Abstract
Despite the proven clinical antineoplastic activity of histone deacetylase inhibitors (HDACI), their effect has been reported to be lower than expected in B-cell lymphomas. Traditionally considered as “epigenetic drugs”, HDACI modify the acetylation status of an extensive proteome, acting as general lysine deacetylase inhibitors (KDACI), and thus potentially impacting various branches of cellular metabolism. Here, we demonstrate through metabolomic profiling of patient plasma and cell lines that the KDACI panobinostat alters lipid metabolism and downstream survival signaling in diffuse large B-cell lymphomas (DLBCL). Specifically, panobinostat induces metabolic adaptations resulting in newly acquired dependency on the choline pathway and activation of PI3K signaling. This metabolic reprogramming decreased the antineoplastic effect of panobinostat. Conversely, inhibition of these metabolic adaptations resulted in superior anti-lymphoma effect as demonstrated by the combination of panobinostat with a choline pathway inhibitor. In conclusion, our study demonstrates the power of metabolomics in identifying unknown effects of KDACI, and emphasizes the need for a better understanding of these drugs in order to achieve successful clinical implementation.
Keywords: DLBCL, Metabolomics, Panobinostat, Choline pathway, PI3K
Highlights
-
•
Lysine deacetylase inhibitor (KDACI) treatment alters choline metabolism in B-cell lymphoma patients.
-
•
KDACI-treated lymphoma cells acquire PI3K pathway dependency via increased choline kinase A (CHKA) activity.
-
•
Targeting the acquired choline dependency improves the anti-lymphoma effect of KDACI.
Pera et al. explored the effects of the lysine deacetylase inhibitor panobinostat in the metabolism of patients with lymphoma. They demonstrated that panobinostat alters choline metabolism leading to PI3K pathway activation. Their findings revealed the mechanism behind the anti-lymphoma activity of dual lysine deacetylase/PI3K inhibitors, and uncovered a novel therapeutic strategy based on targeting choline pathway following panobinostat treatment.
1. Introduction
Despite the promising activity of histone deacetylase inhibitors (HDACI) in the management of T-cell lymphoma subtypes (Apuri and Sokol, 2016), their relatively low efficacy in unselected patients with diffuse large B-cell lymphoma (DLBCL) (Crump et al., 2008) uncovers the need for a better understanding of their biological effects to successfully implement this class of drugs in clinics.
Traditionally considered as “epigenetic drugs” due to the impact of histone acetylation on the modulation of chromatin structure and subsequent transcription regulation (Hebbes et al., 1988), HDACI affect the acetylation of a plethora of non-histone proteins (Choudhary et al., 2009), and can therefore be more appropriately reffered to as lysine deacetylase inhibitors (KDACI). Acetylation of non-histone substrates can have various effects on function, stability and localization of proteins. For example, the administration of KDACI to DLBCL cells can simultaneously result in increased acetylation and inhibition of Hsp90 and BCL6, and furthermore in acetylation and activation of p53 (Bereshchenko et al., 2002, Cerchietti et al., 2010, Rao et al., 2009).
Acetylation state is tightly balanced through reactions catalyzed by lysine acetyltransferases and lysine deacetylases (KDAC). The interplay between acetylation and deacetylation of proteins is crucial for various important mechanisms that maintain cellular and metabolic homeostasis (Choudhary et al., 2014). However, the precise metabolic consequences of changes in the acetylation state of non-histone proteins are still understudied in patients. Given the interconnection between protein acetylation and metabolism, we hypothesized that the characterization of metabolic effects of KDACI in DLBCL patients will uncover pathways relevant for lymphoma survival and potentially identify novel targets for combination therapies.
Metabolomics is a powerful tool to profile small molecules in biological specimens. The metabolome has been regarded as the link between genotype and phenotype (Fiehn, 2002), and represents a readout of the current state of an organism (Kaddurah-Daouk et al., 2008).
Here, by performing metabolomic profiling, we revealed that the KDACI panobinostat administered to DLBCL patients alters lipid metabolism impacting, in particular, choline metabolism. In DLBCL cell lines, panobinostat induces metabolic adaptations that lead to activation of the PI3K pathway inducing a newly acquired dependency on choline metabolism for survival. In a xenograft DLBCL model, we then prove that inhibition of choline or PI3K pathways results in increased anti-lymphoma effect of panobinostat.
2. Materials and Methods
2.1. Metabolomics
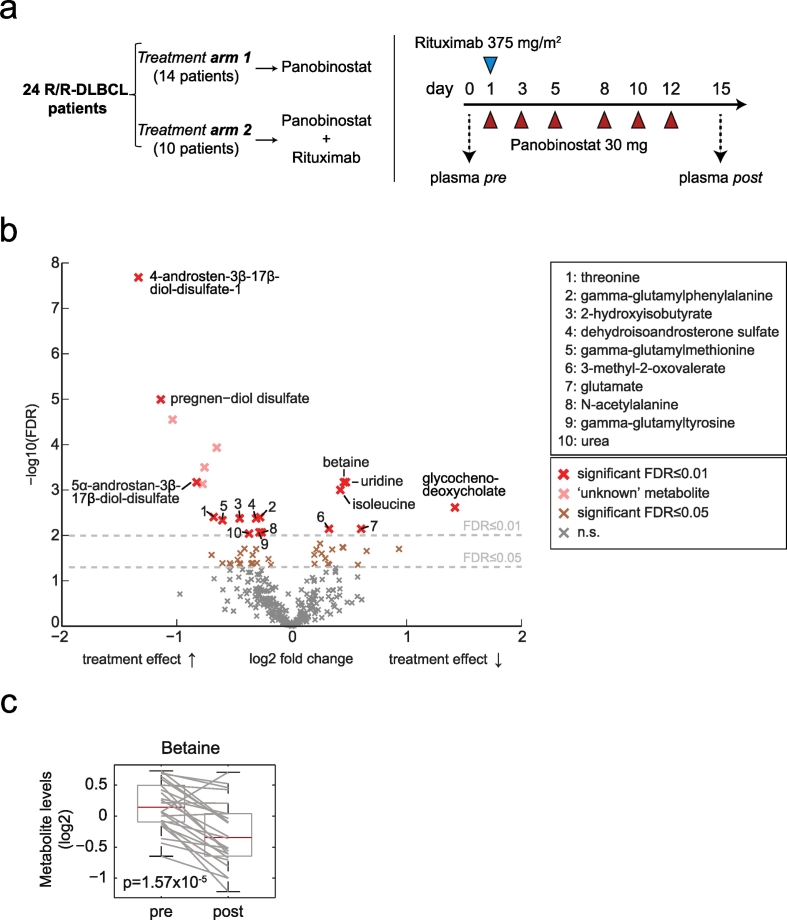
We analyzed plasma from a sub-set of 24 DLBCL patients enrolled in the phase 2 trial NCT01238692 with the approval of the Weill Cornell Medicine IRB. Untargeted metabolomics measurements on plasma were performed by Metabolon Inc., NC, USA. Briefly, measurements were performed by ultra-high-performance liquid-phase chromatography and gas-chromatography separation, coupled with tandem mass spectrometry. Each metabolite was annotated with a) one of seven major biochemical super-pathways (“amino acid”, “peptide”, “lipid”, “energy”, “carbohydrate”, “nucleotide” and “cofactors & vitamins”), and b) one sub-pathway (Table S1). Metabolites classified as “xenobiotic” and those with unknown identity were omitted for this study. Treatment effects for each metabolite were assessed using paired t-tests. False discovery rate (FDR) control according to Benjamini and Yekutieli (Benjamini and Yekutieli, 2001) was used to correct for multiple hypothesis testing. Volcano plots were generated by plotting the log2 fold change of metabolite levels against FDR values for the differences.
For cell metabolomics, the DLBCL cell line OCI-Ly1 was treated with 1 μM of panobinostat for 12 h. Metabolomics analysis was performed on the same LC/GC–MS platform as the plasma data. Each sample was normalized by dividing raw values by cell protein contents, assessed via Bradford assays performed by Metabolonc Inc. Statistical analysis was performed analogously to the plasma analysis, with two-sample t-tests to compare treated and untreated groups (Table S2).
2.2. Reagents
Panobinostat, AZD8186, GDC-0941, CUDC-907 and the compounds from the screened library (Table S3) were from Selleckchem. CK37 was from EMD Millipore. Phosphatidic acid was from Avanti Lipids. Choline chloride was from Sigma.
2.3. Cell Culture
Human diffuse large B-cell lymphoma (DLBCL) cell lines OCI-Ly1 and OCI-Ly7 were grown in Iscove's modified Eagle medium and 10% fetal bovine serum (FBS) (20% for OCI-Ly7) supplemented with 1% penicillin G and streptomycin, 1% l-glutamine, and 1% HEPES. Toledo and SU-DHL-8 were grown in medium containing 90% RPMI-1640 and 10% FCS supplemented with 1% penicillin G and streptomycin, 1% l-glutamine, and 1% HEPES. Cells were maintained in a 37 °C, 5% CO2, humidified incubator. All cell lines were purchased from the American Type Culture Collection or obtained from the Ontario Cancer Institute, and regular testing for Mycoplasma sp. and other contaminants and cell identification by single nucleotide polymorphism were conducted.
2.4. Cell Viability Assays
Growth inhibition 50 (GI50) was determined by a fluorescence assay using 7-hydroxy-3H-phenoxazin-3-one 10-oxide (CellTiter-Blue, Promega) according to the manufacturer's protocol. Cell viability was determined by employing a fluorescence-based assay that relies in live-cell protease activity (CellTiter-Fluor, Promega) following the manufacturer's protocol. All fluorescence measurements were performed in a Synergy4 microplate reader (BioTek).
Caspase assays. Caspase-3 and -7 activity was assessed using the Apo-ONE caspase 3/7 assay (Promega) following the manufacturer's instructions with measurement of fluoresence emission in a Synergy4 microplate reader (BioTek). Caspase activity was normalized by the cell number determined by CellTiter-Fluor (Promega).
2.5. Transient Transfection
OCI-Ly1 cells (2.5 × 106 cells/well) were transfected by electroporation (Amaxa, Lonza AG) in presence of 100 nM of CHKA siRNA (CHKAHSS140690 and CHKAHSS140691, Invitrogen).
2.6. Real-Time Reverse Transcriptase-PCR
Total RNA was purified using TRIzol Reagent (Thermo Fisher Scientific) following manufacturer's instructions and resuspended in RNAse-free water. cDNA was synthesized using high capacity RNA-to-cDNA kit (Applied Biosystems). SYBR Green FastMix was from Quanta BioSciences. Primer sequences can be found in Table S4.
2.7. Immunoblotting
Protein concentrations were determined using the BCA kit (Pierce Biotechnology) according to the manufacturer's instructions. Protein lysates (15–40 μg) were electrophoretically resolved by SDS/PAGE, transferred to PVDF (polyvinylidene difluoride) membrane, and probed with the indicated primary antibodies: Anti-Choline Kinase α (D5X9W) (1:500, 13,422; Cell Signaling), Anti-Acetyl-Histone H3 (Lys9/Lys14) (1:1000, 9677; Cell Signaling), Anti-Phosphate Cytidylyltransferase 1 (1:1000, 109,263, Abcam). Membranes were then incubated with a 1:5000 dilution of a peroxidase conjugated corresponding secondary antibody (sc-2004 and sc-2005, Santa Cruz Biotechnology). Equal loading of the protein samples was confirmed by α-tubulin (1:25,000, ab4074; Abcam) blotting. We used ECL Western Blotting Substrate (Pierce Biotechnology) according to the manufacturer's instructions and the blots were visualized by autoradiography. Quantitative densitometry analysis of western blot bands was performed employing Image J version 10.2 (NIH). The normalized relative densities were calculated relative to the expression of α-tubulin.
2.8. Screening Analysis
Compound screening was conducted in a 96 well format and the effect with or without panobinostat pretreatment was evaluated against 425 targeted compounds. Viability was assessed after 48 h based on rezazurin reduction by cells using Cell titer blue. The data was linearized, normalized to in-plate controls, and the differential effect was computed as the difference between the observed effect with panobinostat and the effect with vehicle, i.e. positive differential effect denotes compounds that are more effective with panobinostat pretreatment. In order to gauge the degree of the effect, meaning, to determine whether an effective drug becomes more effective (higher effect), or a non-effective drug becoming effective we computed a deviation score. The deviation score is defined as the mean-normalized variance between vehicle and panobinostat treatments, approximating to the CV, computed based on the equation below:
where DAB is the deviation score of panobinostat vs. vehicle, DifAB is the defferential effect, and μAB is the mean of the effects observed in panobinostat vs. vehicle treated.
2.9. Mice Studies
All animal procedures were approved by The Research Animal Resource Center of the Weill Cornell Medicine Adult (6- to 8-week-old, male, weighting average of 20 g) severe combined immunodeficiency (SCID) mice were purchased from Charles River Laboratory and subcutaneously injected in the left flank with 10 × 106 low-passage human DLBCL OCI-Ly1 cells. Tumor size was monitored every other day employing electronic digital calipers. Tumor volume was calculated using the equation: tumor volume = (lenght × width2) × 0.5. Treatment schedules are described in Results section. The panobinostat dose (6.2 mg/kg) was determined considering the oral dose administered to the patients in the clinical trial (30 mg) (Assouline et al., 2016) by calculating the animal equivalent dose (AED) as described in (Nair and Jacob, 2016). The CK37 dose was chosen in the light of the work published by Xiong et al. where 4 mg/kg of CK37 was administered daily for 8 days, showing a tumor reduction effect on a murine xenograft T-lymphoma model without signs of gross toxicity (Xiong et al., 2015). Panobinostat was dissolved in DMSO and administered intraperitoneally in 5% dextrose in water in a 1:4 ratio (final volume of 200 μL). CK37 was dissolved in DMSO and administered intraperitoneally (final volume of 50 μL). Mice were weighed every other day. At the end of the experiment, the mice were euthanized by CO2 inhalation.
3. Results
3.1. Panobinostat Induces Changes in Circulating Metabolites in Lymphoma Patients
To determine the metabolic effects of KDACI in cancer patients we conducted metabolomics of lymphoma patients treated with panobinostat. Plasma samples were obtained from refractory or relapsed DLBCL patients enrolled in a phase II study (Assouline et al., 2016) that aimed to evaluate the efficacy of panobinostat vs. panobinostat and rituximab administered according to the schedule shown in Fig. 1a. Untargeted metabolomics was performed in plasma samples (i.e. exo-metabolomics) taken before and after the first cycle of treatment (Fig. 1a, right). After normalization, we compared pre- and post-panobinostat samples for differentially abundant metabolites. We identified 53 metabolites that significantly changed between these time points with a false discovery rate (FDR) < 0.05 (Fig. 1b and Table S1). Among the up-regulated metabolites we identified several sulfated steroids and bile acids, which are part of xenobiotic metabolism pathway. Similarly, we found an increase in gamma-glutamyl aminoacids that could represent the transferring of gamma-glutamyl functional groups to amino acid acceptors in the gamma-glutamyl cycle of xenobiotic detoxification (Williamson et al., 1982). These changes of endogenous plasma metabolites are likely the result of the extensive biotransformation induced by panobinostat (Clive et al., 2012) and KDACI-dependent induction of membrane transporters (Hauswald et al., 2009). Among the down-regulated metabolites, we identified several amino acids such as the branched chain amino acids (BCAA) glutamate and aspartate that are involved in liver transamination reactions, as well as the atypical amino acid betaine. The significant decrease in plasma betaine (Fig. 1c), a non-proteinogenic amino acid, suggests a potential alteration in lipid metabolism (Lever et al., 2011). Betaine derives from the oxidation of choline, which is a component of the phosphatidylcholine pathway, a key phospholipid constituent of cell membranes (Li and Vance, 2008). Since it has previously been demonstrated that panobinostat alters the lipid metabolism of Niemann-Pick mutant fibroblasts in vitro (Pipalia et al., 2011), we therefore investigated the effect of panobinostat on phospholipid metabolism in DLBCL cells.
Fig. 1.
Metabolic profiling reveals changes in choline metabolism of DLBCL patients treated with panobinostat. (a) Treatment arms (left) and treatment schedule (right) of the phase 2 trial. Blood samples were collected prior and post panobinostat in both arms. (b) Volcano plot showing circulating metabolites that significantly changed their levels after panobinostat treatment. (c) Box plot of plasma betaine determined for each patient pre and post panobinostat treatment.
3.2. Panobinostat Induces Choline Pathway Dependency in DLBCL Cells
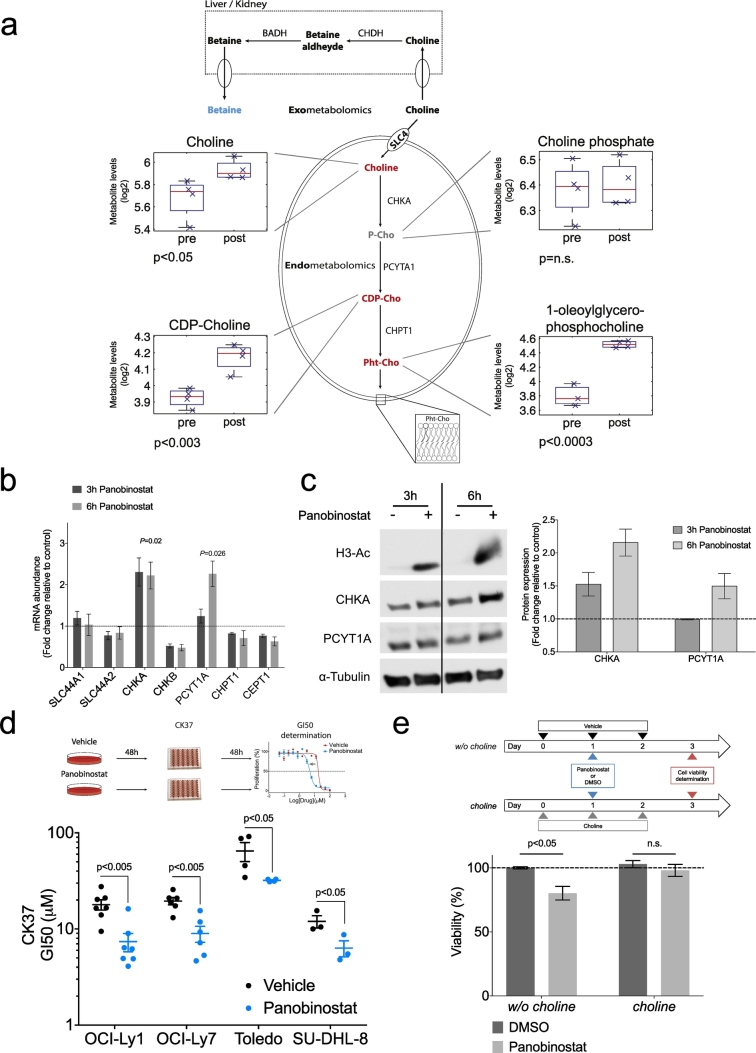
Alteration of plasma betaine in patients receiving panobinostat could impact systemic lipid metabolism, thereby potentially affecting DLBCL cells. To characterize the metabolic effects of panobinostat in lymphoma cells, we conducted cellular metabolomics (i.e. endo-metabolomics) in the DLBCL cell line OCI-Ly1 before and 12 h after panobinostat treatment. We found that panobinostat induced changes in amino acid metabolism (histidine, lysine, polyamine and creatine metabolism), carbohydrate metabolism (glycogen and fructose, manose, galactose, starch and sucrose metabolism), nucleotide metabolism (pyrimidine metabolism), and lipid metabolism (polyunsaturated fatty acid, glycogen, long chain fatty acid, monoacylglycerol and lysolipid metabolisms). At pathway level, the entire group of lipid metabolism was significantly changed (p < 0.02 and FDR < 0.1, Fig. S1 and Table S2). Specifically in the betaine-choline pathway, panobinostat induced up-regulation of choline (p < 0.05), cytidine diphosphate (CDP)-choline (p < 0.003) and the phosphatidylcholine derivative 1-oleoylglycerophosphocholine (p < 0.0003, Fig. 2a). Choline phosphate levels did not change significantly after treatment (Fig. 2a). These changes in cellular metabolomics mirrored those identified in the plasma of DLBCL patients, suggesting that the decrease in betaine plasma levels in patients is likely due to an increased demand of choline metabolites in lymphoma cells.
Fig. 2.
Panobinostat treatment prompts choline pathway dependency on DLBCL cell lines. (a) Levels of choline intracellular metabolites measured pre and post panobinostat treatment in the DLBCL cell line OCI-Ly1. P-Cho phosphocholine, CDP-Cho cytidine 5′-diphosphocholine, Pht-Cho phosphatidilcholine, SLC4 Solute Carrier Family 44, CHKA choline kinase alpha, PCYT1A Phosphate Cytidylyltransferase 1A, CHPT1 Choline Phosphotransferase 1, CHDH Choline Dehydrogenase, BADH Betaine-aldehyde dehydrogenase. (b) Changes in mRNA levels of choline-related enzymes inOCI-Ly1 cells treated with 120 nM of panobinostat (c) Representative western blots for choline kinase alpha (CHKA), phosphate cytidylyltransferase 1 (PCYT1A) and acetyl-histone H3 (H3-Ac) from OCI-Ly1 cells treated with 120 nM panobinostat for 3 and 6 h (left). Quantification of the CHKA and PCYT1A protein levels after treatment (right). (d) Change of GI50 of the CHKA inhibitor CK37 in four DLBCL cell lines after exposure for 48 h to vehicle (black dots) or panobinostat (blue dots) accordingly to the schedule shown on top. (e) Change in cell growth of OCI-Ly1 cells in medium with and without choline upon treatemnt with vehicle (DMSO) or panobinostat 10 nM according to the schedule shown on top. In panels B, C, D and E, data are represented as median ± SEM.
To investigate whether lymphoma cells are more avid on choline metabolites after KDAC inhibition, we first measured messenger RNA expression of key enzymes belonging to the choline pathway in OCI-Ly1 panobinostat-treated cells. We found an increase of choline kinase alpha (CHKA) expression at 3 and 6 h and of phosphocholine cytidytransferase (PCYT1A) at 6 h, while the other enzymes did not significantly change under these conditions (Fig. 2b). Accordingly, protein expression of CHKA increased at 3 and 6 h while PCYT1A protein expression increased at 6 h (Fig. 2c).
To determine whether these changes induce a functional dependence on choline metabolism for survival, we treated a panel of DLBCL cell lines (OCI-Ly1, OCI-Ly7, Toledo and SU-DHL-8) with vehicle or panobinostat for 48 h followed by the CHKA inhibitor CK37 (Clem et al., 2011) for another 48 h (Fig. 2d, top). We reasoned that if panobinostat-treated cells were more reliant on CHKA for survival, we should observe a decrease in the GI50 of CK37. Indeed, DLBCL cell lines showed an increased sensitivity to the CHKA inhibitor after treatment with panobinostat (Fig. 2d, bottom), indicating that panobinostat treatment induced a newly acquired dependency on the choline pathway in the fraction of surviving DLBCL cells. To determine if this dependency relied on the uptake of exogenous choline, we assessed growth of OCI-Ly1 cells in tissue culture medium with or without choline for 24 h followed by 10 nM of panobinostat (or vehicle) and we determined the viability after 48 h. Panobinostat was more effective in cells grown in medium without choline, which suggests that activation of the choline pathway relies on the uptake of exogenous choline (Fig. 2e).
3.3. Panobinostat Induces PI3K Pathway Dependency in DLBCL Cells Via CHKA Activity
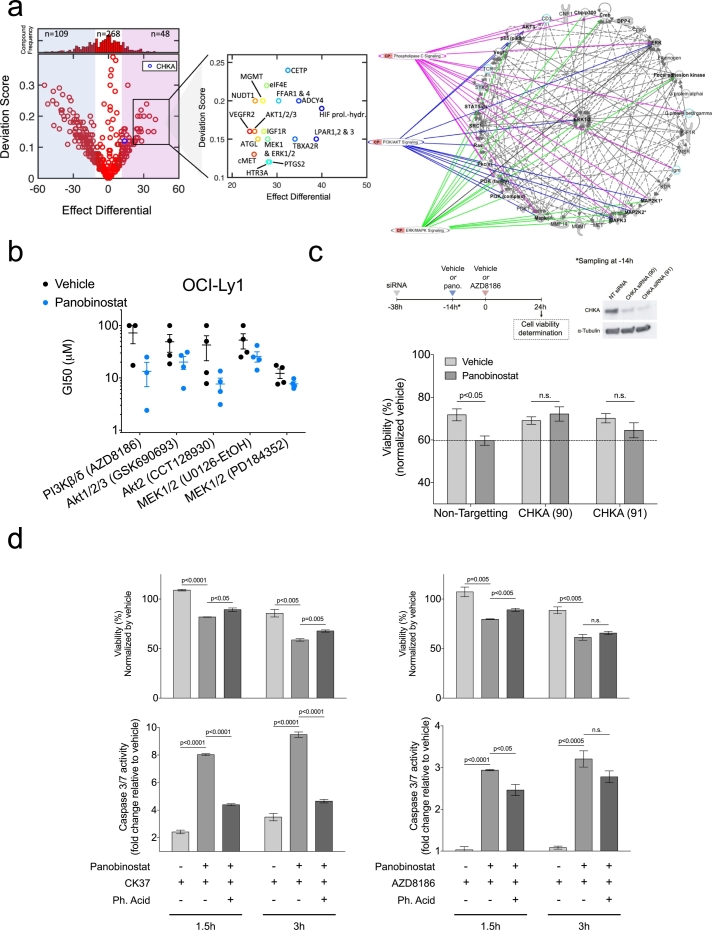
To identify possible pathways fueled by the increased choline pathway activity, we first screened for pathway co-dependency. To this end we treated OCI-Ly1 cells with panobinostat for 48 h followed by exposure to a library of 425 targeted compounds covering 162 target families (Table S3). We calculated and ranked the differential effect, and evaluated the influence on cell proliferation of these compounds in cells treated with panobinostat vs. vehicle. Analysis showed that 109, 268 and 48 compounds respectively, were less, equal and more effective in panobinostat-treated cells (Fig. 3a, left). Among the 48 compounds with increased effect (i.e. with potential co-dependency with choline) in panobinostat-treated cells we found inhibitors of the phosphatidylinositide 3-kinase (PI3K) and the mitogen-activated protein kinase (MAPK) pathways such us the AKT1/2/3 inhibitor GSK690693, and the MEK1 and ERK1/2 inhibitor Selumetinib, respectively. In addition, several other inhibitors were found to target pathways intimately related to the PI3K and/or MAPK pathways (HIF prolyl-hydroxylase inhibitors, antagonists of the lysophpsphatidic acid receptor, inhibitors of the interaction eIF4E/eIF4G, insulin like growth factor receptor inhibitors, cMet inhibitors and VEGFR2 inhibitors). The library of 425 compounds contains 7 related to the PI3K/MAPK pathways (1.65%). From the 48 active compounds, 4 related to the PI3K/MAPK pathways (8.3%), representing a significant enrichment of these targets (Fisher's test p = 0.0003). Interestingly, the screening also uncovered an increase in the effect of molecules that target lipid metabolism (cholesteryl ester transfer protein, free fatty acid receptor and adipose triglyceride lipase inhibitors), or inhibitors with targets of lipidic nature (e.g. the thromboxane A2 synthetase inhibitor Ozagrel) (Fig. 3a, center). Finally, we selected the targets of those hits with a differential effect and deviation score greater than or equal to those established by the CHKA inhibitor CK37 (differential effect ≥ 13.5; deviation score ≥ 0.12). This list of targets were uploaded into Ingenuity Pathway Analysis, which resulted in a network illustrating the interrelation of targets highly represented in the PI3K/AKT, ERK/MAPK and phospholipase C (PLC) pathways (Fig. 3a, right).
Fig. 3.
Choline pathway dependency is linked to the activation of the PI3K pathway in the DLBCL cell line OCI-Ly1. (a) Deviation score vs. the effect differential illustrates the distribution and the number of compounds having enhanced susceptibility with (magenta) and without (blue) panobinostat pretreatment (left). Distribution and number of compounds within each category are denoted above the plot. Inset shows key targets representing high differential effect and deviation score (center). Network illustrating key interactions based on the targets of the 48 compounds identified to have greater effect than the CHKA inhibitor CK37 (right). Panobinostat pretreatment consisted in 10 nM exposure for 48 h. All compounds of the library were tested at 10 μM final concentration. (b) GI50 at 48 h of PI3K and MAPK inhibitors in OCI-Ly1 cells pretreated with vehicle (black dots) or 10 nM panobinostat (blue dots) for 48 h. (c) Viability (bottom) of OCI-Ly1 cells transfected with CHKA siRNA, pretreated with vehicle or 10 nM panobinostat, and exposed to 100 μM of the PI3K inhibitor AZD8186 for 24 h after pretreatment (top). (d) Viability (top) and caspase 3/7 activity (bottom) of OCI-Ly1 cells pretreated with vehicle or 10 nM panobinostat for 48 h, and then treated with 10 μM of the CHKA inhibitor CK37 (CHKAi) (left) or 100 μM of the PI3K inhibitor AZD8186 (right), with or without 100 μM phosphatidic acid (Ph. Acid) for the specified times. Data are represented as median ± SEM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We independently analyzed the acquisition of PI3K and MAPK pathway dependencies by comparing the GI50 of inhibitors of AKT (CCT128930 and GSK690693) and MEK1/2 (PD184352 and U0126) in OCI-Ly1 cells previously treated with 10 nM of panobinostat (vs. vehicle) for 48 h. In addition, we tested the differential effect of inhibitors of PI3Kβ/δ (AZD8186), since δ is a highly expressed isoform in lymphoid tissues (Vanhaesebroeck et al., 1997) and found critical for activation, proliferation and survival of B cells (Okkenhaug et al., 2002). We found a more pronounced differential effect for the PI3Kβ/δ and AKT2 inhibitors (Fig. 3b), suggesting that panobinostat induces mostly PI3K dependency.
Next, we determine whether CHKA activity is required for acquiring PI3K dependency upon KDAC inhibition. Thus, we administered panobinostat to OCI-Ly1 cells transfected with CHKA siRNA and treated them with the PI3K inhibitor AZD8186. Indeed, AZD8186 decreased viability of cells treated with panobinostat in siRNA control, but this effect was completely rescued in cells transfected with siRNA CHKA (Fig. 3c), demonstrating that choline kinase activity is required to induce PI3K dependency upon KDAC inhibition. Choline kinase commits choline to integration into phosphatidylcholine (Ueland, 2011), which in turn it can be cleaved by phospholipases to produce several lipids second messengers such as phosphatidic acid (Buchanan et al., 2005). Since it is known that phosphatidic acid is a key activator of the PI3K pathway in cancer cells (Yalcin et al., 2010), we speculated that the acquired PI3K dependency could be triggered by the synthesis of phosphatidic acid through the choline pathway. Indeed, we were able to rescue both choline and PI3K pathway dependencies by exogenous phosphatidic acid addition, determined by viability and caspase-3/-7 activation (Fig. 3d).
Remarkably, the co-dependencies revealed by the screening were in accordance with our cell metabolomics data. The already-mentioned alterations in lipid metabolism detected in panobinostat treated cells were also uncovered by the screening, and since PI3K-AKT signaling plays an important role in stimulating de novo lipid synthesis (Lien et al., 2016), we speculate that this alteration in lipid metabolism could be a readout of PI3K pathway activation. Additionally, the alteration detected in pyrimidine metabolism, and the significant decrease observed in glycogen metabolism (Fig. S1 and Table S2) could be also the result of a metabolic reprograming executed by the PI3K pathway (Ben-Sahra et al., 2013, Lien et al., 2016).
3.4. The Anti-Lymphoma Effect of the Dual Inhibitor CUDC-907 Relies on the Induction of PI3K Dependency through Choline Pathway Activation
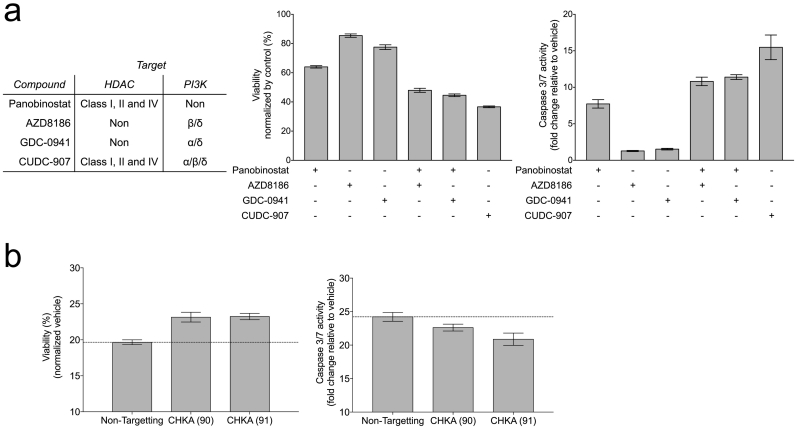
Our data demonstrates that DLBCL cells surviving panobinostat administration are more reliant on the choline and PI3K pathways, providing a mechanistic background for the synergistic antitumor activity known for the combination of KDAC and PI3K inhibitors (Ozaki et al., 2010, Wozniak et al., 2010). Moreover, it could explain the rationale for testing KDAC/PI3K dual inhibitors, such as CUDC-907 (Qian et al., 2012). Specifically, we postulate that CUDC-907 induces a choline pathway dependency through its KDACI moiety, which is then therapeutically exploited by the inhibition of PI3K. To test this, we determined the effect on OCI-Ly1 viability of the PI3Kβ/δ inhibitor AZD8186, the PI3Kα/δ inhibitor GDC-0941 (which contains the scaffold present in the CUDC-907), and CUDC-907 (Fig. 4a, left), administered either alone or in combination with panobinostat. Panobinostat was administered concurrently with the PI3K inhibitor instead of pretreating the cells first with the KDACI, in order to mimic the effect of CUDC-907. Both PI3K inhibitors decreased the viability of OCI-Ly1 cells when given in combination with panobinostat but not when given alone, and the effect was comparable to the hybrid inhibitor CUDC-907 (Fig. 4a, center). The decrease in viability associated with increase in the activity of caspases 3 and 7 (Fig. 4a, right). This suggests that, at least in terms of cell viability, the PI3K inhibitory moiety of CUDC-907 acts similar to the either PI3K inhibitor. Finally, silencing choline pathway in OCI-Ly1 cells by employing siRNA CHKA resulted in a loss of CUDC-907 activity, determined by viability and caspase-3/-7 activation (Fig. 4b). Thus, our data suggests that the anti-lymphoma effect of the hybrid molecule CUDC-907 partially relies on the activation of the choline pathway.
Fig. 4.
CUDC-907 effect relies on the induction of PI3K dependency through choline pathway activation. (a) Viability (center) and caspase-3/7 activity (right) of OCI-Ly1 cells after 24 h at the specified treatments (120 nM panobinostat or 10 μM of AZD8186, GDC-0941 or CUDC-907). (b) Viability (left) and caspase-3/7 activity (right) of OCI-Ly1 cells transfected with CHKA siRNA and treated for 24 h with 100 μM of CUDC-907. Data are represented as ± SEM.
3.5. Therapeutic Exploitation of the Choline Pathway Dependency Results in DLBCL Reduction in Mice
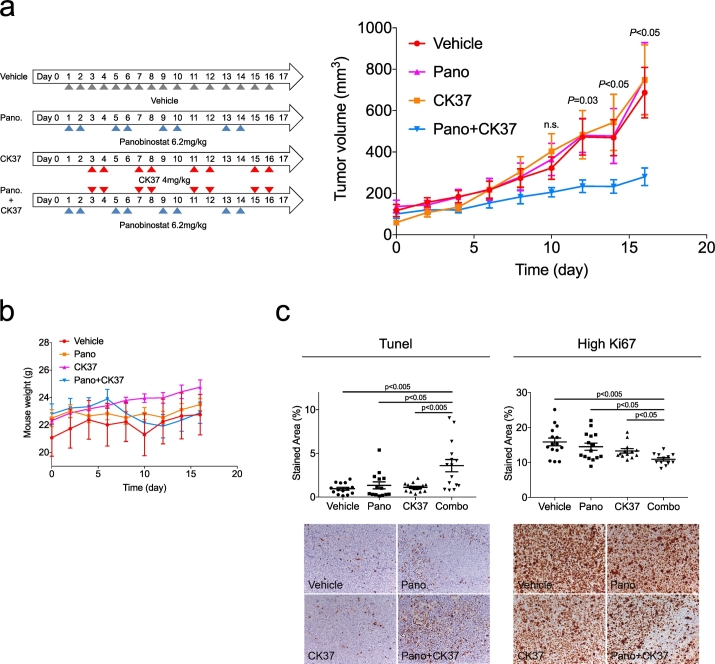
Aiming to explore the therapeutic potential of the revealed choline pathway dependency induced by panobinostat in DLBCL, we administered vehicle (n = 5), panobinostat 6.2 mg/kg (equivalent to a 30 mg oral dose administered to the patients) (n = 5), CK37 4 mg/kg (n = 5), or their combination (n = 5) to mice bearing chemotherapy refractory DLBCL OCI-Ly1 cell line xenografts. When xenografts reached around 100 mm3, mice were randomized into four groups and treated following the schedule shown on Fig. 5a. Compared to the vehicle-treated mice, panobinostat and CK37 alone did not show any effect on tumor volume. Interestingly, their combination significantly suppressed tumor growth (p = 0.03, 0.03 and 0.01 at days 12, 14 and 16, respectively, Fig. 5a). There was no evidence of toxicity based on body weight (Fig. 5b).
Fig. 5.
Combined treatment with panobinostat and CK37 results in tumor growth reduction on OCI-Ly1 xenografts. (a) Treatment doses and schedule (left) and tumor growth curves (right) for mice receiving vehicle, panobinostat, CK37 or both drugs. (b) Mouse weight curves for all the four cohorts. (c) Quantitative plots (top) and representative images (bottom) from the OCI-Ly1 mice tumors at day 16, assayed for apoptosis by TUNEL (left) and for proliferation by Ki67 immunostaining (right). Data are represented as median ± SEM.
We assessed apoptosis and proliferation by TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay) assay and immunohistochemistry for Ki67, respectively, on tumor specimens. The stained area with apoptotic cells was significantly higher in mice treated with the combination compared to the other cohorts (Fig. 5c, left). The number of cells with high Ki67 was lower in mice treated with the combination vs. the other groups (Fig. 5c, right). Overall, these data suggest that targeting the choline pathway with a CHKA inhibitor could be an effective strategy to increase the anti-lymphoma effect of KDACIs.
4. Discussion
Lysine deacetylase inhibitors have emerged as a promising approach to improve current lymphoma treatment. While our understanding of their effects on chromatin status has improved over the past years, whether and how these enzymes affect cellular functions beyond transcription remains uncharted territory. Due to the activity of KDAC on proteins involved in metabolic pathways, we investigated metabolic changes induced by the KDACI panobinostat in DLBCL patients. We postulated that metabolomic profiling of KDACI effects in patients would provide further insights into the biological effects of this class of drugs, potentially revealing pathways relevant for lymphoma survival and therapeutic exploitation.
As a first discovery step of metabolic changes, we measured plasma metabolite levels in DLBCL patients before and after panobinostat-treatment. We identified a significant decrease in the concentration of circulating betaine, a metabolite from the choline pathway. This pointed to a higher requirement of choline in the tumor cells and thus a possible change in lipid metabolism through a higher activity of the choline pathway. Cell metabolomics experiments confirmed this notion.
Although activated choline metabolism is acknowledged as metabolic hallmark of cancer cells (Glunde et al., 2011), little is known about KDACI effects on this pathway. Aiming to find biomarkers of response to KDACI by employing noninvasive imaging (typically magnetic resonance spectroscopy), an increase in the phosphocholine levels has previously been detected when treating cells and/or xenografts with KDACI such as LAQ824, SAHA or belinostat (Sankaranarayanapillai et al., 2006, Chung et al., 2008, Beloueche-Babari et al., 2012, Ward et al., 2013). These represent interesting observations that run counter to conventional knowledge, since typically an increase of choline-containing compounds has been associated with cell transformation while a decrease is observed after cytotoxic treatment. Despite the controversial nature of these findings, only little mechanistic exploration had been performed. For example, some authors described an increase in choline uptake due to increased expression of the transporter SLC44A1 after KDACI-treatment (Ward et al., 2013), and/or an increase in the CHKA expression (Beloueche-Babari et al., 2012, Ward et al., 2013).
In our work, we revealed that this alteration in choline metabolism represents a survival mechanism to panobinostat through activation of the PI3K pathway in lymphoma cells. This finding is of translational relevance, as it points towards a strategy of targeting the choline pathway after KDACI treatment. In this regard, our in vivo DLBCL model supported this therapeutic strategy, as demonstrated by a significant reduction of tumor growth in mice treated with panobinostat and the CHKA inhibitor CK37. Additionally, we demonstrated that this dependency might represent the molecular mechanism behind the anticancer activity of KDAC/PI3K hybrid inhibitors.
In conclusion, by applying metabolomics profiling we partially uncovered the mechanism behind the well-documented synergistic activity of KDAC and PI3K inhibitors in vitro and in vivo (Rahmani et al., 2003, Rahmani et al., 2005, Ozaki et al., 2010, Wozniak et al., 2010, Rahmani et al., 2014) and we were thus able to shed light on the molecular basis behind the activity of the dual-inhibitor CUDC-907 (Qian et al., 2012). With all, our work reveals the impact of KDACI on metabolism, and uncovers the need to investigate the biological effects, beyond translation, of this class of drugs in order to be able to successfully implement them in the clinical setting.
The following are the supplementary data related to this article.
Metabolite super and subpathway changes determined pre vs. post-treatment with panobinostat in B-cell lymphoma patients (top). Volcano plot showing intracellular metabolites that significantly changed their levels in OCI-Ly1 cells treated with panobinostat (bottom).
Complete list of plasma metabolites from B-cell lymphoma patients treated with panobinostat included in the volcano plot shown in Fig. 1
Complete list of intracellular metabolites from B-cell lymphoma cells treated with panobinostat included in the volcano plot shown in Fig. S1
Results from the compound library screening shown in Fig. 3
List of qRT-PCR primers.
Funding Sources
J.K. is a junior investigator of the German Helmholtz Association. K.K.M. is a Chercheur-boursier and S.E.A. is a Chercheur-boursier clinicien of the Fonds de recherche du Québec. L.C. is the Weill Cornell Raymond and Beverly Sackler Scholar. This work was supported by the Irma T. Hirschl Trust Award (L.C.) (Award number: 2012-2017), and the Doris Duke Charitable Foundation (L.C.) (2012070).
Conflict of Interest
S.E.A. has received research funding and has acted as consultant for Novartis Canada (makers of panobinostat) and Hofman-LaRoche Canada (makers of rituximab). The remaining authors declare no competing financial interests.
Author Contributions
B.P. and J.K. performed experimental design and execution, data interpretation and wrote the manuscript. B.P. and J.K. are considered equal co-first authors in this study. R.M. performed experimental design and participated in the data analysis. J.P., L.R. and J.M.P. helped in performing experiments. K.K.M. and S.A.E. collected patient samples and conducted the clinical trial. L.C. conceived the study, participated in its design, analyzed and interpreted the data, and wrote the manuscript.
Acknowledgements
None. The funders have not had any role in the study design, data collection, data analysis, interpretation or writing of this report.
References
- Apuri S., Sokol L. An overview of investigational Histone deacetylase inhibitors (HDACis) for the treatment of non-Hodgkin's lymphoma. Expert Opin. Investig. Drugs. 2016;25:687–696. doi: 10.1517/13543784.2016.1164140. [DOI] [PubMed] [Google Scholar]
- Assouline S.E., Nielsen T.H., Yu S., Alcaide M., Chong L., Macdonald D., Tosikyan A., Kukreti V., Kezouh A., Petrogiannis-Haliotis T., Albuquerque M., Fornika D., Alamouti S., Froment R., Greenwood C.M., Oros K.K., Camglioglu E., Sharma A., Christodoulopoulos R., Rousseau C., Johnson N., Crump M., Morin R.D., Mann K.K. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood. 2016;128:185–194. doi: 10.1182/blood-2016-02-699520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloueche-Babari M., Arunan V., Troy H., TE Poele R.H., TE Fong A.C., Jackson L.E., Payne G.S., Griffiths J.R., Judson I.R., Workman P., Leach M.O., Chung Y.L. Histone deacetylase inhibition increases levels of choline kinase alpha and phosphocholine facilitating noninvasive imaging in human cancers. Cancer Res. 2012;72:990–1000. doi: 10.1158/0008-5472.CAN-11-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Ben-Sahra I., Howell J.J., Asara J.M., Manning B.D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereshchenko O.R., Gu W., Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- Buchanan F.G., Mcreynolds M., Couvillon A., Kam Y., Holla V.R., Dubois R.N., Exton J.H. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1638–1642. doi: 10.1073/pnas.0406698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti L.C., Hatzi K., Caldas-Lopes E., Yang S.N., Figueroa M.E., Morin R.D., Hirst M., Mendez L., Shaknovich R., Cole P.A., Bhalla K., Gascoyne R.D., Marra M., Chiosis G., Melnick A. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J. Clin. Invest. 2010 doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Chung Y.L., Troy H., Kristeleit R., Aherne W., Jackson L.E., Atadja P., Griffiths J.R., Judson I.R., Workman P., Leach M.O., Beloueche-Babari M. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts. Neoplasia. 2008;10:303–313. doi: 10.1593/neo.07834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem B.F., Clem A.L., Yalcin A., Goswami U., Arumugam S., Telang S., Trent J.O., Chesney J. A novel small molecule antagonist of choline kinase-alpha that simultaneously suppresses MAPK and PI3K/AKT signaling. Oncogene. 2011;30:3370–3380. doi: 10.1038/onc.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clive S., Woo M.M., Nydam T., Kelly L., Squier M., Kagan M. Characterizing the disposition, metabolism, and excretion of an orally active pan-deacetylase inhibitor, panobinostat, via trace radiolabeled 14C material in advanced cancer patients. Cancer Chemother. Pharmacol. 2012;70:513–522. doi: 10.1007/s00280-012-1940-9. [DOI] [PubMed] [Google Scholar]
- Crump M., Coiffier B., Jacobsen E.D., Sun L., Ricker J.L., Xie H., Frankel S.R., Randolph S.S., Cheson B.D. Phase II trial of oral vorinostat (suberoylanilide hydroxamic acid) in relapsed diffuse large-B-cell lymphoma. Ann. Oncol. 2008;19:964–969. doi: 10.1093/annonc/mdn031. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Glunde K., Bhujwalla Z.M., Ronen S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer. 2011;11:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswald S., Duque-Afonso J., Wagner M.M., Schertl F.M., Lubbert M., Peschel C., Keller U., Licht T. Histone deacetylase inhibitors induce a very broad, pleiotropic anticancer drug resistance phenotype in acute myeloid leukemia cells by modulation of multiple ABC transporter genes. Clin. Cancer Res. 2009;15:3705–3715. doi: 10.1158/1078-0432.CCR-08-2048. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne A.W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R., Kristal B.S., Weinshilboum R.M. Metabolomics: a global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Lever M., George P.M., Atkinson W., Molyneux S.L., Elmslie J.L., Slow S., Richards A.M., Chambers S.T. Plasma lipids and betaine are related in an acute coronary syndrome cohort. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vance D.E. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- Lien E.C., Lyssiotis C.A., Cantley L.C. Metabolic reprogramming by the PI3K-Akt-mTOR pathway in cancer. Recent Results Cancer Res. 2016;207:39–72. doi: 10.1007/978-3-319-42118-6_3. [DOI] [PubMed] [Google Scholar]
- Nair A.B., Jacob S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016;7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., Peskett E., Pearce W., Meek S.E., Salpekar A., Waterfield M.D., Smith A.J., Vanhaesebroeck B. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- Ozaki K., Kosugi M., Baba N., Fujio K., Sakamoto T., Kimura S., Tanimura S., Kohno M. Blockade of the ERK or PI3K-Akt signaling pathway enhances the cytotoxicity of histone deacetylase inhibitors in tumor cells resistant to gefitinib or imatinib. Biochem. Biophys. Res. Commun. 2010;391:1610–1615. doi: 10.1016/j.bbrc.2009.12.086. [DOI] [PubMed] [Google Scholar]
- Pipalia N.H., Cosner C.C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., Maxfield F.R. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-pick type C1 mutant human fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C., Lai C.J., Bao R., Wang D.G., Wang J., Xu G.X., Atoyan R., Qu H., Yin L., Samson M., Zifcak B., Ma A.W., Dellarocca S., Borek M., Zhai H.X., Cai X., Voi M. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin. Cancer Res. 2012;18:4104–4113. doi: 10.1158/1078-0432.CCR-12-0055. [DOI] [PubMed] [Google Scholar]
- Rahmani M., Yu C., Reese E., Ahmed W., Hirsch K., Dent P., Grant S. Inhibition of PI-3 kinase sensitizes human leukemic cells to histone deacetylase inhibitor-mediated apoptosis through p44/42 MAP kinase inactivation and abrogation of p21(CIP1/WAF1) induction rather than AKT inhibition. Oncogene. 2003;22:6231–6242. doi: 10.1038/sj.onc.1206646. [DOI] [PubMed] [Google Scholar]
- Rahmani M., Reese E., Dai Y., Bauer C., Payne S.G., Dent P., Spiegel S., Grant S. Coadministration of histone deacetylase inhibitors and perifosine synergistically induces apoptosis in human leukemia cells through Akt and ERK1/2 inactivation and the generation of ceramide and reactive oxygen species. Cancer Res. 2005;65:2422–2432. doi: 10.1158/0008-5472.CAN-04-2440. [DOI] [PubMed] [Google Scholar]
- Rahmani M., Aust M.M., Benson E.C., Wallace L., Friedberg J., Grant S. PI3K/mTOR inhibition markedly potentiates HDAC inhibitor activity in NHL cells through BIM- and MCL-1-dependent mechanisms in vitro and in vivo. Clin. Cancer Res. 2014;20:4849–4860. doi: 10.1158/1078-0432.CCR-14-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R., Lee P., Fiskus W., Yang Y., Joshi R., Wang Y., Buckley K., Balusu R., Chen J., Koul S., Joshi A., Upadhyay S., Tao J., Sotomayor E., Bhalla K.N. Co-treatment with heat shock protein 90 inhibitor 17-dimethylaminoethylamino-17-demethoxygeldanamycin (DMAG) and vorinostat: a highly active combination against human mantle cell lymphoma (MCL) cells. Cancer Biol Ther. 2009;8:1273–1280. doi: 10.4161/cbt.8.13.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanapillai M., Tong W.P., Maxwell D.S., Pal A., Pang J., Bornmann W.G., Gelovani J.G., Ronen S.M. Detection of histone deacetylase inhibition by noninvasive magnetic resonance spectroscopy. Mol. Cancer Ther. 2006;5:1325–1334. doi: 10.1158/1535-7163.MCT-05-0494. [DOI] [PubMed] [Google Scholar]
- Ueland P.M. Choline and betaine in health and disease. J. Inherit. Metab. Dis. 2011;34:3–15. doi: 10.1007/s10545-010-9088-4. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Welham M.J., Kotani K., Stein R., Warne P.H., Zvelebil M.J., Higashi K., Volinia S., Downward J., Waterfield M.D. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.S., Eriksson P., Izquierdo-Garcia J.L., Brandes A.H., Ronen S.M. HDAC inhibition induces increased choline uptake and elevated phosphocholine levels in MCF7 breast cancer cells. PLoS One. 2013;8:e62610. doi: 10.1371/journal.pone.0062610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J.M., Boettcher B., Meister A. Intracellular cysteine delivery system that protects against toxicity by promoting glutathione synthesis. Proc. Natl. Acad. Sci. U. S. A. 1982;79:6246–6249. doi: 10.1073/pnas.79.20.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M.B., Villuendas R., Bischoff J.R., Aparicio C.B., Martinez Leal J.F., De La Cueva P., Rodriguez M.E., Herreros B., Martin-Perez D., Longo M.I., Herrera M., Piris M.A., Ortiz-Romero P.L. Vorinostat interferes with the signaling transduction pathway of T-cell receptor and synergizes with phosphoinositide-3 kinase inhibitors in cutaneous T-cell lymphoma. Haematologica. 2010;95:613–621. doi: 10.3324/haematol.2009.013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Bian J., Wang L., Zhou J.Y., Wang Y., Zhao Y., Wu L.L., Hu J.J., Li B., Chen S.J., Yan C., Zhao W.L. Dysregulated choline metabolism in T-cell lymphoma: role of choline kinase-alpha and therapeutic targeting. Blood Cancer J. 2015;5:287. doi: 10.1038/bcj.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A., Clem B., Makoni S., Clem A., Nelson K., Thornburg J., Siow D., Lane A.N., Brock S.E., Goswami U., Eaton J.W., Telang S., Chesney J. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 2010;29:139–149. doi: 10.1038/onc.2009.317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolite super and subpathway changes determined pre vs. post-treatment with panobinostat in B-cell lymphoma patients (top). Volcano plot showing intracellular metabolites that significantly changed their levels in OCI-Ly1 cells treated with panobinostat (bottom).
Complete list of plasma metabolites from B-cell lymphoma patients treated with panobinostat included in the volcano plot shown in Fig. 1
Complete list of intracellular metabolites from B-cell lymphoma cells treated with panobinostat included in the volcano plot shown in Fig. S1
Results from the compound library screening shown in Fig. 3
List of qRT-PCR primers.