Abstract
Immune checkpoints are important targets for immunotherapies. However, knowledge on the epigenetic modification of immune checkpoint genes is sparse. In the present study, we investigated promoter methylation of CTLA4, PD-L1, PD-L2, and PD-1 in diffuse lower-grade gliomas (LGG) harboring isocitrate dehydrogenase (IDH) mutations with regard to mRNA expression levels, clinicopathological parameters, previously established methylation subtypes, immune cell infiltrates, and survival in a cohort of 419 patients with IDH-mutated LGG provided by The Cancer Genome Atlas.
PD-L1, PD-L2, and CTLA-4 mRNA expression levels showed a significant inverse correlation with promoter methylation (PD-L1: p = 0.005; PD-L2: p < 0.001; CTLA-4: p < 0.001). Furthermore, immune checkpoint methylation was significantly associated with age (PD-L2: p = 0.003; PD-1: p = 0.015), molecular alterations, i.e. MGMT methylation (PD-L1: p < 0.001; PD-L2: p < 0.001), ATRX mutations (PD-L2: p < 0.001, PD-1: p = 0.001), and TERT mutations (PD-L1: p = 0.035, PD-L2: p < 0.001, PD-1: p < 0.001, CTLA4: p < 0.001) as well as methylation subgroups and immune cell infiltrates. In multivariate Cox proportional hazard analysis, PD-1 methylation qualified as strong prognostic factor (HR = 0.51 [0.34–0.76], p = 0.001).
Our findings suggest an epigenetic regulation of immune checkpoint genes via DNA methylation in LGG. PD-1 methylation may assist the identification of patients that might benefit from an alternative treatment, particularly in the context of emerging immunotherapies.
Keywords: PD-1, PD-L1, CTLA-4, DNA methylation, Lower-grade glioma, Prognosis
Highlights
-
•
PD-L1, PD-L2, and CTLA4 DNA methylation in lower-grade gliomas is inversely correlated with mRNA expression levels.
-
•
PD-1, PD-L1, PD-L2, and CTLA4 DNA methylation in lower-grade gliomas is associated with age and mutation status.
-
•
PD-1 methylation in lower-grade gliomas is correlated with infiltrating immune cells (B, CD8+ T, and dendritic cells).
-
•
PD-1 methylation is an independent prognostic biomarker for overall survival in lower-grade glioma patients.
Immune checkpoint inhibitors have exhibited dramatic antitumor activity in various malignancies; however, little is known about the epigenetic regulation of immune checkpoint genes in tumors of the central nervous system. In the present study, PD-L1, PD-L2, and CTLA-4 mRNA expression was found to correlate inversely with promoter methylation. Furthermore, PD-1 methylation was prognostic for survival in lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations supporting the pivotal role of DNA methylation in the regulation of immune checkpoint genes in gliomas. Prognostic biomarkers may assist the identification of patients that might benefit from an alternative treatment, particularly in the context of immunotherapies.
1. Introduction
Gliomas are the most common primary brain tumors accounting for approximately 80% of all brain malignancies in the United States (Ostrom et al., 2016). Diffuse lower-grade gliomas (LGG) often present with very variable clinical appearances and survival rates before fatally progressing to glioblastoma multiforme (Cancer Genome Atlas Research Network et al., 2015). Recent developments in genomic profiling have led to a paradigm shift in the classification of gliomas. As a consequence, the 2016 World Health Organization (WHO) classification includes the molecular characterization of primary brain tumors (e.g. isocitrate dehydrogenase (IDH) mutations and codeletions of chromosome arms 1p and 19q (1p/19q co-deletion)) (summarized by Louis et al., 2016). Although the implementation of genetic signatures has led to a better understanding of underlying molecular pathways and more reliable diagnostic criteria, these findings do not fully explain why some LGG patients have far worse courses of disease than others. Recent evidence suggests that DNA methylation profiles might shed light on significantly differing outcomes. Unsupervised cluster analysis of 1122 grade II-III-IV gliomas from The Cancer Genome Atlas (TCGA) identified six methylation groups (LGm1–6) that were in part associated with IDH status and further discovered an epigenetic signature that segregated a subgroup of IDH-mutant diffuse lower-grade gliomas with unfavorable clinical outcome (Ceccarelli et al., 2016). Mutations in the IDH1 and IDH2 genes have previously been identified to lead to a downstream neomorphic enzymatic activity and an accumulation of the onco-metabolite D-2-hydroxyglutarate (D-2HG) in IDH-mutant cells (Dang et al., 2009). As D-2HG inhibits key enzymes involved in histone- and DNA-demethylation, excess D-2HG results in DNA hypermethylation. Gliomas harboring IDH mutations consequently display a CpG island methylator phenotype (G-CIMP), which is characterized by DNA hypermethylation in CpG-rich domains (Turcan et al., 2012) and has been shown to constitute a subset of tumors with a distinct biology and clinical behavior (Noushmehr et al., 2010). These findings emphasize the relevance of epigenetic alterations as an underlying and therapeutically relevant mechanism in glioma.
Gliomas have long been recognized to induce local and systemic immunosuppression, thereby limiting the innate defense against tumor growth (Gousias et al., 2010). Currently emerging immunomodulatory therapies have therefore generated an increasing interest in these novel therapies as potential treatment options for gliomas. Particularly treatments targeting the immune checkpoints programmed cell death 1 receptor (PD-1)/PD-1 ligand 1 (PD-L1) pathway and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) have exhibited dramatic antitumor efficacy in various tumor entities (Chan et al., 2015, Brahmer et al., 2012, Brahmer et al., 2015, Topalian et al., 2012, Hamid et al., 2013, Wolchok et al., 2013, Larkin et al., 2015, Margolin et al., 2012, Berger et al., 2008, Ribas et al., 2016, Garon et al., 2015). Several clinical trials are currently ongoing to determine the potential of PD-1/PD-L1 and CTLA-4 targeted therapies in high-grade gliomas yielding conflicting results (Omuro et al., 2017, Reardon et al., 2016). Furthermore, several studies have been conducted to determine the prognostic value of PD-L1 in gliomas; however, the results so far have been inconsistent (Xue et al., 2017). The regulation of immune checkpoint genes in glioma, particularly on the epigenetic level, seems to be complex and is only poorly understood. Elucidating the regulatory machinery of immune checkpoints might help to improve patient's treatment, particularly in the view of emerging immunotherapeutic strategies. Recently, inverse correlations between immune checkpoint mRNA levels and promoter methylation indicative of an epigenetic regulation as well as significant associations of immune checkpoint methylation levels with survival have been reported for several hematopoietic and solid neoplasms including acute myeloid leukemia (AML), prostate cancer, colorectal adenocarcinomas, and head and neck squamous cell carcinomas (HNSCC) (Franzen et al., 2018, Gevensleben et al., 2016, Goltz et al., 2016a, Goltz et al., 2016b, Goltz et al., 2017a, Goltz et al., 2017b). However, epigenetic association studies regarding tumors of the central nervous system are lacking so far.
In the present study, we investigated DNA promoter methylation of the immune checkpoints genes PD-1 (Human Genome Organisation (HUGO) gene symbol: PDCD1), PD-L1 (CD274), PD-L2 (PDCD1LG2), and CTLA-4 (CTLA4) in patients with LGG harboring IDH mutations with regard to mRNA expression, clinicopathological parameters, previously established methylation subtypes, immune cell infiltrates, and survival.
2. Materials and Methods
2.1. Patients and Clinical Endpoints
The results shown are entirely based on gene methylation data created by the TCGA Research Network (http://cancergenome.nih.gov/). The cohort comprised fresh-frozen tissues from 419 patients with histologically confirmed LGG from several international centres involved in the TCGA project. Clinical, cytological, and mutational data were obtained from the TCGA Research Network. Additional information on methylation subtypes was taken from Ceccarelli et al. (2016). Patients' characteristics are described in detail in Table 1. Overall survival (OS) was defined as time to death or last follow-up. The mean OS was 24.81 months. The TCGA Research Network acquired written informed consent from all participants. All experiments were carried out according to the World Medical Association Declaration of Helsinki.
Table 1.
Association of clinicopathological parameters with PD-L1, PD-L2, PD-1, and CTLA4 promoter methylation in diffuse lower-grade glioma patients (n = 419).
| Variable | All patients | [%] | Mean PD-L1 methylation [%] | p-Value | Mean PD-L2 methylation [%] | p-Value | Mean PD-1 methylation [%] | p-Value | Mean CTLA4 methylation [%] | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| All patients | 419 | 100.0 | 36.11 | 64.49 | 47.61 | 91.97 | ||||
| Gender | ||||||||||
| Male | 231 | 55.1 | 36.56 | 0.31c | 64.91 | 0.57c | 48.8 | 0.081c | 92.31 | 0.24c |
| Female | 187 | 44.6 | 35.57 | 63.88 | 46.53 | 91.55 | ||||
| Unknown | 1 | 0.2 | ||||||||
| Age [years] | ||||||||||
| Mean | 40.87 | |||||||||
| Median | 39 | |||||||||
| ≤ 41 years | 245 | 58.5 | 35.95 | 0.94c | 62.75 | 0.003c | 49.71 | 0.015c | 91.87 | 0.43c |
| > 41 years | 173 | 41.3 | 36.36 | 66.87 | 45.05 | 92.12 | ||||
| WHO classification (2016) | ||||||||||
| IDH-mut, 1p/19q-codel | 169 | 40.3 | 36.43 | 0.44c | 74.89 | < 0.001c | 44.76 | 0.002c | 92.52 | 0.028c |
| IDH-mut, 1p/19q-non-codel | 250 | 59.7 | 35.9 | 57.46 | 49.96 | 91.61 | ||||
| Methylation subgroupsa | ||||||||||
| LGm1 | 45 | 10.7 | 30.59 | < 0.001b | 50.57 | < 0.001b | 40.48 | 0.001b | 88.18 | < 0.001b |
| LGm2 | 251 | 59.9 | 36.72 | 60.82 | 50.06 | 92.32 | ||||
| LGm3 | 123 | 29.4 | 36.9 | 77.06 | 46.07 | 92.66 | ||||
| MGMT promoter statusa | ||||||||||
| Methylated | 389 | 92.84 | 36.43 | < 0.001c | 65.33 | < 0.001c | 47.44 | 0.068c | 92.12 | 0.11c |
| Unmethylated | 30 | 7.2 | 31.9 | 53.58 | 53.29 | 90.06 | ||||
| ATRX status | ||||||||||
| Mutant | 181 | 43.2 | 36.19 | 0.79 | 57.86 | < 0.001c | 51.15 | 0.001c | 91.71 | 0.63c |
| Wildtype | 238 | 56.8 | 36.05 | 69.53 | 45.36 | 92.17 | ||||
| TERT promoter status | ||||||||||
| Mutant | 93 | 22.2 | 36.94 | 0.035c | 73.53 | < 0.001c | 45.47 | < 0.001c | 93.22 | < 0.001c |
| Wildtype | 143 | 34.1 | 36.69 | 58.19 | 52.6 | 92.45 | ||||
| Unknown | 183 | 43.7 |
Data taken from Ceccarelli et al. (2016).
Data taken from Kruskal-Wallis test.
Data taken from Wilcoxon-Mann-Whitney test.
2.2. Promoter Methylation Analyses
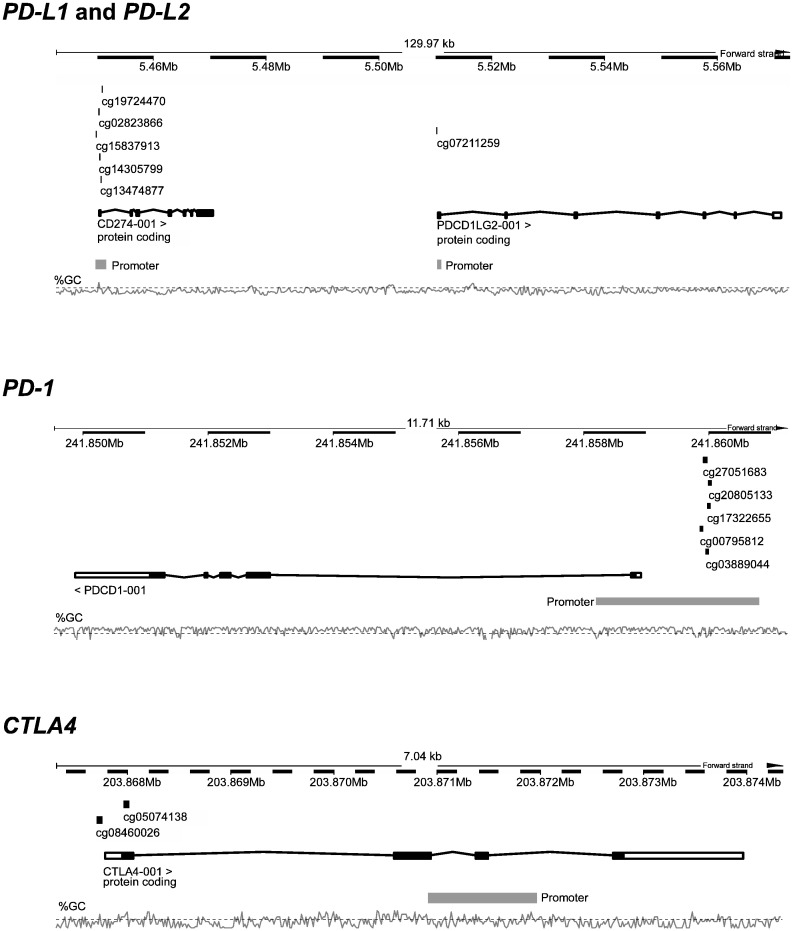
TCGA methylation data were generated using the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA). Relative DNA methylation levels were calculated as previously described for each locus (Meller et al., 2016). In brief, HumanMethylation450 data of level 2 including background-corrected methylated (Intensity_M) and unmethylated (Intensity_U) summary intensities (beads cg15837913, cg02823866, cg14305799, cg13474877, cg19724470 [PD-L1]; cg07211259 [PD-L2]; cg20805133, cg00795812, cg27051683, cg17322655, cg03889044 [PD-1]; cg05074138 and cg08460026 [CTLA4]) were downloaded and extracted by the R package ‘methylumi’. Methylation values for each bead were calculated with the formula: methylation [%] = 100% × Intensity_M / (Intensity_M + Intensity_U). Results from all beads from one gene were mean averaged. The genomic organization of the genes and the target regions of the analyzed beads are shown in Fig. 1.
Fig. 1.
Genomic location and organization of CD274 (PD-L1) and PDCD1LG2 (PD-L2) on chromosome 9, PDCD1 (PD-1) on chromosome 2, and CTLA4 on chromosome 2. Analyzed cg-beads from the Illumina Infinium HumanMethylation450 BeadChip are illustrated. Figure information is based on the Genome Reference Consortium Human Build 38 patch release 7 (GRCh38.p7) illustrated by http://www.ensembl.org.
2.3. mRNA Expression Analyses
mRNA data generated by the TCGA Research Network using the Illumina HiSeq 2000 RNA Sequencing Version 2 analysis (Illumina, Inc., San Diego, CA, USA) were obtained from the TCGA webpage and included normalized gene expression results. Counts per gene were calculated with the RSEM algorithm using the SeqWare framework (Li and Dewey, 2011).
2.4. Statistical Analyses
Statistical analyses were performed using SPSS, version 23.0 (SPSS Inc., Chicago, IL). Mean values are given ± standard deviation. Comparisons of mean values between groups were performed applying one way ANOVA with Bonferroni post-hoc testing, the Wilcoxon-Mann-Whitney test, and the Kruskal-Wallis test. Spearman's ρ rank correlations between mRNA and methylation levels were performed. Survival was defined as OS. Hazard ratios (HR) were calculated using univariate and multivariate Cox proportional hazards models with stratification. For survival analyses, continuous methylation data were logarithmized to base 2. Survival analyses were performed using the Kaplan-Meier method, and differences between the groups were tested using the log-rank test. For Kaplan-Meier survival analysis, methylation data were dichotomized using the median methylation level as cut-off. p-Values < 0.05 were considered as statistically significant.
3. Results
3.1. Correlation of PD-1, PD-L1, PD-L2, and CTLA4 DNA Promoter Methylation With mRNA Expression
The Infinium HumanMethylation450 BeadChip contains 27 beads targeting the PD-1 gene locus (Chr2:241849445–241860885; Reference Consortium Human Build 38 patch release 7 (GRCh38.p7)), 10 beads in the region of the adjacent genes PD-L1 and PD-L2 (Chr9:5445953–5572886), and seven beads probing the CTLA4 gene locus (Chr2:203865714–203874906). In order to avoid multiple testing errors, we have reduced the analyses to pre-specified loci identified in previous studies (Fig. 1) (Franzen et al., 2018, Gevensleben et al., 2016, Goltz et al., 2016a, Goltz et al., 2016b, Goltz et al., 2017a, Goltz et al., 2017b). We found a significant inverse correlation between gene methylation and mRNA expression levels for PD-L1, PD-L2 and CTLA-4 (PD-L1: ρ = − 0.136, p = 0.005; PD-L2: ρ = − 0.642, p < 0.001; CTLA-4: ρ = − 0.249, p < 0.001), while no correlation was present for PD-1 (ρ = 0.020, p = 0.68).
3.2. Association of PD-1, PD-L1, PD-L2, and CTLA4 DNA Promoter Methylation With Clinicopathological Features and Molecular Targets
For a detailed association analysis of promoter methylation of immune checkpoints with clinicopathological characteristics and molecular targets see Table 1. In brief, immune checkpoint promoter methylation was significantly associated with patients' age (PD-L2: p = 0.003; PD-1: p = 0.015), O6-methylguanine DNA methyltransferase(MGMT) methylation (PD-L1: p < 0.001; PD-L2: p < 0.001), ATRX mutations (PD-L2: p < 0.001, PD-1: p = 0.001), telomerase reverse transcriptase (TERT) mutations (PD-L1: p = 0.035, PD-L2: p < 0.001, PD-1: p < 0.001, CTLA4: p < 0.001), and methylation subgroups (LGm1, LGm2 and LGm3; PD-L1: p < 0.001, PD-L2: p < 0.001, PD-1: p = 0.001, CTLA4: p < 0.001).
3.3. PD-1, PD-L1, PD-L2, and CTLA4 DNA Promoter Methylation in Glioma Methylation Subgroups
In a pan-glioma unsupervised cluster analysis, Ceccarelli et al. (2016) previously identified specific methylation subtypes LGm1/LGm2/LGm3, which carried IDH1 or IDH2 mutations, were enriched for lower-grade gliomas, and presented with a genome-wide hypermethylation compared to other methylation clusters. As our association analysis had already revealed a significant association with these methylation clusters (Table 1), we further analyzed the relationship of immune checkpoint methylation with methylation subtypes.
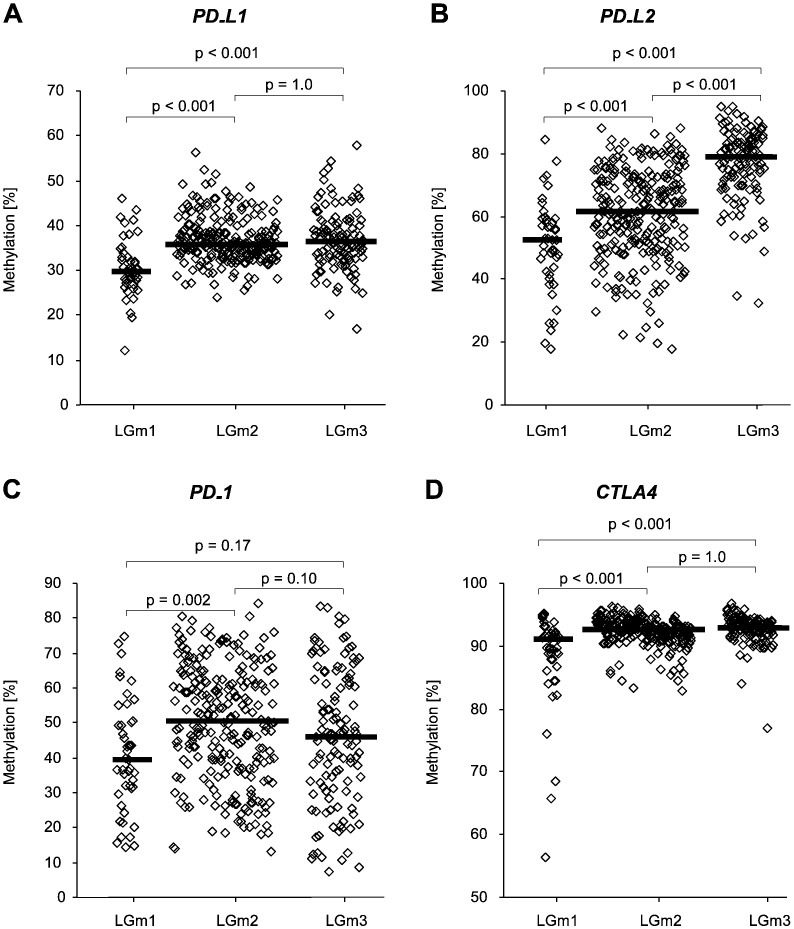
Mean promoter methylation of both PD-L1 and PD-L2 was significantly lower in LGm1 (PD-L1: 30.6% ± 6.4%; PD-L2: 50.6% ± 1.5%) compared to LGm2 and LGm3 (PD-L1: 36.7% ± 5.0%, p < 0.001 for LGm1 vs. LGm2; 36.9% ± 6.6%, p < 0.001 for LGm1 vs. LGm3, Fig. 2A; PD-L2: 60.8% ± 14.5%, p < 0.001 for LGm1 vs. LGm2; 77.1% ± 11.2%, p < 0.001 for LGm1 vs. LGm3; Fig. 2B). Additionally, PD-L2 levels were shown to be lower in LGm2 compared to LGm3 (p < 0.001). Mean PD-1 promoter methylation was significantly lower in LGm1 (40.5% ± 16.7%) compared to LGm2 (50.1% ± 16.4%, p = 0.002; Fig. 2C). No significant differential promoter methylation was seen for LGm3 (46.1% ± 19.2%). Mean CTLA4 promoter methylation was significantly lower in LGm1 (88.2% ± 7.8%) compared to both LGm2 and LGm3 (92.3% ± 22.2%, p < 0.001 for LGm1 vs. LGm2; 92.7% ± 2.3%, p < 0.001 for LGm1 vs. LGm3; Fig. 2D). Of note, the heterogeneity of values was shown to be significantly higher for PD-1 and CTLA4 methylation possibly indicating that methylation levels do not reflect the rather homogenous tumor tissue but might be distorted by PD-1 and CTLA-4 expressing infiltrating immune cells.
Fig. 2.
Methylation of immune checkpoint genes PD-L1 (A), PD-L2 (B), PD-1 (C), and CTLA4 (D) in diffuse lower-grade glioma patients (n = 419) with respect to methylation subtypes (LGm1, LGm2, LGm3). Bars indicate median. p-Values refer to ANOVA with Bonferroni Post hoc test.
3.4. Correlation of PD-1, PD-L1, PD-L2, and CTLA4 DNA Promoter Methylation With Immune Cell Infiltrates
Since PD-1 and CTLA-4 expression has been mainly observed in immune cells (Buchbinder and Desai, 2016), differential PD-1 and CTLA4 promoter methylation may reflect changes in the lymphocyte and antigen presenting cell compartment. Subtypes of tumor infiltrating lymphocytes in the TCGA cohort as assessed by Li et al. (2016) were correlated with PD-1 and CTLA4 promoter methylation. Tumor infiltrating B lymphocytes as well as CD8 positive (CD8+) T lymphocytes and dendritic cells correlated inversely with PD-1 (r = − 0.178; p < 0.001 for B lymphocytes, r = − 0.234; p < 0.001 for CD8+ T lymphocytes, and r = − 0.171; p < 0.031 for dendritic cells, n = 419 for all). For CTLA4 methylation, no significant association with immune cells was observed. Promoter methylation of PD-L1 significantly correlated with PD-1 methylation in tumor samples (r = 0.293; p < 0.001; n = 419). Further, PD-L1 methylation significantly and inversely correlated with infiltrating CD4 positive (CD4+) T lymphocytes (r = − 0.109; p = 0.026; n = 419) and dendritic cells (r = − 0.099; p = 0.043; n = 419). Promoter methylation of PD-L2 significantly correlated with PD-1 methylation in tumor samples (r = 0.186; p < 0.001; n = 419). However, no association was found with immune cells.
3.5. Prognostic Impact of PD-1, PD-L1, PD-L2, and CTLA4 DNA Promoter Methylation
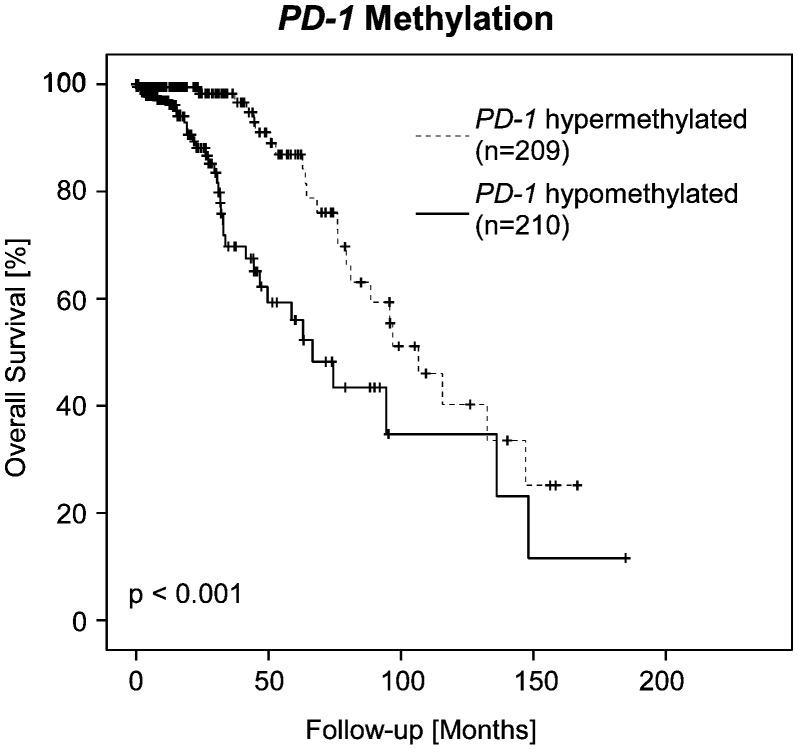
Subsequently, we analyzed whether promoter methylation of immune checkpoints allowed for a risk stratification of LGG patients. Since the DNA methylation clusters had been shown to have an impact on survival, Cox proportional hazard analysis was stratified according to methylation subtypes (LGm1, LGm2 and LGm3). In univariate and multivariate Cox proportional hazard analysis, PD-1 methylation qualified as a strong prognostic factor together with age (univariate Cox proportional hazard analysis: HR = 0.44 [0.30–0.66], p < 0.001; multivariate Cox proportional hazard analysis: HR = 0.51 [0.34–0.76], p = 0.001; Table 2). The prognostic value of dichotomized PD-1 methylation was further confirmed by Kaplan-Meier analysis (Χ2 = 13.04, p < 0.001 for hypomethylated PD-1 and hypermethylated PD-1, respectively; Fig. 3). No prognostic impact was observed for PD-L1, PD-L2, and CTLA4 methylation.
Table 2.
Univariate and multivariate Cox proportional hazard analysis of immune checkpoint methylation, sex, and age. Methylation was analyzed as logarithmized continuous variable. Cox analysis was stratified by methylation subtype (LGm1, LGm2 and LGm3)a.
| Variable | Univariate cox proportional hazards analysis |
Multivariate cox proportional hazards analysis |
||
|---|---|---|---|---|
| p | HR [95% CI] | p | HR [95% CI] | |
| Age at initial diagnosis (> 41 vs. ≤ 41 years) | < 0.001 | 3.83 [2.04–7.17] | < 0.001 | 3.19 [1.69–6.02] |
| Sex (male vs. female) | 0.62 | 1.15 [0.67–1.96] | 0.75 | 1.09 [0.64–1.87] |
| PD-1 methylation | < 0.001 | 0.44 [0.30–0.66] | 0.001 | 0.51 [0.34–0.76] |
| PD-L1 methylation | 0.25 | 0.57 [0.22–1.48] | ND | |
| PD-L2 methylation | 0.16 | 0.63 [0.33–1.20] | ND | |
| CTLA4 methylation | 0.26 | 0.17 [0.01–3.69] | ND | |
Data taken from Ceccarelli et al. (2016); ND: not determined.
Fig. 3.
Kaplan-Meier analysis of overall survival in diffuse lower-grade glioma patients (n = 419) stratified by promoter methylation of PD-1.
4. Discussion
In the present study, DNA promoter methylation of the immune checkpoint PD-1 was shown to serve as highly significant prognostic factor for overall survival in patients with LGG. In its key role as an immune checkpoint PD-1 promotes self-tolerance by suppressing T cell activity. Upon binding to its ligands PD-L1 or PD-L2, PD-1 fosters apoptosis in antigen specific T cells while simultaneously reducing programmed cell death in regulatory T cells (Tregs) (Francisco et al., 2009, Francisco et al., 2010). PD-1 is mainly expressed on activated CD4+ and CD8+ T cells as well as on B cells and inhibits effector T cell activity at later-stage immune responses in peripheral tissues (Francisco et al., 2010). The interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells has been identified as an important mechanism of immune evasion of tumor cells with high PD-1 expression consequently leading to depleted antitumor immune responses (reviewed by Zhang et al., 2017). Recent publications have further provided evidence for an epigenetic promoter control of PD-1 expression in human T lymphocytes (Youngblood et al., 2011, McPherson et al., 2014).
As gliomas have long been recognized as immunosuppressive neoplasms that are characterized by the activation of various immune escape mechanisms (reviewed by Razavi et al., 2016), our findings indicate that high levels of PD-1 promoter methylation might result in functional tumor-specific T cells effectively driving antitumor immune responses, therefore leading to a favorable course of disease. In addition, promoter methylation of PD-L1 also significantly correlated with PD-1 methylation, suggesting that epigenetic regulation of the PD-1 receptor may be paralleled by PD-L1 induction in tumor tissue. PD-1 methylation further inversely correlated with tumor infiltrating B lymphocytes, CD8+ T lymphocytes, and antigen presenting dendritic cells in our study, suggesting a role of PD-1 methylation as surrogate marker for immune cell infiltration. In addition, PD-L1 methylation inversely correlated with infiltrating CD4+ T lymphocytes and dendritic cells, adding to the seemingly reciprocal relationship of infiltrating immune cells and immune checkpoint methylation in LGG.
Tumor-infiltrating immune cells are part of a complex microenvironment that is known to regulate tumor development and growth in gliomas (reviewed by Domingues et al., 2016); however, data on the role of immune cells and their influence on survival have been conflicting. While several studies have demonstrated that high numbers of intratumoral effector T cells are significantly correlated with a better survival in grade IV gliomas (Lohr et al., 2011, Kmiecik et al., 2014), other groups have reported that specific molecular subtypes (e.g. the mesenchymal subtype) are characterized by pro-inflammatory immune signatures and immunosuppression, associating immune infiltrates to a higher risk and poor survival (Doucette et al., 2013, Cheng et al., 2016). Of note, neither tumor infiltrating lymphocytes, nor antigen presenting cells added prognostic information in our study (data not shown). Although further mechanistic studies are clearly warranted in order to fully characterize the role of PD-1 expression in LGG, our results imply that the densities of B and CD8+ T lymphocytic infiltrates as well as antigen presenting dendritic cells might be estimated via PD-1 methylation. This might be of significance for the potential therapeutic application of immunotherapies in LGG patients in the future.
A publication by Ceccarelli et al. (2016) has recently emphasized the relevance of DNA methylation for the clinical classification and biological behavior of gliomas. Unsupervised cluster analysis of 1122 diffuse glioma patients identified six DNA methylation subtypes with distinct molecular and clinical features. In this study, the LGm1/LGm2/LGm3 subgroups harbored IDH1 or IDH2 mutations (449 of 450, 99%), were enriched for LGG (421/454, 93%), and showed genome-wide hypermethylation compared to LGm4–6 clusters, corroborating the association between IDH mutation and increased DNA methylation (Turcan et al., 2012). Interestingly, further analysis between the two discovered IDH mutant-non-codel DNA methylation clusters allowed for the identification of a low-methylation subgroup (G-CIMP-low) which was enriched in the heterogeneous subgroup of LGm1 tumors. As PD-1, PD-L1, PD-L2, and CTLA4 promoter methylation was significantly lower in the LGm1 subgroup compared to LGm2 and LGm3 in the present study, our results might therefore very well reflect the hypermethylated phenotype in LGm2 and LGm3 as opposed to a low-methylated subgroup enriched in LGm1. In line with previous results, Ceccarelli et al. (2016) also reported the G-CIMP-low subgroup to be associated with a worse survival compared to hypermethylated tumors. In our study, methylation of PD-L2 additionally distinguished between LGm2 and LGm3, thus suggesting differential methylation between these two subgroups independent from cluster-related hypermethylation.
Some of the differences in mean methylation found in our study were remarkably small. However, a strength of our study is the large number of included patients allowing for the detection of even small differences with high statistical significance. The biological significance appears to be supported by the finding of a strong inverse correlation between mRNA expression and methylation levels for PD-L2, CTLA4, and PD-L1. CTLA4 methylation in TERT-mutated versus wildtype tumors, for example, seems minor at first glance (93.22% versus 92.45% methylation). However, this difference is equivalent to 6.78% (100%–93.22%) versus 7.55% (100%–92.45%) “unmethylation” which represents a remarkable increase of “unmethylation” by 11%. These findings indicate that a small subgroup of (unmethylated) cells is responsible for the mRNA expression. A major limitation of our study, however, is that we were not able assign methylation and expression levels to specific cell types. This needs to be done in further studies. A second limitation of our study is the relatively short follow-up. LGG patients harboring an IDH mutation and 1p/19q codeletion for example, representing 40.3% of the cohort under investigation, have a relatively good prognosis with a median overall survival of 8 years (Cancer Genome Atlas Research Network et al., 2015). Hence, long follow-up periods are required in order to detect statistically significant survival differences. The high prognostic power of PD-1 promoter methylation presented in our study, even in the absence of long follow-up, however, indicates the high relevance of this gene in LGG. The lack of prognostic power of PD-L1, PD-L2, and CTLA4 methylation, on the other hand, might be due to the limited follow-up, and the prognostic potential of these markers might need to be investigated in a study with sufficient clinical follow-up. The lack of a validation study in our analysis further prompted us to analyze only CpG-sites which have been shown to be of significance in earlier studies (Franzen et al., 2018, Gevensleben et al., 2016, Goltz et al., 2016a, Goltz et al., 2016b, Goltz et al., 2017a, Goltz et al., 2017b). As previously reported, we used mean methylation values of these CpG-sites. Such approach reduces the risk of multiple testing errors since only a limited number of (predefined) features is analyzed. On the other hand, the prognostic performance might be underestimated since other or single individual CpG-sites might be more informative than the predefined and averaged ones. This needs to be addressed in further studies.
Inhibition of the PD-1/PD-L1 and CTLA-4 immune checkpoints is a promising therapeutic approach for the treatment of primary brain tumors. Several clinical trials are currently ongoing and evaluating the effects of immune checkpoint inhibition in high-grade glioma with nivolumab, pembrolizumab and ipilimumab. First study results, however, so far have not painted a clear picture. While the PD-1 checkpoint inhibitor nivolumab recently failed to demonstrate a survival benefit in a first randomized clinical phase III trial (CheckMate 143, ClinicalTrials.gov Identifier: NCT02017717) in high-grade glioma, pembrolizumab appears to have a durable benefit for patients with recurrent PD-L1-positive glioblastomas (Omuro et al., 2017, Reardon et al., 2016). In the context of rapidly developing immunotherapies, e.g. checkpoint inhibitors, robust and clinically applicable biomarkers are needed to estimate the potential effects and identify patients eligible for treatment. In the present study, DNA methylation analysis of PD-1 in LGG patients was shown to add independent prognostic information. Furthermore, DNA methylation analysis can be conducted reliably even in small and formalin-fixed samples. We therefore strongly recommend the integration of immune checkpoint promoter methylation analysis in running and future clinical trials in order to test its ability to predict treatment response to immune checkpoint inhibitors.
Funding Sources
No funding was received.
Conflicts of Interest
Dimo Dietrich is inventor and owns a granted patent on methylation of immune checkpoint genes as prognostic and predictive biomarkers (patent application DE102016005947 (B3)). A second patent application on immune checkpoint genes as biomarkers for immunotherapy response prediction is pending.
Author Contributions
LKR analyzed the data and drafted the manuscript. HG and DG coordinated the study, analyzed the data and drafted the manuscript. JD processed TCGA data. FB and JL revised the manuscript critically for important intellectual content. DD designed and supervised the study, analyzed the data and revised the manuscript.
Acknowledgements
None.
References
- Berger R., Rotem-Yehudar R., Slama G., Landes S., Kneller A., Leiba M., Koren-Michowitz M., Shimoni A., Nagler A. Phase I safety and pharmacokinetic study of CT-011, a humanized antibody interacting with PD-1, in patients with advanced hematologic malignancies. Clin. Cancer Res. 2008;14:3044–3051. doi: 10.1158/1078-0432.CCR-07-4079. [DOI] [PubMed] [Google Scholar]
- Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P., Drake C.G., Camacho L.H., Kauh J., Odunsi K., Pitot H.C., Hamid O., Bhatia S., Martins R., Eaton K., Chen S., Salay T.M., Alaparthy S., Grosso J.F., Korman A.J., Parker S.M., Agrawal S., Goldberg S.M., Pardoll D.M., Gupta A., Wigginton J.M. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J., Reckamp K.L., Baas P., Crinò L., Eberhardt W.E., Poddubskaya E., Antonia S., Pluzanski A., Vokes E.E., Holgado E., Waterhouse D., Ready N., Gainor J., Arén Frontera O., Havel L., Steins M., Garassino M.C., Aerts J.G., Domine M., Paz-Ares L., Reck M., Baudelet C., Harbison C.T., Lestini B., Spigel D.R. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder E.I., Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R., Cooper L.A., Rheinbay E., Miller C.R., Vitucci M., Morozova O., Robertson A.G., Noushmehr H., Laird P.W., Cherniack A.D., Akbani R., Huse J.T., Ciriello G., Poisson L.M., Barnholtz-Sloan J.S., Berger M.S., Brennan C., Colen R.R., Colman H., Flanders A.E., Giannini C., Grifford M., Iavarone A., Jain R., Joseph I., Kim J., Kasaian K., Mikkelsen T., Murray B.A., O'Neill B.P., Pachter L., Parsons D.W., Sougnez C., Sulman E.P., Vandenberg S.R., Van Meir E.G., von Deimling A., Zhang H., Crain D., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton T., Sherman M., Yena P., Black A., Bowen J., Dicostanzo K., Gastier-Foster J., Leraas K.M., Lichtenberg T.M., Pierson C.R., Ramirez N.C., Taylor C., Weaver S., Wise L., Zmuda E., Davidsen T., Demchok J.A., Eley G., Ferguson M.L., Hutter C.M., Mills Shaw K.R., Ozenberger B.A., Sheth M., Sofia H.J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J.C., Ayala B., Baboud J., Chudamani S., Jensen M.A., Liu J., Pihl T., Raman R., Wan Y., Wu Y., Ally A., Auman J.T., Balasundaram M., Balu S., Baylin S.B., Beroukhim R., Bootwalla M.S., Bowlby R., Bristow C.A., Brooks D., Butterfield Y., Carlsen R., Carter S., Chin L., Chu A., Chuah E., Cibulskis K., Clarke A., Coetzee S.G., Dhalla N., Fennell T., Fisher S., Gabriel S., Getz G., Gibbs R., Guin R., Hadjipanayis A., Hayes D.N., Hinoue T., Hoadley K., Holt R.A., Hoyle A.P., Jefferys S.R., Jones S., Jones C.D., Kucherlapati R., Lai P.H., Lander E., Lee S., Lichtenstein L., Ma Y., Maglinte D.T., Mahadeshwar H.S., Marra M.A., Mayo M., Meng S., Meyerson M.L., Mieczkowski P.A., Moore R.A., Mose L.E., Mungall A.J., Pantazi A., Parfenov M., Park P.J., Parker J.S., Perou C.M., Protopopov A., Ren X., Roach J., Sabedot T.S., Schein J., Schumacher S.E., Seidman J.G., Seth S., Shen H., Simons J.V., Sipahimalani P., Soloway M.G., Song X., Sun H., Tabak B., Tam A., Tan D., Tang J., Thiessen N., Triche T., Jr., Van Den Berg D.J., Veluvolu U., Waring S., Weisenberger D.J., Wilkerson M.D., Wong T., Wu J., Xi L., Xu A.W., Yang L., Zack T.I., Zhang J., Aksoy B.A., Arachchi H., Benz C., Bernard B., Carlin D., Cho J., DiCara D., Frazer S., Fuller G.N., Gao J., Gehlenborg N., Haussler D., Heiman D.I., Iype L., Jacobsen A., Ju Z., Katzman S., Kim H., Knijnenburg T., Kreisberg R.B., Lawrence M.S., Lee W., Leinonen K., Lin P., Ling S., Liu W., Liu Y., Liu Y., Lu Y., Mills G., Ng S., Noble M.S., Paull E., Rao A., Reynolds S., Saksena G., Sanborn Z., Sander C., Schultz N., Senbabaoglu Y., Shen R., Shmulevich I., Sinha R., Stuart J., Sumer S.O., Sun Y., Tasman N., Taylor B.S., Voet D., Weinhold N., Weinstein J.N., Yang D., Yoshihara K., Zheng S., Zhang W., Zou L., Abel T., Sadeghi S., Cohen M.L., Eschbacher J., Hattab E.M., Raghunathan A., Schniederjan M.J., Aziz D., Barnett G., Barrett W., Bigner D.D., Boice L., Brewer C., Calatozzolo C., Campos B., Carlotti C.G., Jr., Chan T.A., Cuppini L., Curley E., Cuzzubbo S., Devine K., DiMeco F., Duell R., Elder J.B., Fehrenbach A., Finocchiaro G., Friedman W., Fulop J., Gardner J., Hermes B., Herold-Mende C., Jungk C., Kendler A., Lehman N.L., Lipp E., Liu O., Mandt R., McGraw M., Mclendon R., McPherson C., Neder L., Nguyen P., Noss A., Nunziata R., Ostrom Q.T., Palmer C., Perin A., Pollo B., Potapov A., Potapova O., Rathmell W.K., Rotin D., Scarpace L., Schilero C., Senecal K., Shimmel K., Shurkhay V., Sifri S., Singh R., Sloan A.E., Smolenski K., Staugaitis S.M., Steele R., Thorne L., Tirapelli D.P., Unterberg A., Vallurupalli M., Wang Y., Warnick R., Williams F., Wolinsky Y., Bell S., Rosenberg M., Stewart C., Huang F., Grimsby J.L., Radenbaugh A.J., Zhang J. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015;26:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M., Anjum S., Wang J., Manyam G., Zoppoli P., Ling S., Rao A.A., Grifford M., Cherniack A.D., Zhang H., Poisson L., Carlotti C.G., Jr., Tirapelli D.P., Rao A., Mikkelsen T., Lau C.C., Yung W.K., Rabadan R., Huse J., Brat D.J., Lehman N.L., Barnholtz-Sloan J.S., Zheng S., Hess K., Rao G., Meyerson M., Beroukhim R., Cooper L., Akbani R., Wrensch M., Haussler D., Aldape K.D., Laird P.W., Gutmann D.H., Research Network T.C.G.A., Noushmehr H., Iavarone A., Verhaak R.G. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.A., Wolchok J.D., Snyder A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2015;373:1984. doi: 10.1056/NEJMc1508163. [DOI] [PubMed] [Google Scholar]
- Cheng W., Ren X., Zhang C., Cai J., Liu Y., Han S., Wu A. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology. 2016;86:2226–2234. doi: 10.1212/WNL.0000000000002770. [DOI] [PubMed] [Google Scholar]
- Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C., Marks K.M., Prins R.M., Ward P.S., Yen K.E., Liau L.M., Rabinowitz J.D., Cantley L.C., Thompson C.B., Vander Heiden M.G., Su S.M. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues P., González-Tablas M., Otero Á., Pascual D., Miranda D., Ruiz L., Sousa P., Ciudad J., Gonçalves J.M., Lopes M.C., Orfao A., Tabernero M.D. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav. Immun. 2016;53:1–15. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- Doucette T., Rao G., Rao A., Shen L., Aldape K., Wei J., Dziurzynski K., Gilbert M., Heimberger A.B. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol. Res. 2013;1:112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L.M., Salinas V.H., Brown K.E., Vanguri V.K., Freeman G.J., Kuchroo V.K., Sharpe A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L.M., Sage P.T., Sharpe A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen A., Vogt T.J., Müller T., Dietrich J., Schröck A., Golletz C., Brossart P., Bootz F., Landsberg J., Kristiansen G., Dietrich D. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget. 2018;9:641–650. doi: 10.18632/oncotarget.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon E.B., Rizvi N.A., Hui R., Leighl N., Balmanoukian A.S., Eder J.P., Patnaik A., Aggarwal C., Gubens M., Horn L., Carcereny E., Ahn M.J., Felip E., Lee J.S., Hellmann M.D., Hamid O., Goldman J.W., Soria J.C., Dolled-Filhart M., Rutledge R.Z., Zhang J., Lunceford J.K., Rangwala R., Lubiniecki G.M., Roach C., Emancipator K., Gandhi L., KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- Gevensleben H., Holmes E.E., Goltz D., Dietrich J., Sailer V., Ellinger J., Dietrich D., Kristiansen G. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget. 2016;7:79943–79955. doi: 10.18632/oncotarget.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D., Gevensleben H., Dietrich J., Ellinger J., Landsberg J., Kristiansen G., Dietrich D. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1221555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D., Gevensleben H., Dietrich J., Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2016;6 doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D., Gevensleben H., Dietrich J., Schroeck F., de Vos L., Droege F., Kristiansen G., Schroeck A., Landsberg J., Bootz F., Dietrich D. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017;8:41011–41020. doi: 10.18632/oncotarget.17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D., Gevensleben H., Grünen S., Dietrich J., Kristiansen G., Landsberg J., Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia. 2017;31:738–743. doi: 10.1038/leu.2016.328. [DOI] [PubMed] [Google Scholar]
- Gousias K., Markou M., Arzoglou V., Voulgaris S., Vartholomatos G., Kostoula A., Voulgari P., Polyzoidis K., Kyritsis A.P. Frequent abnormalities of the immune system in gliomas and correlation with the WHO grading system of malignancy. J. Neuroimmunol. 2010;226:136–142. doi: 10.1016/j.jneuroim.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Hamid O., Robert C., Daud A., Hodi F.S., Hwu W.J., Kefford R., Wolchok J.D., Hersey P., Joseph R.W., Weber J.S., Dronca R., Gangadhar T.C., Patnaik A., Zarour H., Joshua A.M., Gergich K., Elassaiss-Schaap J., Algazi A., Mateus C., Boasberg P., Tumeh P.C., Chmielowski B., Ebbinghaus S.W., Li X.N., Kang S.P., Ribas A. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik J., Zimmer J., Chekenya M. Natural killer cells in intracranial neoplasms: presence and therapeutic efficacy against brain tumours. J. Neuro-Oncol. 2014;116:1–9. doi: 10.1007/s11060-013-1265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J.J., Cowey C.L., Lao C.D., Schadendorf D., Dummer R., Smylie M., Rutkowski P., Ferrucci P.F., Hill A., Wagstaff J., Carlino M.S., Haanen J.B., Maio M., Marquez-Rodas I., McArthur G.A., Ascierto P.A., Long G.V., Callahan M.K., Postow M.A., Grossmann K., Sznol M., Dreno B., Bastholt L., Yang A., Rollin L.M., Horak C., Hodi F.S., Wolchok J.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Severson E., Pignon J.C., Zhao H., Li T., Novak J., Jiang P., Shen H., Aster J.C., Rodig S., Signoretti S., Liu J.S., Liu X.S. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J., Ratliff T., Huppertz A., Ge Y., Dictus C., Ahmadi R., Grau S., Hiraoka N., Eckstein V., Ecker R.C., Korff T., von Deimling A., Unterberg A., Beckhove P., Herold-Mende C. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-β. Clin. Cancer Res. 2011;17:4296–4308. doi: 10.1158/1078-0432.CCR-10-2557. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Margolin K., Ernstoff M.S., Hamid O., Lawrence D., McDermott D., Puzanov I., Wolchok J.D., Clark J.I., Sznol M., Logan T.F., Richards J., Michener T., Balogh A., Heller K.N., Hodi F.S. Ipilimumab in patients with melanoma and brain metastases: an open-label, phase 2 trial. Lancet Oncol. 2012;13:459–465. doi: 10.1016/S1470-2045(12)70090-6. [DOI] [PubMed] [Google Scholar]
- McPherson R.C., Konkel J.E., Prendergast C.T., Thomson J.P., Ottaviano R., Leech M.D., Kay O., Zandee S.E., Sweenie C.H., Wraith D.C., Meehan R.R., Drake A.J., Anderton S.M. Epigenetic modification of the PD-1 (Pdcd1) promoter in effector CD4 + T cells tolerized by peptide immunotherapy. elife. 2014;3 doi: 10.7554/eLife.03416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller S., Zipfel L., Gevensleben H., Dietrich J., Ellinger J., Majores M., Stein J., Sailer V., Jung M., Kristiansen G., Dietrich D. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11:871–880. doi: 10.1080/15592294.2016.1241931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., Verhaak R.G., Hoadley K.A., Hayes D.N., Perou C.M., Schmidt H.K., Ding L., Wilson R.K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L.M., Buckley J., Herman J.G., Baylin S.B., Laird P.W., Aldape K., Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A., Vlahovic G., Lim M., Sahebjam S., Baehring J., Cloughesy T., Voloschin A., Ramkissoon S.H., Ligon K.L., Latek R., Zwirtes R., Strauss L., Paliwal P., Harbison C.T., Reardon D.A., Sampsonc J.H. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase 1 cohorts of CheckMate 143. Neuro-Oncology. 2017 doi: 10.1093/neuonc/nox208. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom Q.T., Gittleman H., Xu J., Kromer C., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro-Oncology. 2016;18(Suppl. 5):v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi S.M., Lee K.E., Jin B.E., Aujla P.S., Gholamin S., Li G. Immune evasion strategies of glioblastoma. Front. Surg. 2016;3 doi: 10.3389/fsurg.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon D.A., Kim T.M., Frenel J.S., Santoro A., Lopez J., Subramaniam D.S., Siu L.L., Rodon J., Tamura K., Saraf S., Morosky A., Stein K., Soria J.C. Results of the phase Ib Keynote-028 multi-cohort trial of prembrolizumab monotherapy in patients with recurrent PD-L1-positive glioblastoma multiforme (GBM) Neuro-Oncology. 2016;18:vi25–vi26. [Google Scholar]
- Ribas A., Hamid O., Daud A., Hodi F.S., Wolchok J.D., Kefford R., Joshua A.M., Patnaik A., Hwu W.J., Weber J.S., Gangadhar T.C., Hersey P., Dronca R., Joseph R.W., Zarour H., Chmielowski B., Lawrence D.P., Algazi A., Rizvi N.A., Hoffner B., Mateus C., Gergich K., Lindia J.A., Giannotti M., Li X.N., Ebbinghaus S., Kang S.P., Robert C. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315:1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., Leming P.D., Spigel D.R., Antonia S.J., Horn L., Drake C.G., Pardoll D.M., Chen L., Sharfman W.H., Anders R.A., Taube J.M., McMiller T.L., Xu H., Korman A.J., Jure-Kunkel M., Agrawal S., McDonald D., Kollia G.D., Gupta A., Wigginton J.M., Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W., Lu C., Ward P.S., Thompson C.B., Kaufman A., Guryanova O., Levine R., Heguy A., Viale A., Morris L.G., Huse J.T., Mellinghoff I.K., Chan T.A. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok J.D., Kluger H., Callahan M.K., Postow M.A., Rizvi N.A., Lesokhin A.M., Segal N.H., Ariyan C.E., Gordon R.A., Reed K., Burke M.M., Caldwell A., Kronenberg S.A., Agunwamba B.U., Zhang X., Lowy I., Inzunza H.D., Feely W., Horak C.E., Hong Q., Korman A.J., Wigginton J.M., Gupta A., Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Song G., Yu J. The prognostic significance of PD-L1 expression in patients with glioma: a meta-analysis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-04023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B., Oestreich K.J., Ha S.J., Duraiswamy J., Akondy R.S., West E.E., Wei Z., Lu P., Austin J.W., Riley J.L., Boss J.M., Ahmed R. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu S., Li T., Liu Y.J., Chen W., Chen J. Targeting immune checkpoints in malignant glioma. Oncotarget. 2017;8:7157–7174. doi: 10.18632/oncotarget.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]