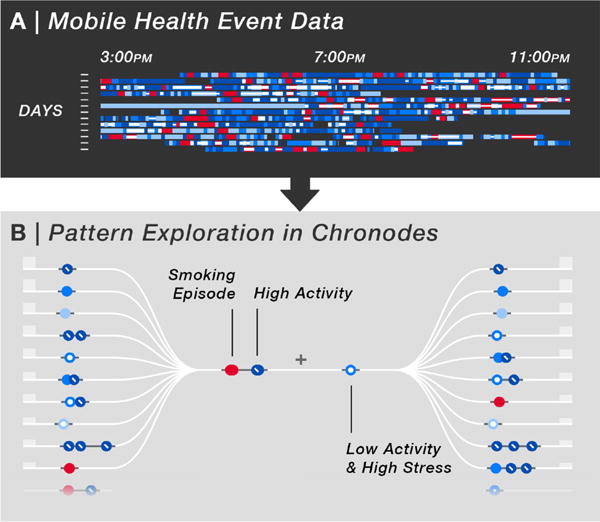
From mobile health (mHealth) event data, Chronodes extracts frequent patterns of events that users can interactively explore. (A) Multiple days of mHealth sensor data are represented as discrete events over time. Only a snapshot of the full dataset is shown. (B) Chronodes mines frequent sequences of events that users can interactively combine to reveal patterns that occur before and after them (left and right). All figures are best viewed in color. Red, smoking episodes; blue, activity intensity.
1.1 Our Contributions
mHealth data introduces unique challenges that existing tools for analyzing EHRs do not adequately address. Chronodes is one of the first attempts to explore how to best represent and explore mHealth data, combining data mining and human-centric visualization techniques to aid data exploration, pattern discovery, and fine-grain analysis of longitudinal mHealth records.
- Chronodes introduces two novel capabilities that help health researchers interactively explore and combine sequences to ask important, complex questions such as “what happens between first lapses that precede high activity, and second lapses that follow low activity but high stress?” and then to generate and explore variations of this question interactively:
- Although existing tools align on multiple occurrences of a single event type, Chronodes allows alignment on two or more event types so that users can see what happens in between (e.g., between a first and second lapse).
- Chronodes’ alignment points can be a single event, or a sequence consisting of multiple events (e.g., smoking lapse → high activity), which can be defined constructively during event sequence exploration.
We conducted an informal study with 20 expert researchers from a spectrum of mHealth related disciplines. By summarizing the insights gained, we outline important open research and design challenges in developing the mHealth field, and offer recommendations and design guidelines for future research.
2 RELATED WORK
Our research builds on prior works in multiple disciplines, from visualization, health research, and data mining.
2.1 Visualization of Multiple Timelines
For EHRs and mobile health data, aggregating the event data of multiple patient timelines into a single timeline can be difficult to make sense of–the order of events from one timeline to another is completely uncorrelated. This challenge is customary in healthcare analysis: apart from needing to understand the relationships between a patient’s various physiological records, analysts must do the same between thousands of patients on end [3]. Principally, Aigner et al. [1] and Brehmer et al. [6] provide a series of typologies and guidelines for visualizing timeline data according to a variety of data types and representational criteria. Meanwhile, approaches to “multifocus” timeline visualization [38, 42] compliment such techniques by enabling users to select multiple segments of time from timeline visualizations, which can then be displayed in a consolidated format and used to find similar patterns [43]. Fundamentally, visualizing dissimilar records side by side proposed in Fouse et al. [13] enables users to identify high-level correlations between the distinct data streams. However, even these visualization approaches become increasingly intractable as we scale to more patients and sensor kinds, which demands new visual and algorithmic techniques for summarizing the data of multiple, concurrent timelines. Accordingly, in an effort to unify many patient timelines, CareFlow [28] visualizes the outcomes of 50,000 patients in a single, tree-like timeline. Although this approach provides an expressive high-level overview of similar patient groups, it has limited support for the specification of behavioral groups and repetitive event sequences. Whereas Chronodes also provides a summary of participant data in this fashion (Figure 1A), its functional concentration is a new sequence-based overview that reveals repeating event patterns at a glance.
2.2 Event Alignment
Tools such as EventFlow [24] and LifeFlow [39], as well as others in non–health-related fields such as Experiscope [17], allow users to select a specific event (e.g., drug A prescribed), thereby displaying the events that occur both before and after the selected event, for all data records. This technique of “event alignment” is useful for extrapolating event causation and chronological trends, and also for organizing event flows around a consistent visual reference point.
Still, even with alignment, the abundance of data presented by these interfaces, especially for large datasets, is complex and difficult to understand at a glance. To mitigate this visual overload, existing tools rely on techniques of simplification, such as Find-and-Replace [24], which can compress multiple user-specified events into one. Although this kind of simplification reduces the number of visual elements on the screen, it hides information as the user is repeatedly working toward an increasingly simplistic representation–it is difficult to recover how the simplified data stream represents the original data [24]. With an interactive strategy similar to event alignment, Chronodes reveals event-based patterns from aggregated data streams without the visual complexity of presenting mHealth data records as is. Then, as opposed to iteratively consolidating events to simplify the visualization of all participant records, Chronodes leverages the properties of frequent event sequences to depict patterns of behaviors that can be constructed and explored interactively.
2.3 Cohort Analysis
An alternate approach to understanding the trends of aggregated mHealth data streams is to consider groups of patients as cohorts, or individuals that share certain properties [4, 23]. In an effort to comparatively assess sequences of events between cohorts, projects such as CoCo (Cohort Comparison) [23] show the chronology of events associated with two user-defined cohorts. Whereas this effectively reveals the different properties and event sequences between specified cohorts, CoCo focuses on specifying and comparing the attributes of two distinctly defined cohorts (e.g., male and female), and does not enable the user to interactively and dynamically redefine cohort groups by arranging sequences of events. Chronodes builds on the cohort comparison methodology by allowing the user to define cohorts constructively: rather than specifying cohorts for comparison and viewing their sequences of events, Chronodes allows cohorts to be specified by an arrangement of the events themselves. By pairing this functionality with event alignment, Chronodes enables the user to fluidly explore cohorts and their associated chains of events in tandem.
2.4 Sequence Mining and Pattern Matching
Building machine learning and pattern matching algorithms into interactive visualizations presents promising opportunities for enhancing human pattern-finding abilities. Hochheiser and Shneiderman [18] and Buono et al. [7] demonstrated how users could select specific patterns in quantitative data streams and see where else they were found. For event-based data, Fails et al. [12] developed a query system to find patterns across multiple event histories. Gschwandtner et al. [16] provide users with more options to specify how these patterns are defined and lends detailed representations of how patterns compare.
Acknowledging the potential of frequent sequence mining in dense event-based data, Click-stream [22] mines frequent sequences of events and visualizes them in clusters according to their similarity. In a more consolidated visual format, ActiviTree [37] renders frequent event patterns found throughout participant data as a visual tree of “common” event sequences. Frequence [29] and Scribe Radar [40] bridge this technique with CareFlow’s [28] representation of events and connecting edges, and then enable specification of cohorts by selecting sequential branches. Allowing users to extend events sequences as they are retrieved, Progressive Insights [36] offers an alternate, constructive methodology to specifying event sequences. Whereas TimeStitch [31] designed new interactive paradigms for specifying and manipulating event sequences in a constructive, nonlinear fashion, Coquito [21] enabled the same constructive definition of behavioral groups for cohorts. Both TimeStitch and Coquito implement kinds of visual query languages, as in Zgraggen et al. [41], that enable the interactive definition of queries on event-based data. Chronodes builds on this foundation of work with a constructive event-based querying system, enabling the exploration and discovery of frequent patterns across multiple continuous timelines of mHealth records.
3 DATASET, RESEARCH QUESTIONS, AND DESIGN MOTIVATIONS
Before beginning development on Chronodes, we obtained real-world mHealth datasets collected through field studies investigating the use of the AutoSense Sensor Suite for inferring a general stress state [11]. In the first of these studies, 6 participants (who smoke) wore the sensors for 3 days, totaling about 400 hours. In a later study, the same format of data was recorded for 52 participants before and after planned smoking abstinence (prequit and postquit, respectively), with each participant’s data totaling approximately 6 days [33]. The sensor suite recorded 40 data streams for each participant, capturing a wide variety of physiological signals, such as electrocardiogram (ECG), galvanic skin response (GSR), and heart rate variability (HRV) measurements. The resulting datasets consisted of more than 750,000 and 4,650,000 data points, respectively (sampled at 1Hz). The AutoSense data represents one of the few mHealth data initiatives available for research use, as collecting good-quality mHealth data is a great technical challenge due to limitations in sensor hardware and battery technologies [9, 11, 15].
Following data collection, we regularly discussed with health researchers on our team, whereby we worked to determine questions about this data that an interactive visualization tool should help to answer. As we developed Chronodes, we showed them iterative designs of our prototype for feedback and improvement suggestions. Accordingly, specialists in health psychology and statistics (see Table 1) identified four initial questions that informed tasks in mHealth data analysis, which primarily concern identifying physiological states, habitual patterns, and external circumstances that can be correlated to smoking lapses, across participant demographics:
Q1 What are the events preceding and proceeding each instance of smoking lapse [5]?
Q2 What habitual events or cues (e.g., smoking every day after lunch) are correlated to smoking lapse [25]?
Q3 What are the correlations between smoking lapse and other physiological factors, such as stress [2, 10]?
Q4 What event patterns are specific to individual participants, and otherwise universal to all participants [26]?
Table 1.
Expert Participants, Their Diverse Domain Expertise, and Years in the Field (12 to 38 Years)
| ID | Domain Expertise | Years in Field |
|---|---|---|
| A | Health psychologist | 13 |
| B | Statistician (focus: clinical design, analytics) | 26 |
| C | Behavioral health researcher | 38 |
| D | Machine learning | 10 |
| E | Health informatics | 31 |
| F | HCI, human-centered computing | 26 |
| G | Sensor and hardware designer | 16 |
| H | Clinician & health informatics | 25 |
| J | mHealth software designer | 13 |
| K | mHealth software architect | 12 |
Note: These experts’ characteristics are representative of the 20 experts in our study.
Using these target questions as our point of departure, we identified existing techniques in visual analytics and mHealth data analysis (as elaborated in Section 2) that would assist in answering these questions with respect to the AutoSense data. Principally, we identified that event alignment would assist in answering Q1 by allowing users to align participant timelines around smoking lapse events. We also determined that an implementation of cohort definition would provide support to answering Q3 and Q4: users should have the capability to select particular groups of participants—according to specific physiological factors or event patterns of interest—that would reveal factors specific to these populations.
Accordingly, in acknowledging that Q1, Q2, and Q3 each addressed the importance of discovering event patterns, we experimented with frequent sequence mining as an approach to defining participant cohorts. To accomplish this, we first produced a series of sketches and graphical mock-ups that envisioned how event patterns could be used to represent cohort groups, with a particular emphasis on the representation of when event patterns occur over time. Concurrently, we produced “dummy” event-based data that reflected the size and dimensions of the AutoSense data, and experimented with a series of frequent sequence mining algorithms to determine whether such an algorithmic approach was tractable and could reveal event patterns preceding and proceeding smoking events.
As an outcome of these experiments, we identified that mining mHealth data for recurring event sequences operates to answer Q2 and Q4: it permits investigations of participant behaviors within days, across days, and between participants. However, to address Q1, we needed to provide users with the capability to select events of interest, such as smoking lapse events. Accordingly, by adapting “event alignment” into “frequent sequence alignment,” we developed an approach to answering Q1: we allow Chronodes users to investigate the frequent sequences that occur before and after aligned sequences of interest. Then, to address Q3—finding correlations between smoking events and physiological factors—we developed an approach to representing physiological factors as events (Section 6). For example, we can consider a high-stress activity and a drive to work as different events, and represent them within frequent sequences once they are mined. As we intend for Chronodes to be applied to a wide variety of health conditions, it was additionally important that the process of converting physiological factors to events was made user adjustable. Accordingly, we enable users to adjust event definitions interactively as in Fails et al. [12], before the events are mined for event sequence patterns. How these events are derived, and the implications of defining events in this way, are discussed in later sections.
4 CHRONODES OVERVIEW
To preface Chronodes’ contributions, we provide a high-level overview of the system’s fundamental features.
4.1 Description of the User Interface
The Chronodes user interface (Figure 2) is comprised of two areas that update to the user’s interactions. The Event Orchestra, fixed to the bottom of the screen (Figure 2B), lists a subset of participants represented by our dataset and displays the series of events that these participants perform over the course of 24-hour days. On the right, a legend indicates the event types (Figure 2C) for smoking episodes [33] (red circles), intensity of physical activity [32] (encoded by shades of blue), and stress probabilities [19] (encoded by size of white dots). In Section 6.1, we describe in detail how these measures are derived and defined. As the user interacts with event sequences, participants and events that are associated with each sequence “light up” correspondingly (mousing over
 in Figure 2). The Event Orchestra demonstrates to new users how derived event sequences relate to the “raw” event data, and also serves as a continuous indicator of the subset of data selected elsewhere in the interface (highlighted). In this way, the Event Orchestra’s primary role is as a guiding frame of reference but is not essential to interacting with Chronodes and can be minimized.
in Figure 2). The Event Orchestra demonstrates to new users how derived event sequences relate to the “raw” event data, and also serves as a continuous indicator of the subset of data selected elsewhere in the interface (highlighted). In this way, the Event Orchestra’s primary role is as a guiding frame of reference but is not essential to interacting with Chronodes and can be minimized.
Fig. 2.
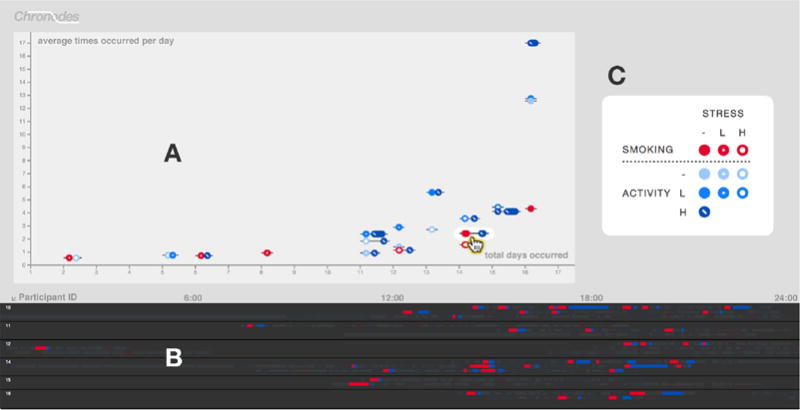
The Chronodes Sequence Stage showing a scatterplot of frequent sequences (A) and the Event Orchestra below it displaying the participant event data from which these sequences were mined (B). Whereas all sequences in the Event Orchestra are grayed out by default, when the user mouses over a frequent sequence on the Sequence Stage, its location throughout the participant data is highlighted in the Event Orchestra. (C) The legend for this pilot dataset is magnified here for better readability.
The Sequence Stage is the primary area of user interaction and accounts for the largest amount of space in the Chronodes interface (Figure 2A). It is within the Sequence Stage that the user selects, manipulates, and compares frequent event sequences. To demonstrate how the user performs these operations, we will describe Chronodes’s functionality through a use case scenario.
4.2 Use Case Walk-Through
This scenario demonstrates how the pilot study data described in Section 1 is used with Chronodes in practice. Our user Jane is a health researcher who intends to discover and understand physiological events commonly associated with smoking episodes. Upon launch, Chronodes displays the events that occur throughout the participants’ days in the Event Orchestra. In the Sequence Stage, Chronodes displays the frequent event sequences derived from this event data. By initially displaying both the raw event data and the derived sequences side by side as in Vrotsou et al. [37], we lend Jane introductory information about how event sequences are derived, what they represent, and where they occur.
Initially, the frequent sequences are displayed in a scatterplot (Figure 2A) that helps Jane identify frequent sequences of interest. The axes of this scatterplot will be described in detail later on; for now, Jane is interested foremost in events surrounding smoking episodes, so she chooses a smoking → high-activity event sequence (
 in Figure 2A) from the scatterplot. As she moves the cursor over this sequence, the Event Orchestra highlights the multiple occurrences of this sequence across multiple participants and days of data. By then clicking the sequence, it becomes a focal sequence and moves to the center of the Sequence Stage. Whereas the Event Orchestra remains fixed in place (Figure 2B), the scatterplot (Figure 2A) moves upward until it is off-screen, which allows Jane to scroll upward to view it at any time. Once the scatterplot is off-screen and the focal sequence is centered, Chronodes reveals the adjacent event sequences that occur most frequently before and after the smoking event sequence that Jane selected (Step 1 of Figure 3).
in Figure 2A) from the scatterplot. As she moves the cursor over this sequence, the Event Orchestra highlights the multiple occurrences of this sequence across multiple participants and days of data. By then clicking the sequence, it becomes a focal sequence and moves to the center of the Sequence Stage. Whereas the Event Orchestra remains fixed in place (Figure 2B), the scatterplot (Figure 2A) moves upward until it is off-screen, which allows Jane to scroll upward to view it at any time. Once the scatterplot is off-screen and the focal sequence is centered, Chronodes reveals the adjacent event sequences that occur most frequently before and after the smoking event sequence that Jane selected (Step 1 of Figure 3).
Fig. 3.
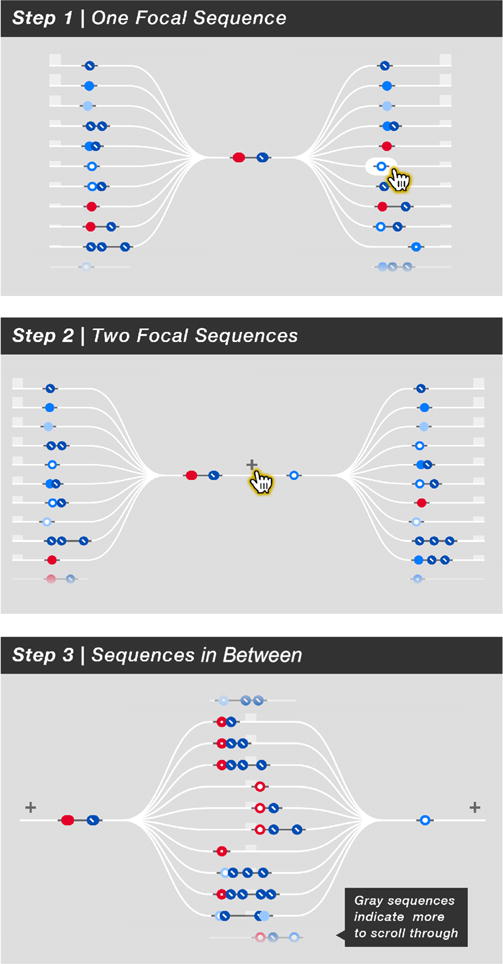
Varying configurations of focal sequences and the sequences of events that occur around them. Step 1 shows one focal sequence with the sequences before and after it. The user clicks a sequence after and it becomes a second focal sequence in Step 2: the two focal sequences with the sequences before and after them. The user clicks the + symbol and the timeline becomes Step 3: the two focal sequences with the sequences between them.
To indicate which of these sequences occurs before and after the focal sequence, Chronodes displays the adjacent sequences as on a timeline: sequences that occur before the focal sequence are placed on the left, and those that occur after are on the right (Figure 3). In the default implementation of Chronodes, the adjacent sequences are ranked vertically by frequency so that the most frequent ones are topmost. However, following from event sequence visualization approaches developed in Perer and Wang [29] and Polack et al. [31], event sequences can also be sorted by duration, cardinality, or the time at which they occur, which gives users more control over the event sequences that are made prominently visible.
Altogether, sequence frequency is not the only thing that matters to Jane; distance in time from the focal sequence is also relevant. To account for this, Chronodes positions sequences horizontally based on their proximity in time to the focal sequence. In other words, sequences that tend to occur immediately before or after the focal sequence are closer to it (notice the horizontal staggering between sequences in Step 1 of Figure 3).
Critically, it is important for our users to understand at a glance that chronology on the Sequence Stage timeline is relative to the focal sequence and not absolute as in a normal timeline. This is distinct from existing healthcare analysis visualizations, which depict participant timelines with linear temporality. To demonstrate this visually, Chronodes displays the frequent sequences on white tracks that depict the flow of time (Figure 3). In this way, every track that Jane can trace with her eyes is a sequence of sequences that occurs somewhere in the participants’ data. The tracks “bottleneck” at the focal sequence because it is the focal sequence that every other sequence has in common in its proximity. In addition, the relative frequency or “volume” of each adjacent frequent sequence is indicated by bars of varying height (Figure 3). The height of each bar, determined by the number of times the frequent sequence occurs in the data, is relative to the topmost adjacent frequent sequence.
To reduce visual clutter, only the top 10 most frequent sequences on each side are displayed, but Jane can scroll vertically through these options to reveal more. Jane sees a low-activity high-stress event (
 in Step 1 of Figure 3) that occurs only sixth most frequently, but immediately, after her focal sequence. She selects it, and it becomes a second focal sequence along the tracks (Step 2 of Figure 3).
in Step 1 of Figure 3) that occurs only sixth most frequently, but immediately, after her focal sequence. She selects it, and it becomes a second focal sequence along the tracks (Step 2 of Figure 3).
Now that Jane has selected two focal sequences, Chronodes displays the sequences that occur only where both focal sequences are present (i.e.,
 ) To illustrate this comprehension to new users, the events satisfied by Jane’s two focal sequences remain dimly lit in the Event Orchestra (as in Figure 2B). Although the adjacent sequences before and after these focal sequences are still visible, another feature now emerges: Jane can click a + symbol in between the focal sequences (Step 2 in Figure 3) to reveal the sequences that occur between two focal sequences (Step 3 in Figure 3). This functionality reveals a fundamental advantage of interactive frequent sequence alignment: the capacity to reveal the trends that occur between two or more events of interest.
) To illustrate this comprehension to new users, the events satisfied by Jane’s two focal sequences remain dimly lit in the Event Orchestra (as in Figure 2B). Although the adjacent sequences before and after these focal sequences are still visible, another feature now emerges: Jane can click a + symbol in between the focal sequences (Step 2 in Figure 3) to reveal the sequences that occur between two focal sequences (Step 3 in Figure 3). This functionality reveals a fundamental advantage of interactive frequent sequence alignment: the capacity to reveal the trends that occur between two or more events of interest.
Jane can clearly see the frequent sequences that occur before, between, and after the two focal sequences, but which participants do these adjacent sequences belong to? What time of the day are they occurring? As before, Jane can simply mouse over any sequence to reveal its associated events in the Event Orchestra. By doing so, Jane discovers that the sequences she is observing are shared by a certain subset of participants. Furthermore, for each of these participants, the events she is highlighting tend to occur in the evening. This shows that by defining these two focal sequences, Jane has effectively navigated to a “cohort” of participants that mutually exhibit the event behaviors she specified. This demonstrates the interactive and expressive capabilities of Chronodes, to use event patterns to discover cohorts of participants.
Finally, Jane can delete one of her focal sequences to return to a previous state of her analysis. As adding more focal sequences defines a more specific cohort because it resembles an AND operation, deleting focal sequences, conversely, broadens the cohort. Jane can continue to explore subsets of her participants’ behaviors in this fashion.
5 CHRONODES CONTRIBUTIONS
Now that an overview of the Chronodes interface has been presented, we proceed in this section to describe these features in terms of the contributions they lend to research in visualization, data mining, and mHealth analysis.
5.1 Visualizing Frequent Sequences for Interaction
Chronodes develops on event-based timelines by replacing events with sequences of events that can be rearranged interactively. Every event sequence, whether discovered by frequent sequence mining or defined interactively by the user, is an interactive element. As in “event alignment,” any event sequence can be designated as a focal sequence so that all events before and after it are displayed chronologically (Figure 3).
Representing mined frequent sequences for interaction is a novel integration of data mining and visualization with a series of conceptual and computational challenges that Chronodes is designed to solve. Fundamentally, it is important for our users to understand that although we represent frequent sequences as singular visual elements, they in actuality represent many event sequences over time. This relationship is demonstrated by the Event Orchestra: when the user mouses over a frequent sequence on the Stage, the corresponding event sequences across any number of participants are highlighted in the Orchestra. In other words, a frequent sequence in the Sequence Stage (Figure 2A) inherently represents many event sequences that frequently occur in the Event Orchestra (Figure 2B).
We represent frequent sequences as a “kebab” (
 ), so as to convey the fact that frequent sequences are a series of events on a timeline, potentially separated by time. Therefore, the length and distancing of events within the frequent sequences (intrasequence, within the
), so as to convey the fact that frequent sequences are a series of events on a timeline, potentially separated by time. Therefore, the length and distancing of events within the frequent sequences (intrasequence, within the
 ) is determined by averaging the timestamps of these events, wherever they occur in the associated event sequences. However, as every frequent sequence represents event sequences that occur at variable times, indicating chronology between sequences (intersequence) is not as simple as averaging the timestamps of the constituent sequences together. Although an average of event sequence timestamps, unlike a median, preserves intersequence chronology, we need to ensure that the average does not oversimplify what our users need to see during alignment. For instance, if event sequence A occurs both before and after focal sequence B, should we place A before, after, or within focal sequence B? As a solution, we consider the sequences before and the sequences after focal points as distinct so that in this scenario, sequence A appears both before and after focal sequence B.
) is determined by averaging the timestamps of these events, wherever they occur in the associated event sequences. However, as every frequent sequence represents event sequences that occur at variable times, indicating chronology between sequences (intersequence) is not as simple as averaging the timestamps of the constituent sequences together. Although an average of event sequence timestamps, unlike a median, preserves intersequence chronology, we need to ensure that the average does not oversimplify what our users need to see during alignment. For instance, if event sequence A occurs both before and after focal sequence B, should we place A before, after, or within focal sequence B? As a solution, we consider the sequences before and the sequences after focal points as distinct so that in this scenario, sequence A appears both before and after focal sequence B.
For Chronodes’ initial scatterplot, retaining intersequence chronology is not an issue, as sequences are positioned according to the axes. The y-axis represents the average number of times an event sequence occurs per day—that is, if a sequence of events tends to occur five times every day, its position on the y-axis is 5. The x-axis indicates the total number of days the event sequence is found: if the event sequence occurs for only one participant’s Monday, Tuesday, and Wednesday, its position on the x-axis is 3. As an emergent result, sequences are distributed into a two-dimensional spectrum of sequence prevalence (Figure 4).
Fig. 4.
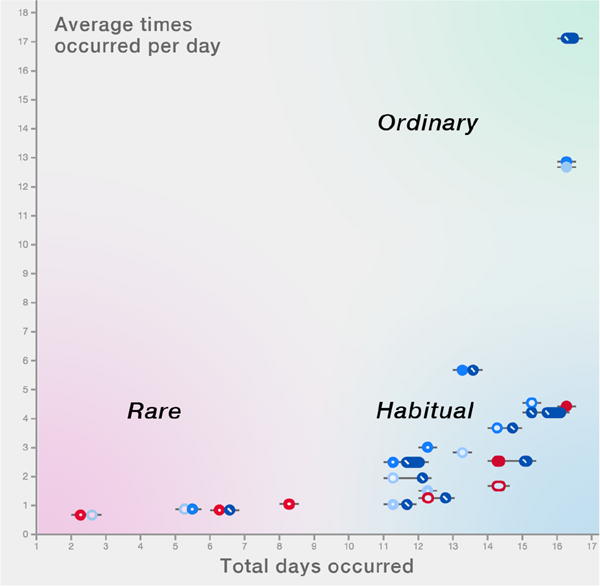
Ordinary sequences occur so frequently that they are not very interesting (e.g., walking). Habitual sequences occur on many days but do not repeat frequently on the days that they occur (e.g., exercising once every day). Rare sequences occur very infrequently.
One consequence of representing frequent event sequences as kebabs is that event icons may occlude one another. Such occlusions could be mitigated by representing events as rectangles, as in the Event Orchestra, or by increasing the horizontal time scale of the kebabs (i.e., each kebab represents a longer duration of time). However, we represent frequent event sequences as kebabs to emphasize the distinction between, on the one hand, chronological event sequences and, on the other, continuous event data that is not averaged across multiple timelines, as reflected in the Event Orchestra. In this way, the kebabs provide a consistent visual iconography to represent event sequences that are averaged and represented on relative—as opposed to absolute—time scales [6]. More generally, the kebab representation on the Sequence Stage helps to differentiate frequent event sequences from continuous events in the Event Orchestra.
5.2 Multiple Sequence Alignments
Unlike existing work on event-based alignment, Chronodes does not limit user interaction to single alignment on single events; instead, it encourages the user to create multiple alignments on any number of events side by side. As exhibited in Figure 3, this also permits the user to find sequences of events between events of interest, unprecedented in existing event alignment techniques.
5.3 Comparing and Cloning Cohorts
When Jane specified the first
 sequence from the Sequence Stage scatterplot, this sequence appeared as a focal sequence with its own tracks and adjacent sequences (Step 1 in Figure 3). As she continued to select more adjacent sequences like the proceeding
sequence from the Sequence Stage scatterplot, this sequence appeared as a focal sequence with its own tracks and adjacent sequences (Step 1 in Figure 3). As she continued to select more adjacent sequences like the proceeding
 , she narrowed down on a more specific subset of the participant data: only days that included all of
, she narrowed down on a more specific subset of the participant data: only days that included all of
 were displayed (Step 2 in Figure 3). In this way, Chronodes provides the functionality to constructively define cohorts using sequences of events. This extends the capabilities of prior work that enables users to display participants that exhibit a specified sequence of events but does not permit the interactive rearrangement of these events once they are set.
were displayed (Step 2 in Figure 3). In this way, Chronodes provides the functionality to constructively define cohorts using sequences of events. This extends the capabilities of prior work that enables users to display participants that exhibit a specified sequence of events but does not permit the interactive rearrangement of these events once they are set.
To return to a broader subset of the participant data, Jane can either remove a focal point or clear the entire current timeline and return to the scatterplot. However, she also has the option to clone the entire timeline (Figure 5), which duplicates it on the Sequence Stage. By modifying the second timeline, she can compare the properties of related cohorts side by side. For example, by adding another smoking event (
 ) to the second timeline, Chronodes displays the differences between the event-based behavior of the original cohort timeline
) to the second timeline, Chronodes displays the differences between the event-based behavior of the original cohort timeline
 and its clone
and its clone
 As demonstrated in Figure 5, once the new
As demonstrated in Figure 5, once the new
 is appended as a focal sequence to the bottom timeline, the adjacent sequences before and after are updated accordingly.
is appended as a focal sequence to the bottom timeline, the adjacent sequences before and after are updated accordingly.
Fig. 5.
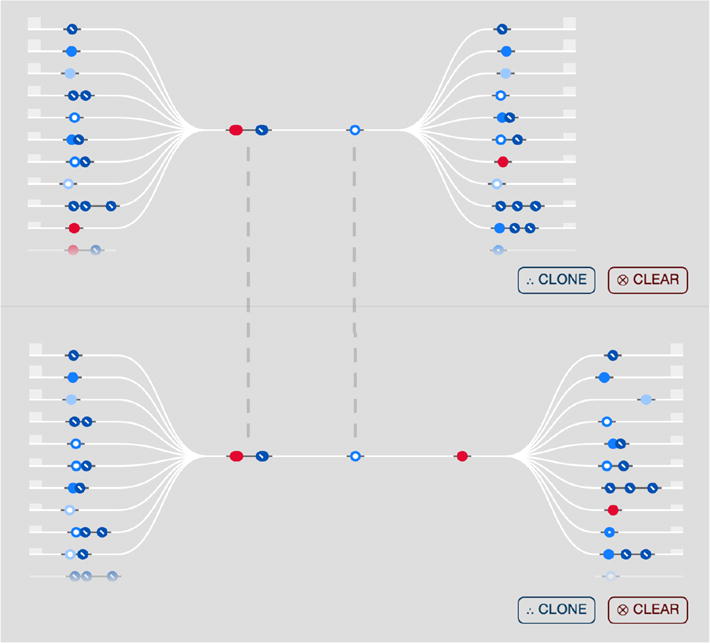
An original timeline of focal sequences (top) that has been cloned and then modified (bottom). By adding another focal sequence to the bottom timeline, the event sequences before and after it are changed. In this way, Chronodes enables users to view the comparative differences between timelines, which each represent a cohort of participants.
6 DERIVING INTERACTIVE EVENT SEQUENCES
Whereas Chronodes presents novel visual and interactive paradigms for analyzing chronological patterns, it is important that the system also be developed for use by mHealth researchers. Here, we highlight Chronodes’ technical contributions in providing new methods of event sequence analysis—namely, the techniques, variables, and considerations involved in the process of deriving events, finding event sequences, and visualizing these sequences for the purposes of interaction.
6.1 Deriving Events From mHealth Sensor Data
Before analyzing mHealth data for chronological patterns to answer our research questions (Q1 through Q4), each participant’s physiological data streams must be represented in a consistent and comparable format. From each participant’s array of AutoSense data streams, we extrapolated timestamped measures of (1) activity from three-axis accelerometer chest sensors [32], (2) probability of stress from physiological sensors (ECG and respiration) [19], and (3) instances of smoking from inertial wrist sensors (three-axis accelerometer and three-axis gyroscope) [33]. Whereas smoking episodes are described by discrete Boolean values of 1 if smoking and 0 if not, activity and probability of stress are variable between participants and need to be normalized before further interpretation.
Once normalized, physiological data streams are classified as being either “none,” “low,” or “high” magnitudes at every 5-minute interval of time (Figure 6). Our selection of this three-phase classification model is informed by Sharmin et al. [35], which suggests that even for experts, a finer-grain classification increases complexity without offering additional analytical benefits. Whereas darkness of the blue indicates activity intensity, the size of the inner white circle indicates probability of stress. At a given point of time, the probability of stress ranges from 0 to 1, or is simply −1 to indicate that data is unavailable (in the occasional event that only the activity sensor is enabled). In addition, for events where activity is high, stress cannot be accurately inferred [19], indicated by a white slash (
 in Figure 6). Adjacent time intervals of the same magnitude are summed into single events so that resulting events are variable in length. As participants may not be wearing the sensors at all times, such as during sleep, consecutive events that are separated by long gaps of no data collection at all are not merged past a user-defined threshold (60 minutes of sensor inactivity by default).
in Figure 6). Adjacent time intervals of the same magnitude are summed into single events so that resulting events are variable in length. As participants may not be wearing the sensors at all times, such as during sleep, consecutive events that are separated by long gaps of no data collection at all are not merged past a user-defined threshold (60 minutes of sensor inactivity by default).
Fig. 6.
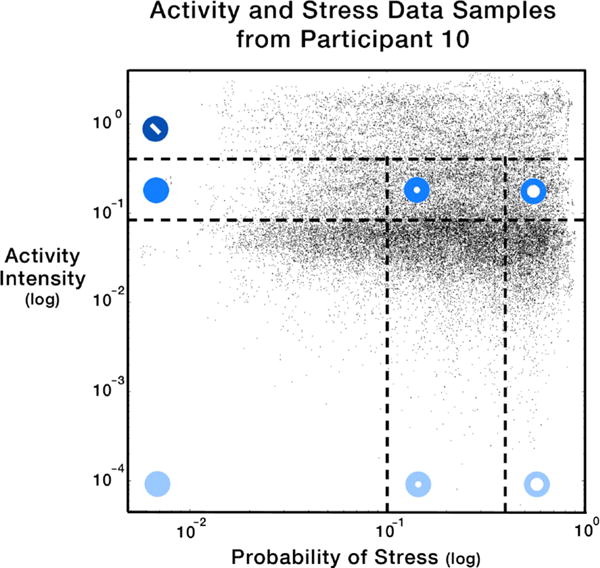
Participants’ activity intensity and probability of stress were sampled and inferred once per second. For Participant 10, this totaled more than 168,000 data points in the scatterplot. These data points were then discretized into events with quartile thresholds (e.g., median). For events with high activity (top), stress cannot be accurately inferred.
Before mining participants’ daily routines for recurrent sequential patterns, we should define what constitutes a “day”: although the majority of participant activity in our study occurred during daytime hours, some instances of irregular sleeping patterns and nighttime activity were present. Some participants even continued to wear the sensors during sleep. This justifies calculations that determine participant waking hours, but presently, for consistency and simplicity, Chronodes defines a day as a hard 24-hour time period from midnight to midnight, accounting for behaviors that repeat at consistent times across days as demanded by Q2.
6.2 Deriving Frequent Sequences From Events
Once participant data streams are represented as a continuous series of events, sequence mining provides Chronodes users with meaningful chronological relationships and sequences of events that can be manipulated interactively. Upon launch, Chronodes finds daily event sequences present throughout all participant events using the PrefixSpan [27] sequence mining algorithm. PrefixSpan retrieves patterns that occur frequently among sequential events and is an efficient algorithm for mining frequent sequences from a large number of individual event strings (i.e., a large number of 24-hour days). Algorithms of this kind are increasingly useful as we scale up to more participants. However, before the mined frequent sequences can be rendered as individual visual components with which the user can interact, we must first modify PrefixSpan to consider variations in repetitive, gapped, and closed sequence mining.
When PrefixSpan runs, it returns a list of frequent sequences ranked by frequency, with their positions in the participant data. Importantly, the algorithm does not mine for repetitive frequent sequences: if a smoking episode occurs three times every day, PrefixSpan only recovers the first occurrence per day. To extend the algorithm to find repetitive sequences, we use the locations of the initially found frequent sequences to search the remainder of the 24-hour day for repetitions.
Also by default, PrefixSpan mines for gapped event sequences: events can be considered as a part of the same sequence even if they are separated by many other events (a “gap”). This is not ideal for scenarios where healthcare analysts are only interested in the events immediately preceding or proceeding smoking relapse; however, preventing gaps entirely would not allow our users to understand relationships between event behaviors that occur hours apart. To suit this range of use cases, we set a maximum gap parameter that Chronodes users can dynamically adjust.
Ultimately, PrefixSpan does not limit mining to closed sequences. For example, it redundantly returns both the closed
 and its subset
and its subset
 Although we can modify PrefixSpan to return only the closed frequent sequences, Chronodes’ design lends interactive capabilities that allow us to do something more comprehensive. By initially providing only the shorter frequent sequences that have a longer variation (in our example, present only
Although we can modify PrefixSpan to return only the closed frequent sequences, Chronodes’ design lends interactive capabilities that allow us to do something more comprehensive. By initially providing only the shorter frequent sequences that have a longer variation (in our example, present only
 ), by selecting this sequence as a focal sequence we can see all of the variations that are related to it. As Chronodes displays the sequences that come after the focal sequence once it is selected (like
), by selecting this sequence as a focal sequence we can see all of the variations that are related to it. As Chronodes displays the sequences that come after the focal sequence once it is selected (like
 ), it is also displaying longer variations of the focal sequence as a side effect (
), it is also displaying longer variations of the focal sequence as a side effect (
 is equivalent to
is equivalent to
 ). In doing so, we enable a new interactive method for exploring event sequences mined with frequent sequence mining, whereby users can constructively narrow in on related sequences. Additionally, as an emergent result, we can keep the frequent sequence “entry points” on the scatterplot simple.
). In doing so, we enable a new interactive method for exploring event sequences mined with frequent sequence mining, whereby users can constructively narrow in on related sequences. Additionally, as an emergent result, we can keep the frequent sequence “entry points” on the scatterplot simple.
6.3 Interacting With Frequent Sequences
Chronodes maps every mined frequent sequence to its recurring location in the event data so that mousing over or selecting each sequence indicates its distribution in the Event Orchestra (Figure 2). In addition, as every frequent sequence is related to a particular cohort of participants (either all or some participants have the sequence), we can use information about the associated cohort to limit our searches for adjacent sequences. Any participant or day outside of the currently specified cohort does not need to be searched for sequences, thereby limiting use of computational resources as we scale to more participants and data streams.
Specific care is taken to ensure that when a frequent sequence is selected in Chronodes, users can understand the effects of their action on the visualization. Upon selection, sequences animate to the center as the nearby sequences fade away, and the new tracks grow outward. Such animations or “visual cues” help principally to direct users to changes in the visualization layout [20], and in the particular case of Chronodes, they indicate that new event sequences have been appended to the Sequence Stage, relative to the focal sequences. After these animations complete, the Event Orchestra updates to indicate the focal sequences and new cohort of participants being observed.
6.4 System Implementation
Chronodes’s front-end visualization component is Web based and written in JavaScript (jQuery,1 React,2 D33), and served by a Python Web server (web.py4). The front-end interface sends API requests to the Web server that returns data processed for display. Timestamped inferences about participant activity, stress, and smoking episodes are stored in an SQLite database5 for its cross-platform compatibility, integration with Python, and support for the dataset size of our pilot study.
7 USER STUDY WITH HEALTH EXPERTS
We conducted an informal pilot investigation with 20 behavioral, biomedical, and computational health experts coming from a large research team to gain insights into the efficacy and limitations of Chronodes. Through the study, we intended to understand how Chronodes may help them with mHealth data exploration, pattern discovery, and decision making. The interdisciplinary nature of the research team presents the unique opportunity for us to gain insight into how Chronodes may be used by experts with diverse backgrounds. Table 1 highlights some of their domain expertise and experience. We refer to these participants as “experts” to avoid confusion with the AutoSense participants (from whom the mHealth data was collected).
7.1 Method
Each study session began with a demonstration of the Chronodes interface. The participants were welcome to ask questions at any time. After the demonstration, the participants were instructed to think aloud in describing their perspectives and criticisms of the interface’s features. Comments were recorded and organized according to which interface feature they pertained: (1) event derivation and representation; (2) frequent sequence derivation, representation, and placement; (3) frequent sequence mining and placement of adjacent sequences around focal sequences; and (4) the use of multiple focal sequences. Due to the nature of our informal study, we did not record audio of the demonstrations. We took notes to capture feedback on the usability, effectiveness, and limitations and possible improvements of the system. Remarks included in this section are paraphrased summaries of this feedback.
8 RESULTS AND IMPLICATIONS FOR FUTURE RESEARCH
In this section, we summarize insights gleaned from the user study, present open challenges, and provide recommendations for future visualization and analysis research with mHealth data.
8.1 Representing mHealth Data
The challenge of analyzing and representing mHealth data is one of variability (continuous quantitative data streams), uncertainty (missing or incorrect measurements), high volume (data from many sensors simultaneously), and high dimensionality (many participants and many data streams). To represent continuous mHealth data streams across many participants effectively, focused investigations into each of these factors are necessary for future research.
8.1.1 Leveraging Temporal Variation During Event Derivation
Drawing on prior works [23, 24], Chronodes represents mHealth data as discrete events, which supports the visualization and analysis of chronological patterns, their frequencies, and their relationships. In EHR scenarios where health data is already described by discrete events (e.g., medications administered in asthma treatment in EventFlow [24] and hospital administration events in CoCo [23]), discretization from continuous data streams to discrete events is not required. However, working with quantitative data streams in mHealth provides the opportunity to define more complex event types than Chronodes initially considered, leveraging temporal variation to define discrete events that capture the ways physiology changes over time.
For example, Expert H described that when an event occurs, health analysts are often interested in the residual effects of the event over time and how these effects might in turn affect our understanding of the events that follow. Additionally, some events are better described as fluctuating, quantitative values altogether. In other words, being able to investigate the underlying causes or emergent effects of an mHealth event is useful for understanding why it is present in the first place and what kinds of implications it has. For this, Expert H provided the example of stress: although we may certainly represent a high-stress episode as happening at a discrete point in time, it may be more effectively represented as a state with a variable magnitude and with variable effects that are sustained until after the high stress is sensed.
Describing mHealth data as temporal features [16] or motifs [8] are potential routes for addressing this challenge. To this end, we enhanced Chronodes with an event derivation system (Figure 7) to discover temporal patterns in mHealth data, which can then be organized in Chronodes. In particular, the event derivation system gives data analysts additional control over the kinds of events presented in Chronodes, which provides an alternative to the default event derivation procedures described in Section 6.1 (Figure 6). To find temporal patterns for use in Chronodes, the event derivation system employs the Symbolic Aggregate approXimation and Vector Space Model (SAX-VSM) [34] to represent the time series in the mHealth data (e.g., stress levels and activity intensities) as temporal segments that are then clustered into groups with k-means [14, 30]. After discovering these groups of trends, or motifs, across participant records, the event derivation system allows users to examine where these motifs occur and select any number of them. Once selected, motifs can be defined as events for use in Chronodes thereafter, represented as glyphs of the sidebar motifs (Figure 7B). Accordingly, whereas the event derivation system enables analysts to identify granular patterns in continuous temporal mHealth data, Chronodes enables analysts to identify chronological relationships between these patterns as such.
Fig. 7.
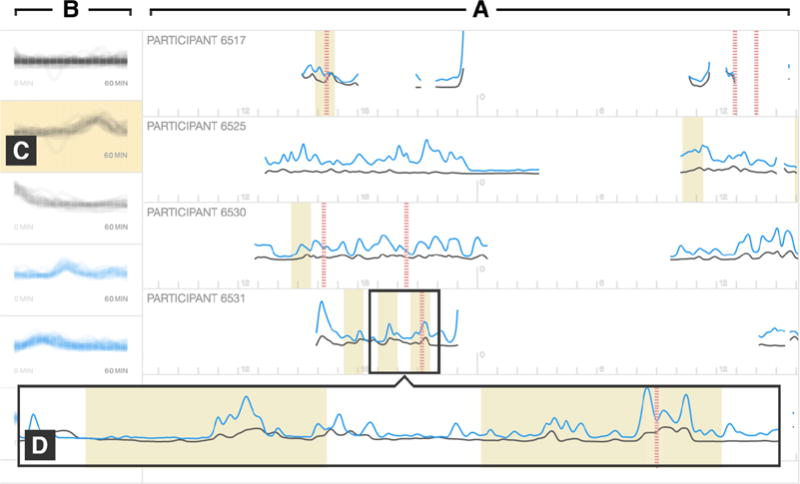
(A) The event derivation interface showing probability of stress (gray), activity intensity (blue), and smoking episodes (red vertical dotted lines) for multiple participants. (B) The sidebar displays common temporal trends (motifs) identified from this data using time series motif mining. (C) The user selects a motif to highlight where it occurs in the data. (D) Zoomed-in view of a 3-hour region showing two instances of the highlighted motif (gray border added to enhance clarity). Motifs selected from the sidebar can be defined as events for use in Chronodes.
8.1.2 Addressing Uncertainty in mHealth Data
mHealth data bestows unique relationships between recorded high-resolution data points and high-level human behaviors that can be inferred from them at varying degrees of certainty. In this way, Expert H was interested in how Chronodes might portray the relationships between ground truth sensor events and the behavioral variables that can be inferred from them (e.g., high stress inferred from increased heart rate). Similarly, prior research suggests that there is a characteristic discrepancy between the actual time of a self-reported event and the time at which it is reported: smokers often report smoking episodes prior to or after the actual smoking event [33]. Designing a temporal visualization that mediates conflicts between self-reported events and sensor-collected objective event points thus introduces novel challenges that need to be further investigated. Recent work has made a first attempt [35], but further research is warranted to find optimal solutions as to how to best integrate self-reported data into sensor recorded data, and to visualize these integrations.
8.1.3 Sense Making From High-Volume mHealth Data
Experts C and J commended Chronodes’ approach to making sense of high-volume event data by way of interaction with event sequences, which was echoed by other experts consistently. These remarks suggested Chronodes’ niche role in exploring mHealth data interactively with event sequences, particularly as these methods relate to other existing healthcare analysis techniques that focus on event-based analysis (e.g., Malik et al. [23] and Monroe et al. [24]). Ultimately, we can use the existing interactive techniques in Chronodes to mine high-volume mHealth data more effectively, such as learning how tools like Progressive Insights [36] permit exploration of data as it is being mined.
8.2 Context and Homologous Events
8.2.1 Instantial Context
Expert D noted that in some cases, events may be identical in terms of their content but different in the context of how they were registered or defined. A specific example given to us was this: how do we denote the difference between a self-reported smoking event and one detected by carbon monoxide sensors? Of course, as these events are instantiated in different ways, we could represent them as visually distinct; however, in developing Chronodes, we would prefer to design a more robust way for users to understand the relationships between, and the existence of, these homologous events. Visually coding events according to their provenance and particular metadata attributes therefore remains an important challenge.
8.2.2 Situational Context
Expert D described the differences between events that might depend on location and spatial context, such as between a smoking event at home and at work. Already we are designing a next iteration of Chronodes to account for these situational contexts. As our frequent sequence mining algorithm supports events that occur simultaneously, we are able to consider and represent “smoking” event (behavioral) occurring “at home” (spatial) at once. As a result, these events in parallel provide context to one another by being present at the same time. However, visualizing multiple contextual events in parallel may confound which events relate to one another in the first place, especially as we scale to more event types. Further research is needed to determine the kinds of computational, visual, and interactive techniques required to indicate how a variety of mHealth events relate to their situational contexts.
8.2.3 Temporal Context
Expert A is working to determine false positives in sensor-detected smoking episodes and indicated a need to understand the differences between events that depend on time, such as between a smoking event in the morning and at night. We are also designing a solution for this kind of temporal context: as the Event Orchestra represents a timeline, we can use it or a minified summary as an interactive filter to specify the time regions in which we are interested. In our scenario, Jane should be able to highlight the morning in the Event Orchestra and choose a smoking episode focal sequence limited to that specified time area. In this way, she would see only the events that occur before and after smoking episodes in the morning. This is a principal update that we are working to develop in our next version of Chronodes.
8.3 Extensibility to Other mHealth Analysis Scenarios
Across the study sessions, Chronodes was deemed generalizable to a variety of uses in healthcare and mHealth data analysis. We describe these applications in this section to motivate future work in this area.
8.3.1 Behavioral Analysis
Expert B commented on the applicability of the Chronodes interface to behavioral scientists and suggested to perform evaluative studies with this research area next. Particularly, after we had explained how timelines represented cohorts and could be cloned, Expert B described the value of imposing constraints on the cohort timelines other than event order. A specific example given was investigating the differences in event chronology between male and female participants. This is a comparative strategy emphasized in prior motivating works such as CoCo [23], and when paired with online event sequence specification is foreseeably powerful.
8.3.2 mHealth Interventions
Expert B also expressed interest in Chronodes’ potential applications in investigating the effects of just-in-time adaptive interventions. Specifically, marking different kinds of interventions as events and specifying them as focal sequences would be a significant step to not only planning interventions but also understanding their outcomes across participant groups. For other end users, Expert J suggested that Chronodes could play an important role in clinicians interacting with these participants, where participants could be asked to recall why they tended to exhibit certain behaviors after a specified event.
8.3.3 Health Sensor Development and Validation
Expert F described Chronodes as a useful tool for mHealth sensor developers who need to understand the relationships between the data elements that they record. Expert C described how Chronodes could be used to help sensor developers identify false positives from sensor alerts and corresponding user reports. With this insight, we are deeply considering Chronodes’ applications not only in end user behavioral analysis but also in the validation of mHealth technologies in the first place.
9 CONCLUSION
We presented Chronodes, a system that unifies data mining and human-centric visualization techniques to support the explorative analysis of longitudinal mHealth data. Chronodes extracts and visualizes frequent behavioral sequences, and promotes them as interactive elements, enabling health researchers to interactively define, explore, and compare groups of participant behaviors using event sequence combinations. We evaluated Chronodes with an informal study with 20 expert health researchers, and determined the application’s utility across a variety of healthcare and computational disciplines. From these insights, we are continuing to develop Chronodes for applications to other health data analysis scenarios, including understanding the health events surrounding congestive heart failure and diabetes management, and observing the effects of just-in-time adaptive interventions from perspectives in behavioral science.
Acknowledgments
This work was supported by the National Institutes of Health under grant 1U54EB020404.
Footnotes
The reviewing of this article was managed by special issue associate editors Yu-Ru Lin and Nan Cao.
Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. Copyrights for components of this work owned by others than ACM must be honored. Abstracting with credit is permitted. To copy otherwise, or republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.
Contributor Information
PETER J. POLACK, JR., Georgia Institute of Technology
SHANG-TSE CHEN, Georgia Institute of Technology.
MINSUK KAHNG, Georgia Institute of Technology.
KAYA DE BARBARO, Georgia Institute of Technology.
RAHUL BASOLE, Georgia Institute of Technology.
MOUSHUMI SHARMIN, Western Washington University.
DUEN HORNG CHAU, Georgia Institute of Technology.
References
- 1.Aigner Wolfgang, Miksch Silvia, Müller Wolfgang, Schumann Heidrun, Tominski Christian. Visualizing time-oriented data—a systematic view. Computers and Graphics. 2007;31(3):401–409. [Google Scholar]
- 2.al’Absi Mustafa, Hatsukami Dorothy, Davis Gary L. Attenuated adrenocorticotropic responses to psychological stress are associated with early smoking relapse. Psychopharmacology. 2005;181(1):107–117. doi: 10.1007/s00213-005-2225-3. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Or A, Healey J, Kontothanassis L, Van Thong JM. Proceedings ofthe 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Vol. 2. IEEE; Los Alamitos, CA: 2004. BioStream: A system architecture for real-time processing of physiological signals; pp. 3101–3104. [DOI] [PubMed] [Google Scholar]
- 4.Basole Rahul C, Braunstein Mark L, Kumar Vikas, Park Hyunwoo, Kahng Minsuk, Horng Duen, Chau Polo, Tamersoy Acar, et al. Understanding variations in pediatric asthma care processes in the emergency department using visual analytics. Journal of the American Medical Informatics Association. 2015;22(2):318–323. doi: 10.1093/jamia/ocu016. [DOI] [PubMed] [Google Scholar]
- 5.Brandon Thomas H, Tiffany Steven T, Obremski Karen M, Baker Timothy B. Postcessation cigarette use: The process of relapse. Addictive Behaviors. 1990;15(2):105–114. doi: 10.1016/0306-4603(90)90013-n. [DOI] [PubMed] [Google Scholar]
- 6.Brehmer Matthew, Lee Bongshin, Bach Benjamin, Riche Nathalie Henry, Munzner Tamara. Timelines revisited: A design space and considerations for expressive storytelling. IEEE Transactions on Visualization and Computer Graphics. 2016;22(1):449–458. doi: 10.1109/TVCG.2016.2614803. [DOI] [PubMed] [Google Scholar]
- 7.Buono Paolo, Aris Aleks, Plaisant Catherine, Khella Amir, Shneiderman Ben. Electronic Imaging 2005. International Society for Optics and Photonics; Bellingham, WA: 2005. Interactive pattern search in time series; pp. 175–186. [Google Scholar]
- 8.Chiu Bill, Keogh Eamonn, Lonardi Stefano. Proceedings of the 9th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; New York, NY: 2003. Probabilistic discovery of time series motifs; pp. 493–498. [Google Scholar]
- 9.Cios Krzysztof J, Moore G William. Uniqueness of medical data mining. Artificial Intelligence in Medicine. 2002;26(1):1–24. doi: 10.1016/s0933-3657(02)00049-0. [DOI] [PubMed] [Google Scholar]
- 10.Cohen Sheldon, Lichtenstein Edward. Perceived stress, quitting smoking, and smoking relapse. Health Psychology. 1990;9(4):466. doi: 10.1037//0278-6133.9.4.466. [DOI] [PubMed] [Google Scholar]
- 11.Ertin Emre, Stohs Nathan, Kumar Santosh, Raij Andrew, al’Absi Mustafa, Shah Siddharth. Proceedings of the 9th ACM Conference on Embedded Networked Sensor Systems. ACM; New York, NY: 2011. AutoSense: Unobtrusively wearable sensor suite for inferring the onset, causality, and consequences of stress in the field; pp. 274–287. [Google Scholar]
- 12.Fails Jerry Alan, Karlson Amy, Shahamat Layla, Shneiderman Ben. Proceedings of the 2006 IEEE Symposium on Visual Analytics Science and Technology. IEEE; Los Alamitos, CA: 2006. A visual interface for multivariate temporal data: Finding patterns of events across multiple histories; pp. 167–174. [Google Scholar]
- 13.Fouse Adam, Weibel Nadir, Hutchins Edwin, Hollan James D. CHI’ 11 Extended Abstracts on Human Factors in Computing Systems. ACM; New York, NY: 2011. ChronoViz: A system for supporting navigation of time-coded data; pp. 299–304. [Google Scholar]
- 14.Goutte Cyril, Toft Peter, Rostrup Egill, Nielsen Finn A, Hansen Lars Kai. On clustering fMRI time series. NeuroImage. 1999;9(3):298–310. doi: 10.1006/nimg.1998.0391. [DOI] [PubMed] [Google Scholar]
- 15.Grimson Jane, Grimson William, Hasselbring Wilhelm. The SI challenge in health care. Communications of the ACM. 2000;43(6):48–55. [Google Scholar]
- 16.Gschwandtner Theresia, Aigner Wolfgang, Kaiser Katharina, Miksch Silvia, Seyfang Andreas. Proceedings of the 2011 IEEE Pacific Visualization Symposium (PacificVis’11) IEEE; Los Alamitos, CA: 2011. CareCruiser: Exploring and visualizing plans, events, and effects interactively; pp. 43–50. [Google Scholar]
- 17.Guimbretière François, Dixon Morgan, Hinckley Ken. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM; New York, NY: 2007. ExperiScope: An analysis tool for interaction data; pp. 1333–1342. [Google Scholar]
- 18.Hochheiser Harry, Shneiderman Ben. CHI’02 Extended Abstracts on Human Factors in Computing Systems. ACM; Los Alamitos, CA: 2002. A dynamic query interface for finding patterns in time series data; pp. 522–523. [Google Scholar]
- 19.Hovsepian K, al’Absi M, Ertin E, Kamarck T, Nakajima M, Kumar S. cStress: Towards a gold standard for continuous stress assessment in the mobile environment. Proceedings of the ACM International Conference on Ubiquitous Computing (UbiComp’15) 2015:493–504. doi: 10.1145/2750858.2807526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong H, Liu Z, Karahalios K. Internal and external visual cue preferences for visualizations in presentations. Computer Graphics Forum. 36(3):515–525. [Google Scholar]
- 21.Krause Josua, Perer Adam, Stavropoulos Harry. Supporting iterative cohort construction with visual temporal queries. IEEE Transactions on Visualization and Computer Graphics. 2016;22(1):91–100. doi: 10.1109/TVCG.2015.2467622. [DOI] [PubMed] [Google Scholar]
- 22.Liu Zhicheng, Wang Yang, Dontcheva Mira, Hoffman Matthew, Walker Seth, Wilson Alan. Patterns and sequences: Interactive exploration of clickstreams to understand common visitor paths. IEEE Transactions on Visualization and Computer Graphics. 2017;23(1):321–330. doi: 10.1109/TVCG.2016.2598797. [DOI] [PubMed] [Google Scholar]
- 23.Malik Sana, Du Fan, Monroe Megan, Onukwugha Eberechukwu, Plaisant Catherine, Shneiderman Ben. Proceedings of the 20th International Conference on Intelligent User Interfaces. ACM; New York, NY: 2015. Cohort comparison of event sequences with balanced integration of visual analytics and statistics; pp. 38–49. [Google Scholar]
- 24.Monroe Megan, Lan Rongjian, Lee Hanseung, Plaisant Catherine, Shneiderman Ben. Temporal event sequence simplification. IEEE Transactions on Visualization and Computer Graphics. 2013;19(12):2227–2236. doi: 10.1109/TVCG.2013.200. [DOI] [PubMed] [Google Scholar]
- 25.Niaura Raymond S, Rohsenow Damaris J, Binkoff Jody A, Monti Peter M, Pedraza Magda, Abrams David B. Relevance of cue reactivity to understanding alcohol and smoking relapse. Journal of Abnormal Psychology. 1988;97(2):133. doi: 10.1037//0021-843x.97.2.133. [DOI] [PubMed] [Google Scholar]
- 26.Patton George C, Carlin John B, Coffey Carolyn, Wolfe Rory, Hibbert M, Bowes Glenn. The course of early smoking: A population-based cohort study over three years. Addiction. 1998;93(8):1251–1260. doi: 10.1046/j.1360-0443.1998.938125113.x. [DOI] [PubMed] [Google Scholar]
- 27.Pei Jian, Han Jiawei, Mortazavi-Asl Behzad, Pinto Helen, Chen Qiming, Dayal Umeshwar, Hsu Mei-Chun. Proceedings of the 17th International Conference on Data Engineering (ICDE’01) IEEE; Los Alamitos, CA: 2001. Prefixspan: Mining sequential patterns efficiently by prefix-projected pattern growth; pp. 215–224. [Google Scholar]
- 28.Perer Adam, Gotz David. CHI’13 Extended Abstracts on Human Factors in Computing Systems. ACM; New York, NY: 2013. Data-driven exploration of care plans for patients; pp. 439–444. [Google Scholar]
- 29.Perer Adam, Wang Fei. Proceedings of the 19th International Conference on Intelligent User Interfaces. ACM; New York, NY: 2014. Frequence: Interactive mining and visualization of temporal frequent event sequences. [Google Scholar]
- 30.Phu Le, Anh Duong Tuan. AI 2011: Advances in Artificial Intelligence. Springer; 2011. Motif-based method for initialization the K-means clustering for time series data; pp. 11–20. [Google Scholar]
- 31.Polack Peter J, Jr, Chen Shang-Tse, Kahng Minsuk, Sharmin Moushumi, Chau Duen Horng. Proceedings of the 2015 IEEE Conference on Visual Analytics Science and Technology (VAST’15) IEEE; Los Alamitos, CA: 2015. TimeStitch: Interactive multi-focus cohort discovery and comparison. [Google Scholar]
- 32.Rahman M, Bari R, Ali A, Sharmin M, Raij A, Hovsepian K, Hossain S, et al. Are we there yet? Feasibility of continuous stress assessment via wireless physiological sensors. Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (BCB’14) 2014:479–488. doi: 10.1145/2649387.2649433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saleheen N, Ali A, Hossain S, Sarker H, Chatterjee S, Marlin B, Ertin E, al’Absi M, Kumar S. puffMarker: A multi-sensor approach for pinpointing the timing of first lapse in smoking cessation. Proceedings of the 2015 ACM International Joint Conference on Pervasive and Ubiquitous Computing (UbiComp’15) 2015:999–1010. [PMC free article] [PubMed] [Google Scholar]
- 34.Senin Pavel, Malinchik Sergey. Proceedings of the 2013 IEEE 13th International Conference on Data Mining (ICDM ’13) IEEE; Los Alamitos, CA: 2013. SAX-VSM: Interpretable time series classification using SAX and Vector Space Model; pp. 1175–1180. [Google Scholar]
- 35.Moushumi Sharmin Andrew, Raij David, Epstien Inbal, Nahum-Shani J, Beck Gayle, Vhaduri Sudip, Preston Kenzie, Kumar Santosh. Proceedings of the 2015 ACM International Joint Conference on Pervasive and Ubiquitous Computing. ACM; NewYork, NY: 2015. Visualization of time-series sensor data to inform the design of just-in-time adaptive stress interventions; pp. 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolper Charles D, Perer Adam, Gotz David. Progressive visual analytics: User-driven visual exploration of in-progress analytics. IEEE Transactions on Visualization and Computer Graphics. 2014;20(12):1653–1662. doi: 10.1109/TVCG.2014.2346574. [DOI] [PubMed] [Google Scholar]
- 37.Vrotsou Katerina, Johansson Jimmy, Cooper Matthew. ActiviTree: Interactive visual exploration of sequences in event-based data using graph similarity. IEEE Transactions on Visualization and Computer Graphics. 2009;15(6):945–952. doi: 10.1109/TVCG.2009.117. [DOI] [PubMed] [Google Scholar]
- 38.Walker James, Borgo Rita, Jones Mark W. TimeNotes: A study on effective chart visualization and interaction techniques for time-series data. IEEE Transactions on Visualization and Computer Graphics. 2016;22(1):549–558. doi: 10.1109/TVCG.2015.2467751. [DOI] [PubMed] [Google Scholar]
- 39.Wongsuphasawat Krist, Gómez John Alexis Guerra, Plaisant Catherine, Wang Taowei David, Taieb-Maimon Meirav, Shneiderman Ben. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM; New York, NY: 2011. LifeFlow: Visualizing an overview of event sequences. [Google Scholar]
- 40.Wongsuphasawat Krist, Lin Jimmy. Proceedings of the 2014 IEEE Conference on Visual Analytics Science and Technology (VAST’14) IEEE; Los Alamitos, CA: 2014. Using visualizations to monitor changes and harvest insights from a global-scale logging infrastructure at Twitter; pp. 113–122. [Google Scholar]
- 41.Zgraggen Emanuel, Drucker Steven M, Fisher Danyel, Deline Robert. (s| qu) eries: Visual regular expressions for querying and exploring event sequences. Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems (CHI’15) 2015:2683–2692. [Google Scholar]
- 42.Zhao Jian, Chevalier Fanny, Balakrishnan Ravin. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM; New York, NY: 2011. Kronominer: Using multi-foci navigation for the visual exploration of time-series data; pp. 1737–1746. [Google Scholar]
- 43.Zhao Jian, Chevalier Fanny, Pietriga Emmanuel, Balakrishnan Ravin. Exploratory analysis of time-series with chronolenses. IEEE Transactions on Visualization and Computer Graphics. 2011;17(12):2422–2431. doi: 10.1109/TVCG.2011.195. [DOI] [PubMed] [Google Scholar]


