Abstract
Pituitary adenomas, arising from the pituitary gland cells, are one of the most frequent tumors found in the sella region. However, the molecular mechanisms involved in the carcinogenesis and progression of pituitary adenomas is still not understood in detail. Long noncoding RNA (lncRNA) colon cancer-associated transcript 2 (CCAT2), a newly identified lncRNA, has been reported to be abnormally expressed in some cancers. In the present study, we found that CCAT2 was significantly upregulated in pituitary adenomas tissues. Elevated CCAT2 expression was correlated with poor prognosis in patients with pituitary adenomas. Moreover, CCAT2 expression was activated by E2F1. Loss-of-function and gain-of-function assays showed that CCAT2 positively regulated pituitary adenoma cell proliferation, migration, and invasion. Further investigation demonstrated that CCAT2 interacted with PTTG1, and promoted its stability. Furthermore, CCAT2 affected the expression of downstream genes regulated by PTTG1, including SOX2, DLK1, MMP2, and MMP13. Cumulatively, CCAT2 functions as an oncogene in pituitary adenomas and its overexpression contributes to pituitary adenoma carcinogenesis and progression.
Keywords: CCAT2, PTTG1, pituitary adenomas
Introduction
The pituitary gland is the central mediator for peripheral endocrine homeostatic regulation via the secretion of tropic hormones such as thyroid stimulating hormone (TSH), growth hormone (GH), follicle-stimulating hormone (FSH), and luteinizing hormone (LH) [1]. Human pituitary adenomas are classified as clinically functioning or non-functioning. Functioning tumors secrete excess anterior pituitary hormones, causing hormone-specific clinical syndromes. Clinically non-functioning adenomas (NFA) account for more than 40% of all diagnosed pituitary tumors, which cannot produce excess functional hormones [2]. Some pituitary adenomas grow rapidly and cause the tumor mass effect i.e., the local compressive effect of large pituitary tumors on brain structures and cranial nerves. They can also invade downwards into the paranasal sinuses, laterally into the cavernous sinuses, and upwards into the parenchyma of the brain [3]. Medical approaches that target the underlying defects of tumor development are not yet available. Furthermore, the molecular mechanisms involved in the carcinogenesis and progression of pituitary adenomas is still not understood in detail.
Long non-coding RNAs (lncRNAs) are RNA molecules whose transcripts are more than 200 nt in length [5]. They cannot encode proteins, but play important roles in regulating gene expression via epigenetic or post-transcriptional mechanisms [6,7]. Although little is known about their carcinogenic mechanisms, many lncRNAs have been found to be abnormally expressed in tumors and play crucial roles in various biological events such as cell cycle distribution, cell differentiation, and tumorigenesis [4]. The differential expression and dysregulation of lncRNAs is believed to be involved in carcinogenesis and cancer progression, recurrence, and metastasis [5]. As a tumor suppressor gene, MEG3 is found to be downregulated in several types of cancers and its decreased expression is significantly correlated with poor prognosis [6,7]. The overexpression of MALAT1 is also confirmed in many malignant cancers and its aberrant elevation is demonstrated to promote tumor progression [8]. In recent years, some reports demonstrated that pituitary adenomas altered the expression profiles of lncRNAs [9]. Nevertheless, the clinical significance, biological function, and mechanisms of lncRNAs in the pathogenesis of pituitary tumors remain largely unknown. Only a small number of lncRNAs in pituitary adenomas have been validated so far.
lncRNA colon cancer-associated transcript 2 (CCAT2), a newly identified lncRNA, has been reported to be abnormally expressed in colon and breast cancers [10,11]. A previous study demonstrated that CCAT2 promoted breast cancer growth by regulating the WNT signaling pathway [11]. CCAT2 also facilitates growth via the upregulation of FOMX1 in hepatocellular carcinoma (HCC) [12]. To date, the functional roles of CCAT2 in pituitary adenomas remain unclear. In the present study, we found that CCAT2 promoted cell proliferation, migration, and invasion in pituitary adenomas cells. Mechanistic investigation demonstrated that CCAT2 was upregulated by E2F1, and interacted with PTTG1. Cumulatively, these results suggest that CCAT2 exerts oncogenic properties in pituitary adenomas.
Materials and methods
Cell culture and tissue samples
The HP75 cell line was purchased from Cell Bank of Chinese Academy of Sciences. The cells were cultured in Dulbecco’s modified eagle’s medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS, Gibco) at 37°C in 5% CO2. The tumor tissues were obtained from Cangzhou Central Hospital. All patients had received definite diagnosis by imaging, surgery, and pathological examination. No treatment has given to any patient before the surgery. All of the patients were provided written informed consent. The use of human specimens in this study was permitted by the local ethics committee at Cangzhou Central Hospital.
Overexpression or knockdown of CCAT2
For knockdown assay, two shRNAs against CCAT2 were designed. The target sequences of CCAT2 were provided as follow: sh1: 5’-GCCAGAGTTAATACCCTCA-3’, sh2: 5’-CTTAGACTGGGCTGTGTAT-3’. The target sequence of E2F1 was provided as follow: shE2F1: 5’-GGACCACCTGATGAATATC-3’. shRNAs against target genes were cloned into shRNA-expressing lentiviral vector pLKO.1. Production of lentiviral particles was performed according to the standard protocols. Cells were transduced with lentiviral constructs for 24 hours with 5 μg/ml polybrene (Sigma). After 48 hours, stable cells were selected by using 1 μg/ml puromycin for 7 days.
For overexpression, full-length human CCAT2 cDNA was cloned into lentiviral expressing vector pLV-puro. Production of lentiviral particles was performed according to the standard protocols. Cells were transfected with lentiviral constructs expressing empty vector or CCAT2 for 24 hours with 5 μg/ml polybrene (Sigma). After 48 hours, stable cells were selected by using 1 μg/ml puromycin for 7 days.
RNA isolation and real-time PCR (RT-PCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen) according to the standard protocol. RNA was reverse-transcribed using a High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s instructions. cDNA was quantified by RT-PCR using an ABI 7500 detection System (Applied Biosystems). RT-PCR was performed using SYBR Green mixture reagents (Roche). GAPDH was used as an internal control. Primers used for RT-RCR are listed as follow: GAPDH-F: GAAGTCCAAGAACCACATCCA, GAPDH-R: GCAGCTGCGTAGTACAGATATT; CCAT2-F: GGTGCTCCAGGCAATAACT, CCAT2-R: CTTCCTACAGGCCCAAACAT.
Cell proliferation assay
3 × 103 cells per well were seeded in the 96-well plate and incubated for different time point, respectively. Cell proliferation was measured with a Cell Counting Kit-8 (CCK-8) (Dojindo) following the manufacturer’s instructions. Absorbance was measured at 450 nm using Elx800 Reader (Bio-Tek Instruments, VT, USA).
Cell cycle assay
The cell cycle was analyzed using an in situ cell proliferation kit (Roche) according to the manufacturer’s instruction. The cell cycle distribution was analyzed by flow cytometry. The data were analyzed by FlowJo software (Tree Star).
Apoptosis assay
Cells were stained with fluorescein isothiocyanate-conjugated Annexin V and 7-AAD (Apoptosis Detection Kit, KeyGEN, Nanjing, China) according to the manufacturer’s instruction. Cells were analyzed with flow cytometer, and the data were studied using FlowJo software (Tree Star).
Western blot
The cells were lysed with RIPA buffer (Beyotime Biothechnology, Beijing, China). Protein lysates were separated by SDS-PAGE and then transferred onto PVDF membranes. The membranes were blocked and incubated with anti-PTTG1 (Abcam) and GAPDH antibody (Proteintech) at 4°C overnight. The membranes were incubated with HRP-conjugated anti-rabbit or -mouse secondary antibody. Signal was detected by an ECL system (Amersham Pharmacia, Piscataway, NJ).
Cell migration and invasion assay
Cells growing in the log phase were treated with trypsin and resuspended as a single-cell solution. A total of 1 × 105 cells in 0.2 mL serum-free DMEM were seeded on an 8-µm pore polycarbonate membrane Boyden chamber insert in a transwell apparatus (Costar, Cambridge, MA, USA), either coated with or without Matrigel (BD Biosciences, San Jose, CA, USA). DMEM containing 20% FBS was added to the lower chamber. After the cells were incubated for 24 h, cells on the top surface of the insert were removed by wiping with a cotton swab. Cells that migrated to the bottom surface of the insert were fixed in 100% methanol for 10 min, stained in 0.5% crystal violet for 10 min and then subjected to microscopic inspection. Values for invasion and migration were obtained by counting ten fields per membrane and representing the average of three independent experiments.
Chromatin immnoprecipitation (ChIP)
ChIP was performed using the EZ ChIPTM Chromatin Immunoprecipitation Kit (Millipore), according to the manufacturer’s instructions. The chromatin was immunoprecipitated using anti-E2F1 (Abcam) antibody. Normal rabbit immunoglobulin G (IgG) was used as a negative control. Primers for CCAT2 promoter regions are listed followed: F: GTGTGGACATAGAAGGCAGATAC, R: GTGGTTGGTTTGAAAGGAAGTG.
RNA immunoprecipitation (RIP)
RIP assays were performed as previously described [13]. Cells were subjected to a RIP assay by using 5 μg PTTG1 antibody (Abcam) or negative control IgG using RNA Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions.
RNA pull-down and mass spectrometry assay
RNA pull-down was performed as previously described [14]. In vitro biotin-labeled RNAs (CCAT2 and antisense CCAT2) were transcribed with the biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche) treated with RNase-free DNase I (Promega) and purified with RNeasy Mini Kit (QIAGEN). Biotinylated RNA was incubated with nuclear extracts of breast cancer cells, and pull-down proteins were run on SDS-PAGE gels. Mass spectrometry followed.
Statistical analysis
The Student’s t test (two-tailed), one-way analysis of variance, and the Mann-Whitney U test were conducted to analyze the in vitro and in vivo data by SPSS 17.0 software (IBM). P values less than 0.05 were considered significant.
Results
Elevated CCAT2 expression predicts poor prognosis in patients with pituitary adenomas
We performed RT-PCR to determine the differential expression of CCAT2 in pituitary adenoma tissues and the corresponding normal tissues from 74 patient samples. As shown in Figure 1A, CCAT2 expression levels were significantly higher in the pituitary adenoma patient samples than in normal pituitary tissues.
Figure 1.
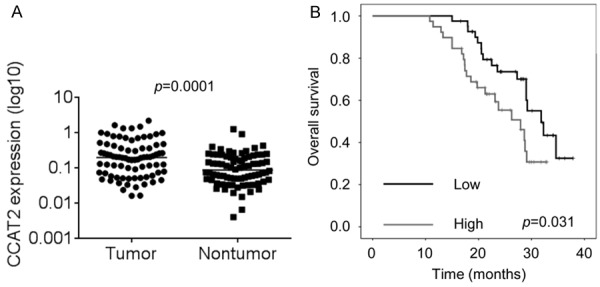
Elevated CCAT2 expression predicts poor prognosis of patients with pituitary adenomas. A. The CCAT2 expression levels in pituitary adenomas tissues and corresponding normal tissues from 74 patients were examined by RT-PCR. B. Kaplan-Meier survival curve and log-rank test were used to evaluate the association of CCAT2 expression with overall survival rate. Patients were segregated into CCAT2-high group and CCAT2-low according to the median of CCAT2 expression in pituitary adenomas tissues.
Next, we used the Kaplan-Meier survival analysis to examine the correlation between CCAT2 expression and the prognosis of patients with pituitary adenoma (Figure 1B). The results showed that patients with higher CCAT2 levels exhibited shorter overall survival time than those with lower CCAT2 levels. These findings suggest that elevated CCAT2 may exert an oncogenic function in pituitary adenomas.
E2F1 activates CCAT2 transcription
Although lncRNA dysregulation has been reported in various cancers, the regulators involved in the dysregulation of these molecules are not properly understood. Using the JASPAR online database, we chose to analyze the transcription factor E2F1, which was predicted to bind to the CCAT2 promoter region with high scores. We transfected HP75 cells with shRNA targeting E2F1. Interestingly, we found that E2F1 knockdown significantly inhibited CCAT2 expression (Figure 2A). Moreover, we designed a primer that covered the E2F1 binding site and performed ChIP assays followed by RT-PCR to validate the ability of E2F1 to bind to this site. We found that E2F1 bound to this site, and that E2F1 knockdown suppressed its binding levels (Figure 2B). Next, we constructed luciferase reporter plasmids containing the CCAT2 promoter region with wild-type or mutant E2F1 binding sites. Dual luciferase reporter assays showed that E2F1 increased the luciferase activity of the wild-type CCAT2 promoter, but had no effect on the CCAT2 promoter with the mutant E2F1 binding site (Figure 2C). These findings indicate that some transcription factors can contribute to human cancer development and progression not only by affecting the expression of the protein coding genes, but also by regulating noncoding genes, such as lncRNA transcription.
Figure 2.
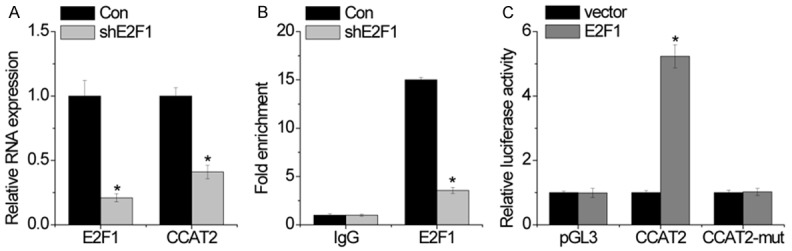
E2F1 activates CCAT2 transcription. A. The relative expression level of CCAT2 in control and E2F1-silencing cells was detected by RT-PCR. B. The binding of E2F1 and CCAT2 promoter was detected by ChIP assay. C. Luciferase assays of the cells indicated that were transfected with pGL3, pGL3-CCAT2, or pGL3-CCAT2-mut vectors, the E2F1 vector, or an empty vector. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
CCAT2 enhances cell proliferation, induces cell cycle progression, and inhibits cell apoptosis in pituitary adenoma cells
To determine the functional role of CCAT2 in pituitary adenomas, we introduced stable CCAT2 knockdown in HP75 cells via two different shRNA-expressing lentiviral particles. The RT-PCR results indicated that the CCAT2 expression was suppressed by both, sh1 and sh2 (Figure 3A). We found that the cell proliferation of HP75 cells with CCAT2 knockdown was significantly decreased when compared to the control cells using CCK-8 assays (Figure 3B). In contrast, we generated HP75 cells that overexpressed CCAT2 (Figure 3C). Overexpression of CCAT2 significantly enhanced cell proliferation (Figure 3D).
Figure 3.
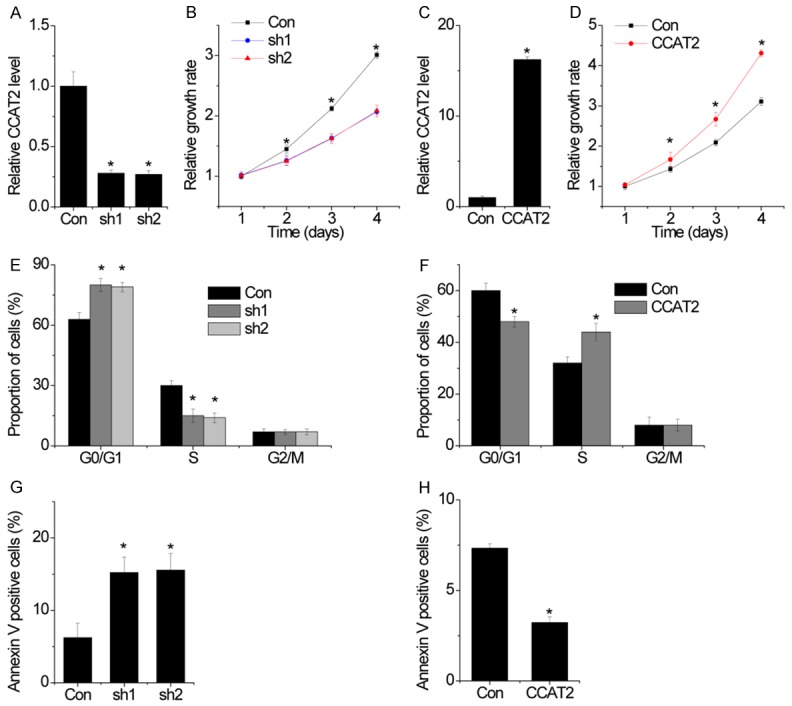
CCAT2 enhances cell proliferation, induced cell cycle progression and inhibits cell apoptosis in pituitary adenomas cells. A. The relative expression of CCAT2 in control and CCAT2-knockdown cells was detected by RT-PCR. B. Growth curves for HP75 cells after transfection with CCAT2 shRNA or the negative control were determined by CCK-8 assays. C. The relative expression of CCAT2 in control and CCAT2-overexpressing cells was detected by RT-PCR. D. Growth curves for HP75 cells after transfection with CCAT2 or empty vector were determined by CCK-8 assays. E. Cells in S-phase population were significantly decreased when CCAT2 was silenced in HP75 cells. F. FACS analysis showing significant increases or decreases of cells in S- or G-1 phase, respectively, in HP75 cells overexpressing CCAT2. G. Cells with silenced CCAT2 expression were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. H. Cells with CCAT2 overexpression were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
To gain insight into the mechanism by which CCAT2 regulated cell proliferation, we performed flow cytometry to analyze the effect of CCAT2 on cell cycle and apoptosis. We found that significant G1/S arrest was observed in CCAT2-silenced cells (Figure 3E), whereas CCAT2 overexpression demonstrated the opposite effect (Figure 3F). Moreover, apoptotic analyses showed that CCAT2-knockdown HP75 cells had a significantly higher percentage of Annexin V-positive cells when compared to the control cells (Figure 3G), whereas protective effects were observed in CCAT2-overexpressing HP75 cells (Figure 3H). Collectively, the data strongly suggested that CCAT2 promoted cell proliferation by facilitating cell cycle progression and inhibiting cell apoptosis.
CCAT2 promotes the migration and invasion of pituitary adenoma cells
To evaluate whether CCAT2 contributed to the progression of pituitary adenomas, we examined the effect of CCAT2 on the migration and invasive behavior of pituitary adenoma cells. Using a transwell assay, we found that the migration and invasive ability of the pituitary adenoma cells was dramatically inhibited following shRNA-mediated suppression of CCAT2 (Figure 4A). Conversely, CCAT2 overexpression increased the migration and invasive ability of the pituitary adenoma cells (Figure 4B). Collectively, the data suggests that CCAT2 plays a crucial role in pituitary adenoma progression.
Figure 4.
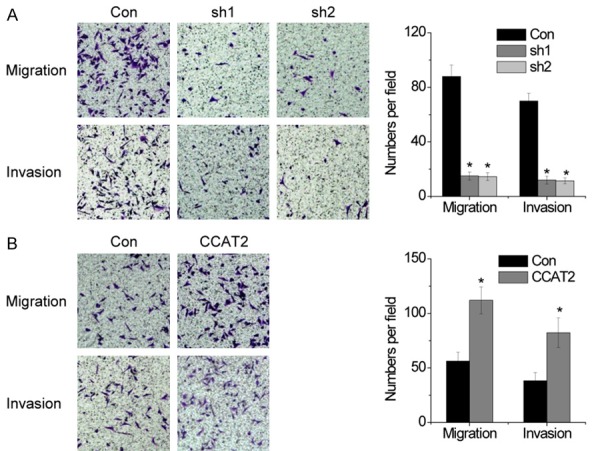
CCAT2 promotes the migration and invasion of pituitary adenomas cells. A. The migration and invasive ability after knockdown of CCAT2 in HP75 cells was assessed using transwell assays. B. The migration and invasive ability after CCAT2 overexpressing in HP75 cells was assessed using transwell assays. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
CCAT2 interacts with PTTG1
lncRNAs are considered to exert their functions through RNA-interacting proteins via various mechanisms. Therefore, we performed an RNA pull-down assay with biotin-labeled CCAT2, followed by mass spectrometry to search for potential CCAT2-associated proteins. The pituitary tumor transforming gene (PTTG1) was identified to potentially interact with CCAT2 in pituitary adenoma cells. Next, we performed the RIP assay and confirmed that CCAT2 directly bound to PTTG1 in HP75 cells (Figure 5A). Moreover, we constructed a series of CCAT2 truncations to map its binding fragment with PTTG1. We found that the 5’-end fragment of CCAT2 (0-600 nt, 1752 nt) was essential for binding PTTG1 (Figure 5B). These results demonstrate a direct interaction between CCAT2 and PTTG1.
Figure 5.
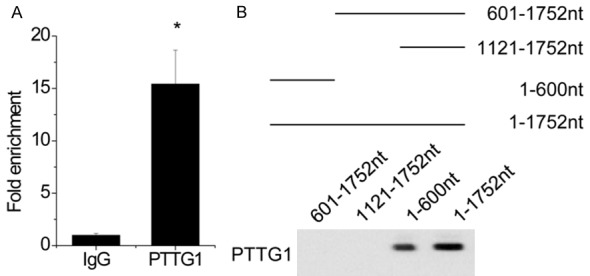
CCAT2 interacts with PTTG1. A. The interaction between CCAT2 and PTTG1 was detected by RIP assay. B. Deletion mapping of PTTG1-binding domain in CCAT2. (Up) The schematic diagram of full-length and deleted fragments of CCAT2; (Down) western blot of PTTG1 in protein samples pulled down by different CCAT2 fragments. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
CCAT2 suppresses the degradation of PTTG1
Next, we explored the functional relationship between CCAT2 and PTTG1. Silencing CCAT2 significantly inhibited the PTTG1 protein level, but had no influence on its mRNA level (Figure 6A). Conversely, overexpressing CCAT2 increased the PTTG1 protein level, but not the mRNA level (Figure 6B). These results strongly indicated that CCAT2 influenced the degradation of PTTG1. To further confirm this regulation, control and CCAT2-silenced HP75 cells were treated with the protein synthesis inhibitor, cycloheximide (CHX). We found that the half-life of PTTG1 was much shorter in CCAT2 knockdown cells than in control cells (Figure 6C). In contrast, CCAT2 upregulation significantly elongated the half-life of PTTG1 (Figure 6D). When MG132 (an inhibitor of proteasome degradation) was used, the PTTG1 protein level in CCAT2-knockdown HP75 cells was markedly upregulated and reached a level that was comparable to that in control cells (Figure 6E). These results suggest that CCAT2 is important for the stability of the PTTG1 protein.
Figure 6.
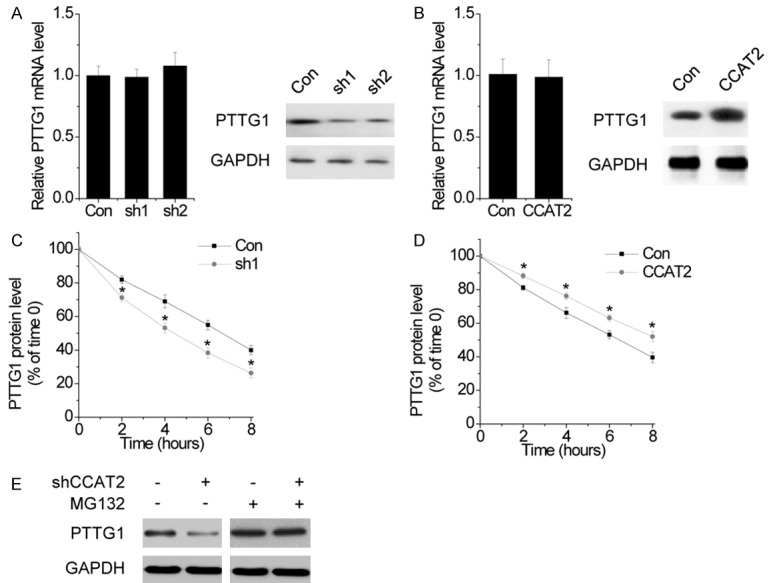
CCAT2 suppresses the degradation of PTTG1. A. The effect of CCAT2 knockdown on PTTG1 mRNA (left) and protein (right) level was determined by RT-PCR and western blot, respectively. B. The effect of CCAT2 overexpression on PTTG1 mRNA (left) and protein (right) level was determined by RT-PCR and western blot, respectively. C. Quantification of PTTG1 protein levels in control and CCAT2 knockdown cells treated with CHX. D. Quantification of PTTG1 protein levels of PTTG1 in control and CCAT2 overexpressing cells treated with CHX. E. Western blot of PTTG1 expression in control and CCAT2 knockdown cells treated with vehicle control or MG132. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
CCAT2 affects the expression of downstream genes regulated by PTTG1
The genes modulated by PTTG1 includes SOX2 [15], DLK1 [16], MMP2 [17], and MMP13 [18]. We hypothesized that CCAT2 probably regulated the expression of these genes through PTTG1. As shown in Figure 7A, overexpression of CCAT2 increased the expression of SOX2, DLK1, MMP2, and MMP13, whereas knockdown of PTTG1 abolished the upregulation of these genes. Moreover, SOX2, DLK1, MMP2, and MMP13 mRNA expression was inhibited by CCAT2 silencing, and this suppression was rescued once PTTG1 was restored (Figure 7B). Our results suggest that CCAT2 affects the expression of PTTG1-regulated genes.
Figure 7.
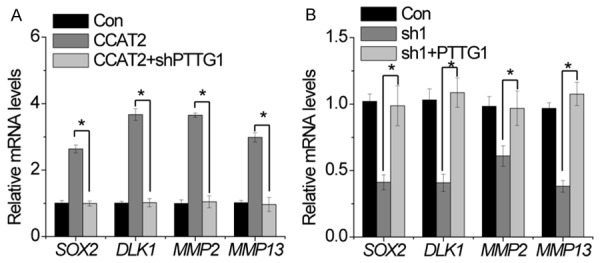
CCAT2 affects the expression of downstream genes regulated by PTTG1. A. RT-PCR of Sox2, DLK1, MMP2 and MMP13 in cells expressing control and CCAT2 with or without PTTG1 shRNA. B. RT-PCR of Sox2, DLK1, MMP2 and MMP13 in cells expressing control and CCAT2 shRNA with or without PTTG1. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
PTTG1 is crucial for CCAT2-mediated oncogenic properties
Next, we performed rescue experiments to determine the role of PTTG1 in CCAT2-mediated malignant phenotypes of pituitary adenoma cells. PTTG1 shRNA was transfected into HP75 cells overexpressing CCAT2. The cellular function assay showed that the enhanced cell proliferation, migration, and invasion induced by CCAT2 were abrogated by PTTG1 knockdown in HP75 cells (Figure 8A and 8B). Overall, we concluded that PTTG1 is crucial for CCAT2-mediated oncogenic properties.
Figure 8.
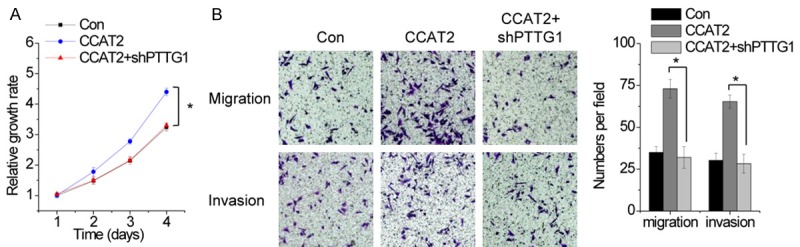
PTTG1 is crucial for CCAT2-mediated oncogenic properties. A. Downregulation of PTTG1 abolished the proliferation increased by CCAT2 overexpression. B. Downregulation of PTTG1 attenuated the migration and invasion induced by CCAT2 overexpression. Error bars indicate mean ± standard errors of the mean. *P < 0.05.
Discussion
lncRNAs play crucial roles in certain types of cancer progression. However, the biological functions and mechanisms of most lncRNAs in human pituitary adenomas still remain unclear. Here, we found that CCAT2 was significantly upregulated in pituitary adenoma tissues. Increased CCAT2 expression was associated with a poor prognosis in patients with pituitary adenomas. Through loss-of-function and gain-of-function assays, we showed that CCAT2 played a crucial role in pituitary adenoma cell proliferation, migration, and invasion. Knockdown of CCAT2 significantly decreased pituitary adenoma cell proliferation, induced cell cycle arrest and apoptosis, and inhibited migration and invasion, whereas overexpression of CCAT2 had the opposite effects. These findings suggest that CCAT2 functions as an oncogene in pituitary adenomas and its overexpression contributes to pituitary adenoma carcinogenesis and progression.
CCAT2, a lncRNA that spans the highly conserved 8q24 region harboring the rs6983267 SNP, was first discovered by Ling et al [19]. It induced chromosomal instability and metastases partly through the MYC and WNT signaling pathways [19], and reprogrammed energy metabolism in an allele-specific manner by interacting with the Cleavage Factor I (CFIm) complex in order to regulate alternative splicing of glutaminase [20]. A recent study demonstrated a positive feedback loop between CCAT2 and FOXM1, which played crucial roles in tumorigenesis of hepatocellular carcinoma [12]. The results suggested that the abnormal expression of CCAT2 potentially contributed to a highly malignant degree of cancer. However, the mechanisms by which CCAT2 exerts its oncogenic function on pituitary adenoma cells still remain elusive. Furthermore, we identified PTTG1 as a potential interacting protein of CCAT2 via RNA pull-down assays followed by mass spectrum analysis.
PTTG1, also known as securin, is a crucial component of the spindle checkpoint that controls faithful chromatid separation. It has also been identified as a proto-oncogene [21]. The PTTG1 protein is found at low levels in most normal adult tissues [3], but is upregulated in various tumors such as pituitary [22], lung [23], colorectal [24], and liver cancers [25]. PTTG1 plays a vital role in tumorigenesis. It also contributes to angiogenesis by transactivating the fibroblast growth factor 2 (FGF-2) and vascular endothelial growth factor (VEGF) via the Src homology 3 (SH3)-interacting domain [26]. As a transcriptional regulatory factor, PTTG1 exerts its transcriptional activity either by directly binding to DNA or by interacting with proteins [27], and is regulated by miRNAs and other transcriptional activators [28,29]. However, the upstream regulatory mechanisms of PTTG1 in pituitary adenomas are still unclear. In the present study, we revealed a novel post-translational method by which CCAT2 regulated PTTG1. RIP and RNA pull-down assays demonstrated a direct interaction between CCAT2 and PTTG1. CCAT2 did not influence the PTTG1 mRNA level, but significantly increased the PTTG1 protein level, suggesting that CCAT2 influenced the stability of PTTG1. CCAT2 and PTTG1 interaction could be used as a potential target for pituitary adenoma therapy. Moreover, our results also showed that CCAT2 affected the expression of downstream genes regulated by PTTG1 such as SOX2, DLK1, MMP2, and MMP13.
The regulators responsible for the abnormal expression of lncRNA in different cancers are still not thoroughly elucidated. Recently, emerging evidence has demonstrated that lncRNA expression can be regulated in a manner similar to protein coding genes. For example, epigenetic regulation modulates the expression of lncRNA SPRY4-IT1 and MEG3 via histone modification and DNA methylation respectively in lung cancer [30,31], and the transcription factor E2F1 activates ANRIL expression [32]. In the current study, we found that CCAT2 expression could also be activated by E2F1 after it bound to its promoter region. These results indicated that the unique lncRNA expression pattern found in different cancers may be associated with the regulation of different regulators.
In summary, these results demonstrate that CCAT2 overexpression plays an important role in human pituitary adenoma development and progression by promoting pituitary adenoma cell proliferation, migration, and invasion depending on the regulation of PTTG1 expression, which occurs after CCAT2 interacts with PTTG1 and suppresses its degradation. Additionally, CCAT2 overexpression in human pituitary adenoma cells occurs partly due to the transcription factor E2F1 binding to its promoter region and promoting transcription. Our findings further the understanding of pituitary adenoma pathogenesis and facilitate the development of therapeutics against this disease.
Disclosure of conflict of interest
None.
References
- 1.Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011;7:257–266. doi: 10.1038/nrendo.2011.40. [DOI] [PubMed] [Google Scholar]
- 2.Chaidarun SS, Klibanski A. Gonadotropinomas. Semin Reprod Med. 2002;20:339–348. doi: 10.1055/s-2002-36708. [DOI] [PubMed] [Google Scholar]
- 3.Asa SL, Ezzat S. The pathogenesis of pituitary tumours. Nat Rev Cancer. 2002;2:836–849. doi: 10.1038/nrc926. [DOI] [PubMed] [Google Scholar]
- 4.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 5.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Shi S, Jiang J, Li X, Lu H, Ren F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312–319. doi: 10.1016/j.biopha.2017.04.085. [DOI] [PubMed] [Google Scholar]
- 7.Hu D, Su C, Jiang M, Shen Y, Shi A, Zhao F, Chen R, Shen Z, Bao J, Tang W. Fenofibrate inhibited pancreatic cancer cells proliferation via activation of p53 mediated by upregulation of LncRNA MEG3. Biochem Biophys Res Commun. 2016;471:290–295. doi: 10.1016/j.bbrc.2016.01.169. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu X, Zhan Y, Liu L, Lin J, Zhou Q, Xu W, Zhao G, Cai Z, Huang W. Tetracycline-inducible shRNA targeting long non-coding RNA PVT1 inhibits cell growth and induces apoptosis in bladder cancer cells. Oncotarget. 2015;6:41194–41203. doi: 10.18632/oncotarget.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Li C, Liu C, Yu S, Zhang Y. Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in non-functioning pituitary adenomas and their relationship to tumor behavior. Pituitary. 2015;18:42–47. doi: 10.1007/s11102-014-0554-0. [DOI] [PubMed] [Google Scholar]
- 10.Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther. 2015;8:2657–2664. doi: 10.2147/OTT.S90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F, Bai G, Li Y, Feng Y, Wang L. A positive feedback loop of long noncoding RNA CCAT2 and FOXM1 promotes hepatocellular carcinoma growth. Am J Cancer Res. 2017;7:1423–1434. [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 14.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ, Yoo KC, Lee GH, Cui YH, Oh YS, Gye MC, Lee YY, Park IC, An S, Hwang SG, Park MJ, Suh Y, Lee SJ. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J Biol Chem. 2012;287:19516–19527. doi: 10.1074/jbc.M111.337428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espina AG, Mendez-Vidal C, Moreno-Mateos MA, Saez C, Romero-Franco A, Japon MA, Pintor-Toro JA. Induction of Dlk1 by PTTG1 inhibits adipocyte differentiation and correlates with malignant transformation. Mol Biol Cell. 2009;20:3353–3362. doi: 10.1091/mbc.E08-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li WH, Chang L, Xia YX, Wang L, Liu YY, Wang YH, Jiang Z, Xiao J, Wang ZR. Knockdown of PTTG1 inhibits the growth and invasion of lung adenocarcinoma cells through regulation of TGFB1/SMAD3 signaling. Int J Immunopathol Pharmacol. 2015;28:45–52. doi: 10.1177/0306419015572073. [DOI] [PubMed] [Google Scholar]
- 18.Lin YH, Tian Y, Wang JS, Jiang YG, Luo Y, Chen YT. Pituitary tumor-transforming gene 1 regulates invasion of prostate cancer cells through MMP13. Tumour Biol. 2015 doi: 10.1007/s13277-015-3796-1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, Fernandez AF, Valledor L, Van Roosbroeck K, Chang S, Shah M, Kinnebrew G, Han L, Atlasi Y, Cheung LH, Huang GY, Monroig P, Ramirez MS, Catela Ivkovic T, Van L, Ling H, Gafa R, Kapitanovic S, Lanza G, Bankson JA, Huang P, Lai SY, Bast RC, Rosenblum MG, Radovich M, Ivan M, Bartholomeusz G, Liang H, Fraga MF, Widger WR, Hanash S, Berindan-Neagoe I, Lopez-Berestein G, Ambrosio ALB, Gomes Dias SM, Calin GA. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell. 2016;61:640. doi: 10.1016/j.molcel.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- 22.Filippella M, Galland F, Kujas M, Young J, Faggiano A, Lombardi G, Colao A, Meduri G, Chanson P. Pituitary tumour transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas: a clinical and immunohistochemical study. Clin Endocrinol (Oxf) 2006;65:536–543. doi: 10.1111/j.1365-2265.2006.02630.x. [DOI] [PubMed] [Google Scholar]
- 23.Honda S, Hayashi M, Kobayashi Y, Ishikawa Y, Nakagawa K, Tsuchiya E. A role for the pituitary tumor-transforming gene in the genesis and progression of non-small cell lung carcinomas. Anticancer Res. 2003;23:3775–3782. [PubMed] [Google Scholar]
- 24.Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- 25.Cho-Rok J, Yoo J, Jang YJ, Kim S, Chu IS, Yeom YI, Choi JY, Im DS. Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver cancer cell growth in vitro and in vivo. Hepatology. 2006;43:1042–1052. doi: 10.1002/hep.21137. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa H, Heaney AP, Yu R, Horwitz GA, Melmed S. Human pituitary tumor-transforming gene induces angiogenesis. J Clin Endocrinol Metab. 2001;86:867–874. doi: 10.1210/jcem.86.2.7184. [DOI] [PubMed] [Google Scholar]
- 27.Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B, Hou Z, Li C, Tong Y. MiRNA-494 inhibits metastasis of cervical cancer through Pttg1. Tumour Biol. 2015;36:7143–7149. doi: 10.1007/s13277-015-3440-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhou C, Wawrowsky K, Bannykh S, Gutman S, Melmed S. E2F1 induces pituitary tumor transforming gene (PTTG1) expression in human pituitary tumors. Mol Endocrinol. 2009;23:2000–2012. doi: 10.1210/me.2009-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, Lu BB, Yin R, Zhang EB, Xu L, De W, Wang ZX. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


