Abstract
The epidermal growth factor receptor (EGFR) is known to play a critical role in non-small cell lung cancer (NSCLC). Constitutively active EGFR mutations, including in-frame deletion in exon 19 and L858R point mutation in exon 21, contribute about 90% of all EGFR-activating mutations in NSCLC. Although oral EGFR-tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib, show dramatic clinical efficacy with significantly prolonged progression-free survival in patients harboring these EGFR-activating mutations, most of these patients will eventually develop acquired resistance. Researchers have recently named genomic instability as one of the hallmarks of cancer. Genomic instability usually involves a transient phase of polyploidization, in particular tetraploidization. Tetraploid cells can undergo asymmetric cell division or chromosome loss, leading to tumor heterogeneity and multidrug resistance. Therefore, identification of signaling pathways involved in tetraploidization is crucial in overcoming drug resistance. In our present study, we found that gefitinib could activate YAP-MKK3/6-p38 MAPK-STAT3 signaling and induce tetraploidization in gefitinib-resistance cells. Using p38 MAPK inhibitors, SB203580 and losmapimod, we could eliminate gefitinib-induced tetraploidization and overcome gefitinib-resistance. In addition, shRNA approach to knockdown p38α MAPK could prevent tetraploidy formation and showed significant inhibition of cancer cell growth. Finally, in an in vivo study, losmapimod could successfully overcome gefitinib resistance using an in-house established patient-derived xenograft (PDX) mouse model. Overall, these findings suggest that losmapimod could be a potential clinical agent to overcome gefitinib resistance in NSCLC.
Keywords: Non-small cell lung cancer (NSCLC), Gefitinib resistance, Tetraploidization, p38 mitogen activating protein kinase (MAPK), Losmapimod
Highlights
-
•
Gefitinib induces tetraploidy formation in gefitinib-resistant NSCLC cells
-
•
YAP-MKK3/6-p38 MAPK signaling is essential for tetraploidization
-
•
Losmapimod, a p38 MAPK inhibitor, overcomes gefitinib-resistance both in vitro and PDX xenograft mode
Gefitinib is a targeted drug therapy in non-small cell lung cancer (NSCLC) which shows dramatic clinical efficacy. However, most of these patients eventually develop drug resistance. Although researchers have identified different mechanisms contributing to the drug resistance, developing a single therapy to overcome the drug resistance remains difficult. In this study, we find that tetraploidization of cancer cells through YAP-MKK3/6-p38 MAPK signaling could be one of the common mechanisms in developing the drug resistance. By using losmapimod, we could eliminate tetraploidization and overcome gefitinib resistance in an animal model suggesting that losmapimod could be a potential clinical agent to overcome gefitinib resistance in NSCLC.
1. Introduction
Lung cancer is one of the most lethal cancers worldwide (Siegel et al., 2016). Non-small cell lung cancer (NSCLC) is the major type of lung cancer and the overall 5-year survival rate is < 10% (Miller et al., 2016). In NSCLC, mutations are frequently observed in the epidermal growth factor receptor (EGFR), including an in-frame deletion in exon 19 and the L858R point mutation in exon 21 (Lynch et al., 2004). These mutations comprise about 90% of all EGFR-activating mutations in NSCLC. Oral EGFR-tyrosine kinase inhibitors (TKIs), such as gefitinib and erlotinib, show dramatic clinical efficacy initially with significantly prolonged progression-free survival in patients harboring these EGFR-activating mutations (Bria et al., 2011). However, most of these patients will eventually develop acquired resistance. Different mechanisms have been identified to contribute to the acquired resistance (Sequist et al., 2011). Among them, the development of a secondary T790 M mutation in exon 20 of the Egfr gene and mesenchymal-epithelial transition factor (Met) amplification are the two main mechanisms, which account for 50% and 20% of acquired resistance, respectively (Engelman et al., 2007, Kobayashi et al., 2005a). Based on this heterogeneity of mechanisms, single-agent treatment strategies to successfully overcome acquired resistance have severe limitations.
In recent years, researchers have identified genomic instability as one of the hallmarks of cancers (Hanahan and Weinberg, 2011). Genomic instability usually involves a transient phase of polyploidization, in particular, tetraploidization (Vitale et al., 2007). In response to DNA damage or cellular stress, cells can undergo cell cycle checkpoints to delay cell cycle progression and allow time for cell repair. G1/S and G2/M are two main cell cycle checkpoints that have been identified. Prolonged G2-arrest can induce G1-state tetraploidization by allowing cells to enter S-phase by endo-reduplication (Lilly and Duronio, 2005). This p53-induced tetraploidy provides a survival mechanism by preventing 4 N cells to undergo mitosis prematurely (Shen et al., 2013). Most importantly, these tetraploid cells can undergo asymmetric cell division or chromosome loss, leading to tumor heterogeneity (Vitale et al., 2011, Zhang et al., 2014). Tetraploid cells have been identified in 37% of early stage cancers and have been demonstrated to promote uncontrolled cancer cell growth (Zack et al., 2013). Moreover, accumulating evidence shows that tetraploid cells are associated with multidrug resistance and lead to poor prognosis (Bakhoum and Compton, 2012, Lee et al., 2011). Therefore, identification of the signaling pathways that are involved in tetraploidization is crucial in overcoming drug resistance.
The p38 MAPK belongs to the mitogen activated protein kinase (MAPK) superfamily and is a major cellular signal transducer of extracellular stress (Han et al., 1994, Schaeffer and Weber, 1999). The p38 MAPK is activated by phosphorylation of the Thr-Gly-Tyr dual phosphorylation motif at residues Thr180 and Tyr182 (Doza et al., 1995). The p38 MAPK family consists of four members: p38α, p38β, p38γ and p38δ (Jiang et al., 1997). Among the four isoforms, the p38α MAPK isoform is the best characterized. The role of the p38 MAPK pathway in regulating cell cycle checkpoints, cell differentiation and cell survival is well-established (Thornton and Rincon, 2009). Both cell cycle checkpoints, G1/S and G2/M, have been reported to be associated with p38 MAPK (Bulavin et al., 2001, Lafarga et al., 2009). However, the role of p38 MAPK in tetraploidization remains largely unknown.
Several p38 MAPK inhibitors are under development and are being evaluated in various phases of clinical trials (Kumar et al., 2003, Triantaphyllopoulos et al., 2010). However, most of these inhibitors have failed because of unacceptable side effects. Losmapimod is the only p38 MAPK inhibitor that has progressed through Phase III clinical trials and has been shown to be well-tolerated in human clinical studies (O'Donoghue et al., 2016). In the current study, we determined whether inhibition of p38 MAPK could overcome gefitinib resistance by using the prototypical p38 MAPK inhibitor, SB203580, or losmapimod. We found that gefitinib could induce tetraploidization by activating YAP-MKK3/6-p38 MAPK-STAT3 signaling in gefitinib-resistant cells. We further used losmapimod to examine whether the ablation of p38 MAPK activation could inhibit tetraploidization and overcome gefitinib-resistance. Our data indicate that ablation of p38 MAPK signaling could eliminate tetraploidy formation induced by gefitinib and significantly inhibit cell proliferation and anchorage-independent cell growth. Knockdown of p38 MAPK also showed substantial inhibition of cancer cell growth. Finally, we found that losmapimod could successfully overcome gefitinib resistance in vivo in an in-house established PDX mouse model. Overall, these findings demonstrate that losmapimod could be a potential clinical agent to overcome gefitinib resistance in NSCLC.
2. Materials and Methods
2.1. Chemicals and Reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. SB203580 was purchased from Selleck Chemicals (Houston, TX) and losmapimod was from Medchemexpress (Princeton, NJ). Gefitinib was obtained from LC Laboratory (Woburn, MA). All the above reagents were dissolved in dimethyl sulfoxide (DMSO), stored at -80 °C, and diluted in culture medium for experiments. Rosewell Park Memorial Institute Medium (RPMI)-1640, DMEM, gentamicin, antibacterial-antimycotic solution, trypsin-EDTA and Opti-MEM were all from Life Technologies, Inc. (Grand Island, NY). Fetal bovine serum (FBS) was obtained from Biological Industries (Beit-Haemek, Israel). The primary antibody against Ki-67 (Thermo Fisher Scientific Cat# PA5-19462, RRID:AB_10981523) was purchased from ThermoScientific (Fremont, CA) and the secondary antibody against rabbit (Santa Cruz Biotechnology Cat# sc-2004, RRID:AB_631746) and mouse (Santa Cruz Biotechnology Cat# sc-2005, RRID:AB_631736) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All other antibodies, including phospho-p38 MAPK (Cell Signaling Technology Cat# 9211, RRID:AB_331641), p38 MAPK (Cell Signaling Technology Cat# 9212, RRID:AB_330713), p38α MAPK (Cell Signaling Technology Cat# 9218S, RRID:AB_10694846), p21 (Cell Signaling Technology Cat# 2947S, RRID:AB_823586), cyclin D1 (Cell Signaling Technology Cat# 2922, RRID:AB_2228523), p-MKK3 (Ser189)/MKK6 (Ser207) (Cell Signaling Technology Cat# 9236S, RRID:AB_491009), MKK3 (Cell Signaling Technology Cat# 8535S, RRID:AB_1122023), MKK6 (Cell Signaling Technology Cat# 8550S, RRID:AB_1122022), p-Stat3 (Tyr705) (Cell Signaling Technology Cat# 9145, RRID:AB_2491009), Stat3 (Cell Signaling Technology Cat# 9139, RRID:AB_331757), p-YAP (Ser109) (Cell Signaling Technology Cat# 46931), p-YAP (Ser127) (Cell Signaling Technology Cat# 13008, RRID:AB_2650553), YAP (Cell Signaling Technology Cat# 14074, RRID:AB_2650491) and GAPDH (Cell Signaling Technology Cat# 2118, RRID:AB_561053) were purchased from Cell Signaling Technology (Danvers, MA).
2.2. Tissue Specimens
A total of 25 primary lung adenocarcinoma tissues and matched non-tumorous adjacent specimens were collected from 25 patients who underwent surgical resection at the Henan Cancer Hospital (Henan, China). The histomorphology and molecular characteristics of all the samples were analyzed and tested by the Department of Pathology at Henan Cancer Hospital. Written informed consent from each patient and institutional review board approval were obtained for the current study.
2.3. Immunohistochemistry (IHC) Staining
Tissue specimens were fixed in 10% (v/v) formaldehyde in phosphate-buffered saline, embedded in paraffin and cut into 5 μm sections. The sections were deparaffinized in xylene solution and rehydrated using gradient ethanol concentrations. Antigen retrieval was performed using sodium citrate and the slides were then incubated with H2O2 to block endogenous peroxidases. Thereafter, primary antibodies: Ki-67 (1:100), phosphorylated (p)-p38 (1:75), and cyclin D1 (1:75) were incubated at 4 °C overnight and the signals were visualized by the indirect avidin biotin-enhanced horseradish peroxidase method according to the manufacturer's instructions (Vector Laboratories, Burlingame, CA). After developing, all sections were observed by microscope (400 ×) and quantitative analysis was performed using the Image-Pro Premier software (v.9.0) program.
2.4. Cell Culture
HCC827 (ATCC Cat# CRL-2868, RRID:CVCL_2063) and H1975 (ATCC Cat# CRL-5908, RRID:CVCL_5908) human lung adenocarcinoma cell lines and the HEK293T (ATCC Cat# CRL-3216, RRID:CVCL_0063) human embryonic kidney cell line were purchased from American Type Culture Collection (ATCC; Manassas, VA). HCC827GR (RRID:CVCL_V620) cells were kindly provided by Professor Pasi A. Jane from Dana-Farber Cancer Institute (Boston, MA). All cells were cytogenetically tested and authenticated before freezing. All cell culture conditions were performed following ATCC's instructions. All lung adenocarcinoma cells were cultured in RPMI-1640, whereas HEK293T cells were cultured in DMEM, supplemented with 10% (v/v) FBS, 2 mM glutamine, 100 units/mL penicillin, and 100 mg/mL streptomycin. Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. Each vial of frozen cells was thawed and maintained in culture for 10 to 20 passages.
2.5. Flow Cytometry for Cell Cycle Analysis
Cells were plated at a density of 3 × 105 cells/dish in 60-mm dishes overnight. The cells were then treated with vehicle, gefitinib, or a combination of gefitinib and a p38 MAPK inhibitor (either SB203580 or losmapimod) for another 24 h. Cells were trypsinized, washed twice with cold PBS and then fixed with 70% ethanol overnight at -20 °C. The cells were stained with propidium Iodide and analyzed with the FACSCalibur flow cytometer (BD Bioscience, San Jose, CA). The data were then analyzed by CellQuest and Modfit LT V4.0 software (Verity Software House, Topsham, ME).
2.6. Western Blot Analysis
Cells were washed with ice-cold PBS twice and disrupted with lysis buffer (20 mM Tris-HCl, pH 7.5, 2 mM EDTA, 100 mM NaCl, 5 mM MgCl2, 1% (v/v) Triton X-100, 5 mM NaF, 10% (v/v) glycerol, 0.5 (v/v) 2-mercaptoethanol, 0.1 mM Na3VO4 and protease inhibitors). Cell lysates were centrifuged at 12000 × g for 5 min at 4 °C and supernatant fractions were collected. Protein extracts (30 to 50 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) in 20 mM Tris-HCl (pH 8.0), containing 150 mM glycine and 20% (v/v) methanol. Membranes were blocked with 5% non-fat milk in TBS containing 0.05% Tween-20 (TBS-T) and incubated with antibodies against p-p38, p38, p38α, p21, cyclin D1 and GAPDH at 4 °C overnight. Blots were washed 3 times in TBS-T buffer, followed by incubation with an appropriate HRP-conjugated secondary antibody for 1 h at room temperature for hybridization. The blots were visualized using the enhanced chemiluminescence (ECL) reagent (Millipore, Billerica, MA).
2.7. Cell Viability Assay
Cytotoxicity of losmapimod and/or gefitinib was evaluated by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays. Briefly, cells (3 × 103 cells/well) were plated in 96-well plates overnight for attachment. The cells were then treated with vehicle, gefitinib, a p38 MAPK inhibitor (either SB203580 or losmapimod), or a combination of gefitinib and p38 MAPK inhibitor for 72 h and MTT (0.3 mg/mL) was added to the media for 1 h at 37 °C. The reaction was terminated by the addition of 100 μL DMSO. The optical density of the MTT formazan formation was read at 490 nm on a microplate reader. Absorbance values were normalized as a percentage of untreated cells (set at 100%).
2.8. Anchorage-Independent Cell Growth Assay
Cells (8 × 103) were suspended in 1 mL RPMI-1640/10% FBS/0.33% agar with vehicle, gefitinib, a p38 MAPK inhibitor (either SB203580 or losmapimod), or a combination of gefitinib and p38 MAPK inhibitor and plated on 3 mL solidified RPMI-1640/10% FBS/0.5% agar with the same concentration of vehicle, SB203580, losmapimod and/or gefitinib in each well of 6-well plates in triplicates and cultured for 3 weeks. Images from 5 independent fields of each well were captured by a microscope using Images-Pro Plus software. Colony numbers > 200 pixels were quantified by Image J software.
2.9. Generation of MAPK14 shRNA Stable Cell Lines
The pLKO.1 (lentiviral backbone) was a gift from Bob Weinberg (Stewart et al., 2003). The psPAX2 (a packaging vector) and pMD2.G (an envelope vector) were gifts from Didier Trono (Addgene plasmid #12259 and #12260). The shRNA sequence of MAPK14 (5′-CTCAGAGTCTGCAAGAAACTA-3′) was cloned into the pLKO.1 backbone at the AgeI and EcoRI restriction sites. The expressing vectors were confirmed by direct DNA sequencing (Genewiz Inc., Beijing, China). The lentiviral vector containing MAPK14-shRNA, packaging vector (psPAX2) and envelope vector (pMD2.G) were triple-transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) and virus-containing supernatant fractions were harvested at 24 and 48 h. The pooled supernatant fractions were then filtered with a 0.45 μm PVDF filter and frozen at − 80 °C for later use. The p.LKO.1 empty backbone served as a control. Gefitinib-resistant cells, both HCC827GR and H1975, were infected with lentivirus and polybrene (8 μg/mL; Millipore, Billerica, MA) for 16 h and, subsequently, 2.5 mg/mL puromycin was used to select the stably-transduced cells.
2.10. The in vivo Gefitinib-Resistant NSCLC PDX Model
This study was performed according to guidelines approved by the Zhengzhou University Institutional Animal Care and Use Committee. Previously, an in vivo gefitinib-resistant NSCLC PDX model was generated in-house (Zhang et al., 2015). The gefitinib-resistant tumor fragments were passaged to 40 mice for an in vivo study. Mice were divided into four groups (n = 10 mice per group). The four groups were: 1) vehicle control; 2) 50 mg/kg gefitinib; 3) 12 mg/kg losmapimod; and 4) 50 mg/kg gefitinib and 12 mg/kg losmapimod (Willette et al., 2009). Once the tumor volumes reached approximately 25 mm3, mice were treated by oral gavage with vehicle control (dimethyl sulfoxide 5%, normal saline 50% and PEG400 50%), gefitinib and/or losmapimod. Body weights and tumor measurements were performed twice a week and tumor volume was calculated based on the formula: length × width2 × 0.5. At the end of the experiment, mice were sacrificed prior to removal of the tumors for further analysis.
2.11. Statistical Analysis
Each experiment was performed 3 times independently. All quantitative data are expressed as mean values ± standard error of the mean (SEM) and significance between groups was determined by one-way ANOVA unless otherwise indicated. (*p-value < 0.05, **p-value < 0.01 and ***p-value < 0.001).
3. Results
3.1. Phosphorylation of p38 MAPK is Elevated in Gefitinib-Resistant NSCLC Tissues
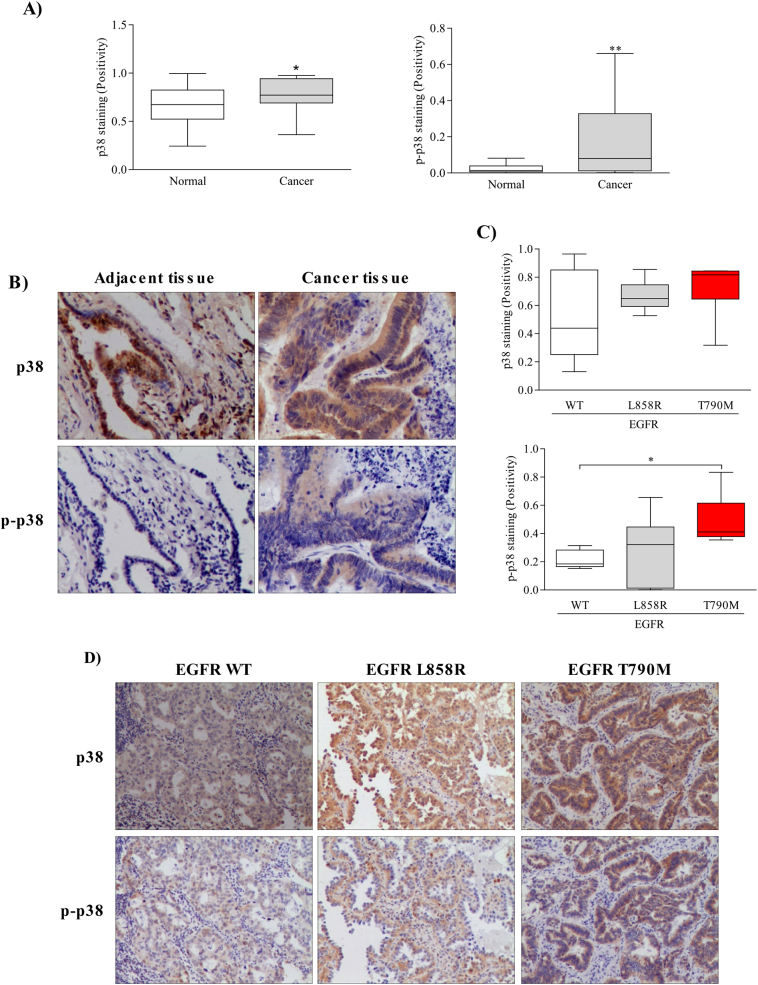
The p38 MAPK was reported to be a stress-activated kinase and is crucial in cell cycle regulation. Therefore, p38 MAPK is considered to be a potential candidate in tetraploidization. We first examined p38 MAPK and phosphorylated (p)-p38 MAPK expression in 25 paired NSCLC specimens. Both p38 and p-p38 MAPK expression were elevated in NSCLC tissues compared with non-tumor adjacent tissues (Fig. 1A, B). Next, we analyzed 18 NSCLC tissues harboring a different EGFR mutation status, including wild type, L858R and T790 M point mutation. Results revealed no significant difference in p38 MAPK expression among all specimens (Fig. 1C). In addition, EGFR wild type and EGFR L858R tissues showed comparable p-p38 expression levels. However, a high p-p38 expression level was observed in tissues containing the gefitinib-resistant mutation, T790 M (Fig. 1C, D). These results suggest that p38 MAPK is overexpressed in NSCLC tissues and most importantly, p38 activity is elevated in gefitinib-resistant tissues.
Fig. 1.
High expression of phosphorylated p38 MAPK in gefitinib-resistant NSCLC tissues. (A) p38 MAPK and p-p38 MAPK expression in 25 matched NSCLC and non-tumorous adjacent clinical samples was assessed by immunohistochemistry. The integrated optical density (IOD) was evaluated using the Image-Pro premier software offline (v9.0) program. (B) Representative images of p38 MAPK and p-p38 expressions in clinical tissues described in A. (C) p38 and p-p38 MAPK expression in 18 NSCLC samples with a different EGFR mutation status including wild type, L858R, and T790 M point mutation was accessed by immunohistochemistry. (D) Representative images from immunohistochemistry described in C. Statistical analysis was performed using a two-tailed paired Student's t-test and significant differences are defined as *p-value < 0.05 and **p-value < 0.01.
3.2. Gefitinib Induces Tetraploidization in Gefitinib-Resistant Lung Cancer Cells and is Associated with YAP-Mediated p38 MAPK Activation
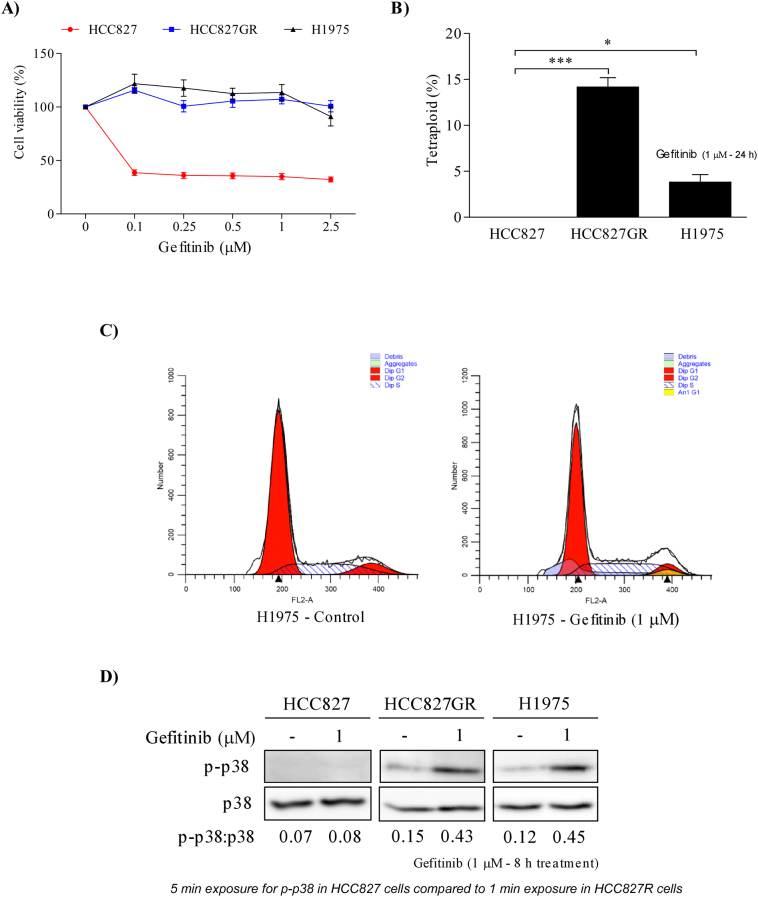
To study the effects of gefitinib on lung adenocarcinoma cells, we analyzed 3 different NSCLC cell lines: HCC827 (gefitinib-sensitive), and HCC827GR and H1975 (gefitinib-resistant). Cells were plated in 96-well plates with various concentrations of gefitinib (0.1–2.5 μM) and incubated for 72 h. As expected, gefitinib inhibited proliferation of HCC827 cells, but not proliferation of HCC827GR and H1975 cells (Fig. 2A). We then determined whether tetraploid formation could be one of the possible mechanisms causing gefitinib resistance. First, we treated all 3 cell lines with gefitinib for 24 h and then subjected cells to cell cycle analysis using propidium iodide staining. Results revealed that all 3 cell lines did not contain any tetraploidy (data not shown). Interestingly, tetraploidy was significantly induced after 24 h of gefitinib treatment in gefitinib-resistant cells, but not in gefitinib-sensitive cells (Fig. 2B and C). In order to determine whether p38 MAPK activation is necessary for tetraploidization, we examined phosphorylation of p38 MAPK after gefitinib treatment. We found that the level of p-p38 in gefitinib-resistant cells was dramatically up-regulated after 10 h of gefitinib treatment, whereas no change was observed in gefitinib-sensitive cells (Fig. 2D). To further determine the signaling molecules of gefitinib-induced p38 MAPK activation, we examined the phosphorylation of YAP, MKK3/6 and STAT3. As shown in Supplementary Fig. 1, phosphorylated YAP and MKK3/6 were observed upon gefitinib treatment in 2 h and 6 h, respectively. At later time points (> 8 h), phosphorylated p38 and STAT3 were also up-regulated. Based on these results, we assumed that tetraploidization induced by YAP-MKK3/6-p38 MAPK-STAT3 signaling might be involved in the survival signaling through which these cells resist gefitinib treatment.
Fig. 2.
Gefitinib induces tetraploidization in gefitinib-resistant NSCLC cells by activating p38 MAPK. (A) Viability of gefitinib-sensitive (HCC827) and -resistant NSCLC cells (HCC827GR and H1975) was determined by MTT assay with different concentrations of gefitinib treatment for 72 h. (B) Tetraploid subpopulations in gefitinib-sensitive and -resistant cells treated with gefitinib (1 μM) for 24 h and cell cycle were assessed by PI staining. The percentage of tetraploid subpopulations were analyzed using the ModFit LT V4.0 software program. (C) FACS analysis of propidium iodide-stained H1975 cells, treated with either vehicle or gefitinib for 24 h. Tetraploid subpopulation was analyzed by ModFit LT V4.0 software. (D) Western blot analysis of p-p38 expression in untreated gefitinib-sensitive and -resistant cells or treated with gefitinib (1 μM) for 10 h. Equal loading is confirmed by total p38 MAPK.
Supplementary Fig. 1.
Gefitinib induces activation of p38 MAPK signaling in gefitinib-resistant cells. Western blot analysis of p-YAP, p-MKK3/6, p-p38 and p-STAT3 expressions in untreated gefitinib-resistant cells (left, HCC827GR and right, H1975) or cells treated for different times (between 1 and 10 h) with gefitinib (1 μM). Total YAP, MKK3, MKK6, p38 MAPK and STAT3 proteins were used as loading controls.
3.3. Inhibition of p38 MAPK Prevents Gefitinib-Induced Tetraploidization
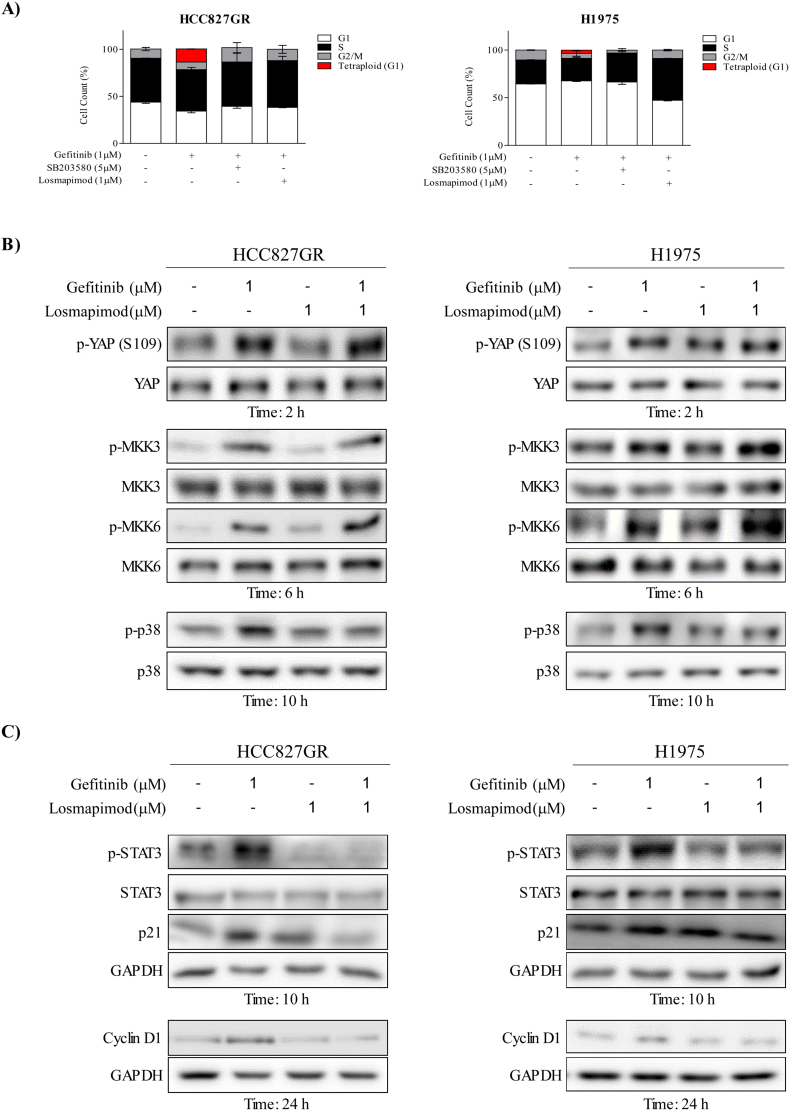
We observed that gefitinib treatment could induce tetraploidization through p38 MAPK signaling in gefitinib-resistant cells. Thus, to further investigate whether inhibition of p38 MAPK could prevent tetraploidization, both HCC827GR and H1975 cell lines were treated with gefitinib and/or p38 MAPK inhibitors (either losmapimod or SB203580) for 24 h. Results showed that each p38 MAPK inhibitors could eliminate gefitinib-induced tetraploidy (Fig. 3A) in both resistant cell lines. In addition, Western blot analyses showed that losmapimod could inhibit p38 MAPK phosphorylation induced by gefitinib treatment. However, gefitinib-induced MKK3/6 phosphorylation was further up-regulated by losmapimod and there was no changes of YAP phosphorylation (Fig. 3B). To further examine the detailed mechanism of gefitinib-induced tetraploidization, we determined p-STAT3, p21 and cyclin D1 expressions after gefitinib and/or losmapimod treatment in gefitinib-resistant cells. Consistent with the results of tetraploidization and p38 MAPK inhibition, Western blotting revealed that phosphorylation of STAT3 and expressions of p21 and cyclin D1 were up-regulated with gefitinib treatment in both resistant cell lines, and that the up-regulations could be inhibited by losmapimod (Fig. 3C). All these findings suggest that gefitinib-induced tetraploidization requires p38 MAPK signaling and could be targeted by p38 MAPK inhibitors.
Fig. 3.
Losmapimod inhibits gefitinib-induced tetraploidization in gefitinib-resistant NSCLC cells. (A) Cell cycle analysis in untreated gefitinib-resistant NSCLC cells (left, HCC827GR and right, H1975) and in cells treated for 24 h with gefitinib or a p38 MAPK inhibitor (SB203580 or losmapimod) either alone or in combination. Tetraploid subpopulations were analyzed by the ModFit LT V4.0 software program. (B) Western blot analysis of p-YAP, p-MKK3/6 and p-p38 expressions in untreated gefitinib-resistant cells (left, HCC827GR and right, H1975) or cells treated for 2, 6 or 10 h with gefitinib (1 μM) or losmapimod (1 μM) either alone or in combination. Total YAP, MKK3/6 and p38 MAPK proteins were used as loading controls. (C) Western blot analysis of p-STAT3, p21 and cyclin D1 expressions in untreated gefitinib-resistant cells (left, HCC827GR and right, H1975) or cells treated for 10 or 24 h with gefitinib (1 μM) or losmapimod (1 μM) either alone or in combination. Total STAT3 and GAPDH proteins were used as loading controls.
3.4. Losmapimod Successfully Overcomes Gefitinib-Resistance in Lung Cancer Cells
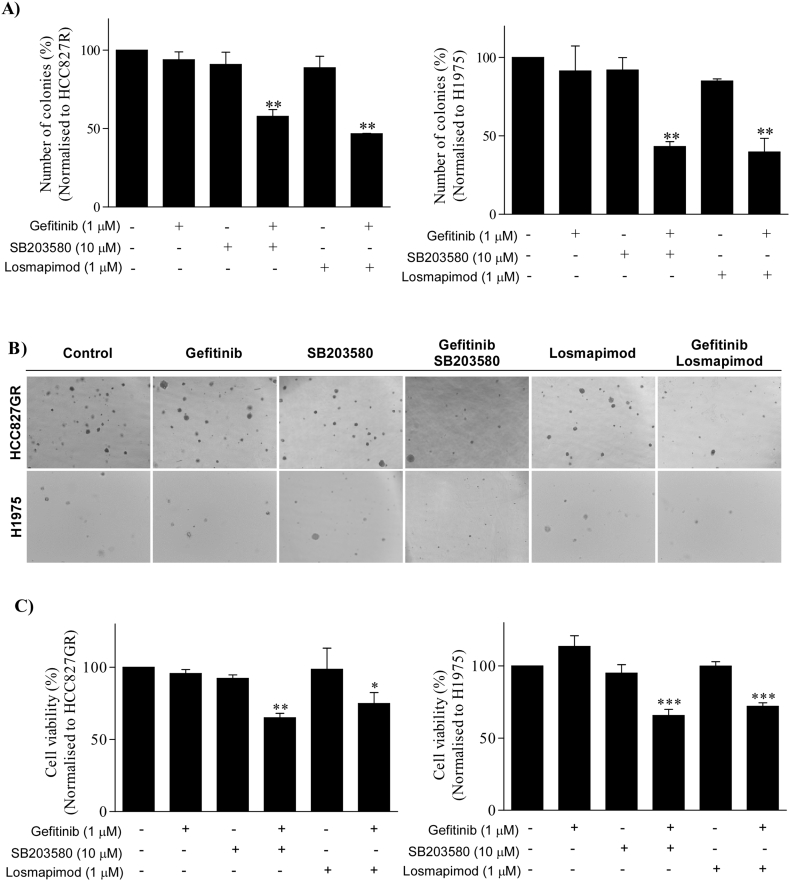
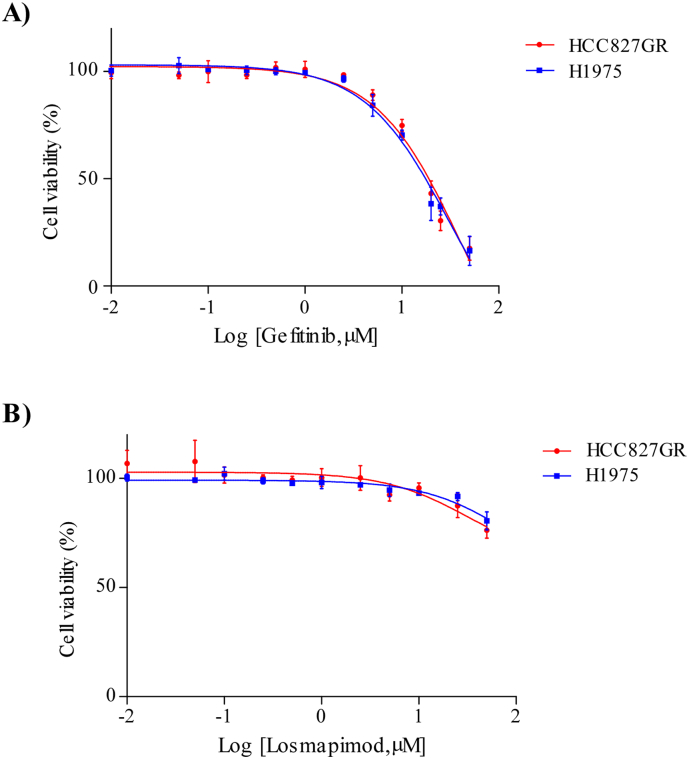
Based on the previous data, we determined whether losmapimod could overcome gefitinib resistance in gefitinib-resistant human lung cancer cells. First, we treated both HCC827GR and H1975 cells with either gefitinib or losmapimod alone and then in combination. Our results revealed that the combined treatment with gefitinib and losmapimod significantly reduced both anchorage-independent cell growth (Fig. 4A, B) and proliferation (Fig. 4C) of gefitinib-resistant cells. However, neither gefitinib nor losmapimod alone could inhibit either type of cell proliferation. In order to examine the combination effect of gefitinib and losmapimod, the IC50 value of gefitinib or losmapimod alone treatment in both cells was first determined (Supplementary Fig. 2 and Table 1) and the combination index was then calculated by using the CompuSyn program. Results showed that co-treatment of gefitinib and losmapimod synergistically inhibited cell proliferation with a combination index of 0.14 for the HCC827GR and 0.22 for the H1975 cell line (Supplementary Table 2). Overall, our data indicates that losmapimod could be a potential drug to overcome gefitinib resistance in NSCLC.
Fig. 4.
Losmapimod overcomes gefitinib resistance in NSCLC cells. (A) Anchorage-independent growth assay in untreated gefitinib-resistant NSCLC cells (left, HCC827GR and right, H1975) and in cells treated with gefitinib, a p38 MAPK inhibitor (SB203580 or losmapimod) or a combination of gefitinib and inhibitor. Images were taken at 21 days and colony numbers with > 200 pixels were quantified using the Image J software program. (B) Representative images from the anchorage-independent growth assay. (C) Cell viability assay was conducted using untreated gefitinib-resistant NSCLC cells (left, HCC827GR and right, H1975) and cells treated for 72 h with gefitinib (1 μM), a p38 MAPK inhibitor (SB203580 or losmapimod) or a combination of gefitinib and inhibitor.
Supplementary Fig. 2.
Dose-response curve of gefitinib and losmapimod in gefitinib-resistant cells. Viability of gefitinib-resistant cells (red, HCC827GR and blue, H1975) was determined by MTT assay with different concentrations of gefitinib or losmapimod treatment for 72 h.
3.5. p38α Knockdown Prevents Tetraploidization and Effectively Inhibits Gefitinib-Resistant Lung Cancer Cells Growth
To test our hypothesis and examine whether p38 MAPK knockdown plays any roles in suppressing cell growth in NSCLC, both HCC827GR and H1975 cells were infected with lentivirus expressing the pLKO.1-mock or pLKO.1-shMAPK14 plasmid. The expression of total p38 MAPK and the p38 MAPKα isoform was then analyzed by Western blotting (Fig. 5A). Next, to investigate whether p38α knockdown could affect gefitinib-induced tetraploidization, p38α knockdown cells were incubated with gefitinib for 24 h and subjected to cell cycle analysis. Our results revealed that p38α knockdown could prevent gefitinib-induced tetraploidization in both HCC827GR and H1975 cells (Fig. 5B). Moreover, we found that p38α MAPK knockdown could significantly inhibit both proliferation (Fig. 5C) and colony formation (Fig. 5D, E) of gefitinib-resistant cells. Proliferation was further reduced when these cells were treated with gefitinib. However, no further reduction in colony formation was observed. Nonetheless, these results suggested that p38α MAPK is essential to gefitinib-induced tetraploidization in gefitinib-resistant cells.
Fig. 5.
p38α knockdown prevents tetraploidization and inhibits gefitinib-resistant NSCLC cell growth. (A) Western blot analysis of p38α isoform expression in HCC827GR and H1975 cells transduced with p38α shRNA or empty vector (as a control). (B) Cell cycle analysis of untreated p38α shRNA-transduced cells (left, HCC827GR and right, H1975) or cells treated for 24 h with gefitinib (1 μM). Tetraploid subpopulations were analyzed by the ModFit LT V4.0 software program. (C) A cell viability assay was conducted in untreated p38α shRNA-transduced cells (left, HCC827GR and right, H1975) or cells treated for 72 h with gefitinib (1 μM). Results were normalized to corresponding parental cells. (D) An anchorage-independent growth assay was conducted in untreated p38α shRNA-transduced cells (left, HCC827GR and right, H1975) and cells treated with gefitinib (1 μM), a p38 MAPK inhibitor (SB203580 or losmapimod) or in combination. Images were taken at 21 days and colony numbers with > 200 pixels were quantified using the Image J software program. (E) Representative images from the anchorage-independent growth assay.
3.6. Losmapimod Suppresses Tumor Growth in a Gefitinib-Resistant NSCLC PDX Model
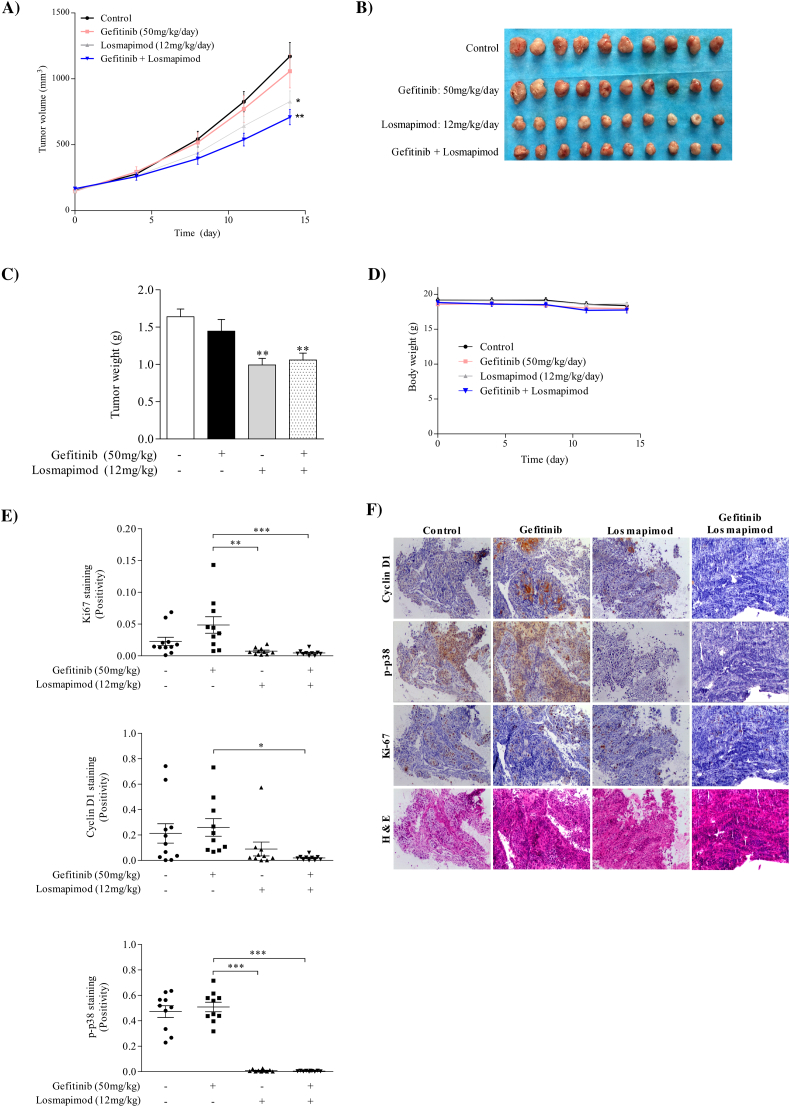
To further investigate the chemotherapeutic effect of losmapimod and its clinical relevance, we conducted in vivo experiments using an in-house generated gefitinib-resistant PDX model. Results showed that losmapimod alone or in combination with gefitinib markedly reduced gefitinib-resistant NSCLC PDX tumor volume and weight, whereas gefitinib alone had no effect (Fig. 6A-C). In addition, no changes in mouse body weight were observed, suggesting that toxicity was not associated with the different treatments (Fig. 6D). In addition, immunohistochemical analysis of harvested PDX tumors were conducted to evaluate the expression level of cyclin D1, p-p38 and Ki-67 (Fig. 6E, F). Our results showed that cyclin D1, p-p38 and Ki-67 were significantly reduced in both losmapimod-treated and groups treated with a combination of losmapimod and gefitinib compared with the vehicle- or gefitinib-treated group. These data provide strong evidence that losmapimod could be used as a promising candidate to overcome gefitinib-resistant lung cancers clinically.
Fig. 6.
Losmapimod inhibits tumor growth in a gefitinib-resistant NSCLC patient derived xenograft (PDX) mouse model. (A) A tumor growth kinetic of untreated gefitinib-resistant PDX or tumors treated with gefitinib and/or losmapimod for 14 days. Tumor size was measured twice a week and calculated based on the formula: length × width2 × 0.5. (B) Tumor size and (C) tumor weight. (D) Body weight of mice during the experiment. (E) Cyclin D1, p-p38 MAPK and Ki-67 expressions and HE staining in xenograft tissues were assessed by immunohistochemistry. The integrated optical density (IOD) was evaluated using the Image-Pro premier software offline (v9.0) program. (F) Representative photographs of immunohistochemistry analysis for each antibody and each group are shown.
4. Discussion
In this study, we have provided the first evidence that p38 MAPK activation could be involved in gefitinib resistance in NSCLC. We show that both p38 MAPK and phosphorylated p38 MAPK are overexpressed in NSCLC tissues compared with corresponding non-tumor tissues. Moreover, we showed that clinical samples expressing the EGFR T790 M mutation, a gefitinib resistant mutation, elevates p38 MAPK activity compared with samples expressing either EGFR wild type or EGFR L858R, a gefitinib-sensitive mutation.
EGFR is one of the most frequently mutated genes in NSCLC and tyrosine kinase inhibitors against mutated EGFR have shown dramatic clinical efficacy. Unfortunately, most of the patients eventually develop acquired resistance through different mechanisms. Here, we have determined that p38 MAPK phosphorylation is increased with gefitinib treatment in two gefitinib-resistant NSCLC cell types (HCC827GR and H1975), but not in gefitinib-sensitive cells. These two resistant cell lines represent the two most common TKI-acquired resistance mechanisms. HCC827GR cells have been reported to acquire resistance through MET amplification whereas H1975 exhibits a secondary T790 M EGFR mutation (Engelman et al., 2007, Kobayashi et al., 2005b). We also showed that treatment with a combination of gefitinib and losmapimod synergistically inhibited both cell proliferation and anchorage-independent growth in gefitinib-resistant cells. In addition, pharmacological inhibition of p38 MAPK using losmapimod also overcame gefitinib resistance in a patient derived xenograft model. This result suggests that p38 MAPK activation may act as a general gefitinib resistant signaling mechanism and is necessary for the survival of gefitinib-resistant cells.
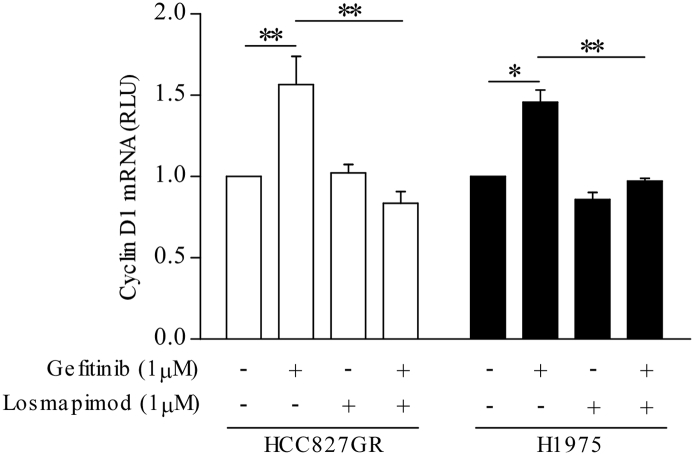
The involvement of p38 MAPK in gefitinib resistance may be explained by its role in cell cycle checkpoints regulation (Bulavin et al., 2001). Recent research suggests that tumor cells with elevated genomic content, such as tetraploidy and aneuploidy, are associated with increased drug resistance and poor prognosis (Lee et al., 2011). Interestingly, our data demonstrated that a tetraploid subpopulation is induced after gefitinib treatment in both gefitinib-resistant cell lines. Different signaling pathways have been reported to be involved in tetraploidization. It is shown that YAP promotes polyploidy in hepatocytes and that YAP is also necessary for p38 activation (Wang et al., 2016, Zhang et al., 2017). Our results demonstrated that gefitinib could activate YAP, followed by MKK3/6-p38 MAPK and then STAT3 in both gefitinib-resistant cell lines. This finding is consistent with the findings of Chaib et al. who showed that co-activation of YAP and STAT3 signaling is an immediate response in lung cancer cells upon TKI treatment (Chaib et al., 2017). However, the activation profiles, particularly p-YAP (S127) and p-p38 MAPK, and the amounts of gefitinib-induced tetraploid subpopulation between these two cells appear to be slightly different suggesting that these two cells may consist of different p38 signaling profile and efficiency. Another most studied mechanism is the p53 pathway (Taylor and Stark, 2001). Prolonged p53-p21 signaling in G2-arrested cells may drive these cells into a tetraploid-G1 state and that G1-state tetraploidy is characterized by increased expression of the G1 marker, cyclin D (Nelsen et al., 2005, Shen and Maki, 2010, Shen et al., 2008). The p38 MAPK can activate and stabilize p53 by phosphorylation of the Ser15 and Ser392 residues (Huang et al., 1999, She et al., 2000). Therefore, the p53 pathway might possibly be involved in the p38 MAPK-mediated tetraploid formation. Here, we also showed that both p21 and cyclin D1 expression and cyclin D1 mRNA level (Supplementary Fig. 3) were up-regulated with gefitinib treatment in both gefitinib-resistant cells. We then treated both cells with either SB203580 or losmapimod to further explore the role of p38 MAPK activation in tetraploidization following gefitinib treatment. The addition of p38 MAPK inhibitors could inhibit phosphorylation of p38 MAPK and STAT3, but not YAP and MKK3/6, upon gefitinib treatment. Indeed, the phosphorylation of MKK3/6 were augmented suggesting a possible negative feedback mechanism. Moreover, both inhibitors could eliminate tetraploidization and repress expression of p21 and cyclin D1 expression induced by gefitinib treatment.
Supplementary Fig. 3.
Losmapimod inhibits gefitinib-induced Cyclin D1 mRNA expression in gefitinib-resistant NSCLC cells. Total RNA was extracted in untreated gefitinib-resistant cells (left, HCC827GR and right, H1975) or cells treated for 24 h with gefitinib (1 μM) or losmapimod (1 μM) either alone or in combination. Relative Cyclin D1 mRNA expression was evaluated by comparative RT-PCR. The endogenous control was GAPDH.
Recently, Vitale et al. showed that p38α is essential to generate tetraploidy in p53−/− cells (Vitale et al., 2010). Losmapimod and SB203580 have been reported to inhibit both the p38 MAPK α and β isoforms (Willette et al., 2009). To further validate whether the gefitinib-induced tetraploidy is isoform specific, lentivirus-based shRNA delivery was used to knockdown the p38α isoform in gefitinib-resistant cells. We showed that p38α knockdown could prevent tetraploidy formation and inhibit growth of gefitinib-resistant cells. Overall, we demonstrated that both HCC827GR and H1975 cells seems to survive the growth inhibitory effect of gefitinib through p38 MAPK-mediated tetraploidization and, most importantly, this process could be targeted by p38 MAPK inhibitors. Finally, to further translate our findings into clinical relevance, we utilized a gefitinib-resistant PDX model to predict the effect of losmapimod in overcoming gefitinib resistance. Our results clearly showed that a combination of losmapimod and gefitinib could effectively reduce gefitinib-resistant patient-derived tumor size. Immunohistochemistry analyses also showed that phosphorylation of p38 MAPK and cyclin D1 level were significantly inhibited in the group of co-administration of gefitinib and losmapimod. These data strongly indicate that losmapimod could overcome gefitinib resistance in NSCLC treatment.
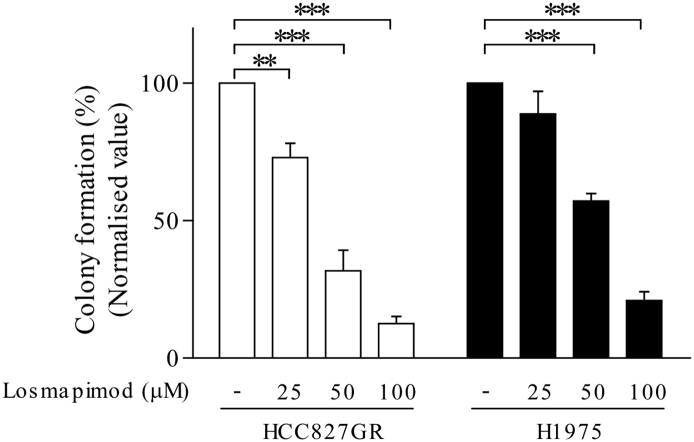
There are few limitations in our study, for example the difference of losmapimod efficacy between in vitro and in vivo studies. Losmapimod alone could significantly suppress PDX tumor growth while it only showed efficacy at high concentration (> 50 μM) in an in vitro assay (Supplementary Fig. 4). Based on immunohistochemistry analysis, it is revealed that there was a high expression of phosphorylated p38 in vehicle-treated PDX tumors. This suggests that losmapimod might also have a potential as the sole agent to target NSCLC depending on the p-p38 MAPK profile. Therefore, a larger pool of PDX models could be used to precisely determine the potential use of losmapimod either alone or in combination with gefitinib in NSCLC patients, particularly gefitinib-resistant patients. Although we have shown here that p-YAP, p-MKK3/6, p-p38 MAPK and p-STAT3 and, ultimately, cyclin D1 transcript are up-regulated upon gefitinib treatment, specific inhibitors or siRNA to each signaling molecules should be used to validate this novel signaling pathway. In addition, since both STAT3 and p21 could regulate cyclin D1 transcription, the relationship between YAP-MKK3/6-p38-STAT3 and p53-p21 signaling has yet to be determined in future studies (Chen et al., 1995, Leslie et al., 2006).
Supplementary Fig. 4.
Losmapimod inhibits anchorage-independent growth in gefitinib-resistant cells. Anchorage-independent growth assay in untreated gefitinib-resistant NSCLC cells (left, HCC827GR and right, H1975) and in cells treated with 25, 50 or 100 μM losmapimod. Images were taken at 21 days and colony numbers with greater than 200 pixels were quantified using the Image J software program.
5. Conclusions
In conclusion, our study shows for the first time that phosphorylation of p38 MAPK is up-regulated in clinical NSCLC tissues, particularly in the presence of the EGFR T790 M mutation. In addition, we have demonstrated a role of p38 MAPK signaling in tetraploidization of gefitinib-resistant NSCLC. Using losmapimod, we successfully abolished gefitinib-induced tetraploidization and overcame gefitinib resistance in NSCLC both in vitro and in vivo. Overall, these data indicate that losmapimod could potentially be used as adjuvant therapy clinically to overcome gefitinib resistance, although further studies using larger cohorts of patients will be necessary.
The following are the supplementary data related to this article.
IC50 value of gefitinib or losmapimod in gefitinib-resistant cells.
Combination index of gefitinib (1 μM) and losmapimod (1 μM) in gefitinib-resistant cells.
Funding
This work was supported by Henan Provincial Government, the National Natural Sciences Foundation of China81650110531, 81372269, 81572812 and US National Institutes of Health grants CA-187027, CA-196639, and CA-166011.
Conflicts of Interest
The authors disclose no conflicts of interest.
Author Contributions
Zigang Dong and Kangdong Liu designed and supervised the experiments; Yiu To Yeung and Ann M. Bode prepared the manuscript; Yiu To Yeung, Shuying Yin, Bingbing Lu, Suyu Fan and Ran Yang performed experiments; Ruihua Bai and Chengjuan Zhang recruited volunteers and collected clinical samples.
Acknowledgements
We thank Ms. Nicki Brickman and Dr. Tia Rai at The Hormel Institute, University of Minnesota for assistance with manuscript submission.
Contributor Information
Kangdong Liu, Email: kdliu@zzu.edu.cn.
Zigang Dong, Email: zgdong@hi.umn.edu.
References
- Bakhoum S.F., Compton D.A. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest. 2012;122(4):1138–1143. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bria E. Outcome of advanced NSCLC patients harboring sensitizing EGFR mutations randomized to EGFR tyrosine kinase inhibitors or chemotherapy as first-line treatment: a meta-analysis. Ann. Oncol. 2011;22(10):2277–2285. doi: 10.1093/annonc/mdq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin D.V. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411(6833):102–107. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- Chaib I. Co-activation of STAT3 and YES-Associated Protein 1 (YAP1) Pathway in EGFR-Mutant NSCLC. J. Natl. Cancer Inst. 2017;109(9) doi: 10.1093/jnci/djx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Bargonetti J., Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55(19):4257–4263. [PubMed] [Google Scholar]
- Doza Y.N. Activation of the MAP kinase homologue RK requires the phosphorylation of Thr-180 and Tyr-182 and both residues are phosphorylated in chemically stressed KB cells. FEBS Lett. 1995;364(2):223–228. doi: 10.1016/0014-5793(95)00346-b. [DOI] [PubMed] [Google Scholar]
- Engelman J.A. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Han J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265(5173):808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Huang C. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J. Biol. Chem. 1999;274(18):12229–12235. doi: 10.1074/jbc.274.18.12229. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p38delta. J. Biol. Chem. 1997;272(48):30122–30128. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kobayashi S. An alternative inhibitor overcomes resistance caused by a mutation of the epidermal growth factor receptor. Cancer Res. 2005;65(16):7096–7101. doi: 10.1158/0008-5472.CAN-05-1346. [DOI] [PubMed] [Google Scholar]
- Kumar S., Boehm J., Lee J.C. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discov. 2003;2(9):717–726. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- Lafarga V. p38 mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 2009;29(16):4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71(5):1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie K. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66(5):2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- Lilly M.A., Duronio R.J. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24(17):2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- Lynch T.J. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Miller K.D. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- Nelsen C.J. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. 2005;280(1):768–776. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- O'Donoghue M.L. Effect of losmapimod on cardiovascular outcomes in patients hospitalized with acute myocardial infarction: a randomized clinical trial. JAMA. 2016;315(15):1591–1599. doi: 10.1001/jama.2016.3609. [DOI] [PubMed] [Google Scholar]
- Schaeffer H.J., Weber M.J. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 1999;19(4):2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist L.V. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q.B., Chen N., Dong Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J. Biol. Chem. 2000;275(27):20444–20449. doi: 10.1074/jbc.M001020200. [DOI] [PubMed] [Google Scholar]
- Shen H., Maki C.G. Persistent p21 expression after Nutlin-3a removal is associated with senescence-like arrest in 4N cells. J. Biol. Chem. 2010;285(30):23105–23114. doi: 10.1074/jbc.M110.124990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Moran D.M., Maki C.G. Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Res. 2008;68(20):8260–8268. doi: 10.1158/0008-5472.CAN-08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. Two 4N cell-cycle arrests contribute to cisplatin-resistance. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0059848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Stewart S.A. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W.R., Stark G.R. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Thornton T.M., Rincon M. Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int. J. Biol. Sci. 2009;5(1):44–51. doi: 10.7150/ijbs.5.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphyllopoulos K. In vitro target validation and in vivo efficacy of p38 MAP kinase inhibition in established chronic collagen-induced arthritis model: a pre-clinical study. Clin. Exp. Rheumatol. 2010;28(2):176–185. [PubMed] [Google Scholar]
- Vitale I. Inhibition of Chk1 kills tetraploid tumor cells through a p53-dependent pathway. PLoS One. 2007;2(12) doi: 10.1371/journal.pone.0001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I. Involvement of p38alpha in the mitotic progression of p53(−/−) tetraploid cells. Cell Cycle. 2010;9(14):2823–2829. [PubMed] [Google Scholar]
- Vitale I. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2011;18(9):1403–1413. doi: 10.1038/cdd.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. SIRT1 increases YAP- and MKK3-dependent p38 phosphorylation in mouse liver and human hepatocellular carcinoma. Oncotarget. 2016;7(10):11284–11298. doi: 10.18632/oncotarget.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette R.N. Differential effects of p38 mitogen-activated protein kinase and cyclooxygenase 2 inhibitors in a model of cardiovascular disease. J. Pharmacol. Exp. Ther. 2009;330(3):964–970. doi: 10.1124/jpet.109.154443. [DOI] [PubMed] [Google Scholar]
- Zack T.I. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wang Y., Zhang S. Asymmetric cell division in polyploid giant cancer cells and low eukaryotic cells. Biomed. Res. Int. 2014:432652. doi: 10.1155/2014/432652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. 244-MPT overcomes gefitinib resistance in non-small cell lung cancer cells. Oncotarget. 2015;6(42):44274–44288. doi: 10.18632/oncotarget.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2. Cancer Cell. 2017;31(5) doi: 10.1016/j.ccell.2017.04.004. (669-684.e7) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IC50 value of gefitinib or losmapimod in gefitinib-resistant cells.
Combination index of gefitinib (1 μM) and losmapimod (1 μM) in gefitinib-resistant cells.