Abstract
This study was designed to investigate the significance of the effect of miR-34a on 5-fluorouracil (5-FU) sensitivity in vitro and in vivo. miR-34a expression in tumor tissues or serum was determined by quantitative polymerase chain reaction. CRC cell lines HCT116 and SW480 were used to evaluate cell viability, cell apoptosis, and the cell cycle using a cell proliferation assay, flow cytometry, and Western blotting, respectively. For the in vivo studies, xenografts derived from SW480 cells were established to assess the antitumor activity between miR-34a and 5-FU. Patients with high levels of miR-34a expression were found to benefit more from 5-FU-based chemotherapy than patients with low levels of miR-34a expression, regardless of disease stage. Ectopic expression of miR-34a alone or 5-FU alone was found to inhibit CRC cell growth in vitro and in vivo. Moreover, cell growth in vitro and in vivo was further inhibited when miR-34a combined with 5-FU through increasing the rate of cell apoptosis. The potential targets of miR-34a, including CREB1, Bcl-2, Notch 1, Sirt1, and E2F3, were predicted and preliminarily validated and merit further study. Conclusion: miR-34a might function as a predictor of fluorouracil chemosensitivity in CRC, and a combination strategy of miR-34a with fluorouracil was expected to be more beneficial for CRC patients.
Keywords: miR-34a, colorectal cancer, 5-fluorouracil, chemosensitivity
Introduction
Although the incidence of colorectal cancer (CRC) has gradually increased worldwide, patient prognosis has improved over the past few years [1]. According to CRC guidelines issued by the NCCN, chemotherapy is necessary for patients with stage III or IV CRC and some patients with stage II CRC. Fluorouracil-based chemotherapy has been the most common regimen for CRC over the past fifty years [2,3]. Allegra et al. reported that the 3-year disease-free survival rates (3-year DFS) of stage II and III CRC patients who received fluorouracil-based adjuvant chemotherapy were 85.4% and 71.7%, respectively [4]. Among stage IV CRC patients, about 55.0% responded to systematic chemotherapy [5]. These data suggested that there was readily visible heterogeneity between patients [4-8], and that predictive markers were needed to guide adjuvant or systematic chemotherapy, followed by optimizing strategies to enhance sensitivity to chemotherapy.
microRNAs (miRNAs) constitute a class of non-coding RNA molecules of approximately 18-25 nucleotides in length [9,10]. Increasing amounts of evidence have indicated that microRNAs (miRNAs) play various roles in multiple cancers, including CRC [9-13]. miRNAs can be easily detected from a variety of samples, such as tissues, plasma, serum, and stools [14]. Our previous study demonstrated that miR-34a was downregulated in CRC tumor tissues, and miR-34a expression was positively correlated with disease-free survival (DFS) in stage II/III CRC patients [15]. In our study we found that patients with low levels of miR-34a tend to early recurrence than patients with high levels of miR-34a among patients who received fluorouracil-based adjuvant chemotherapy (detailed results are shown in the text). It has been suggested that miR-34a has some synergistic effect on fluorouracil.
Akao Y et al reported similar findings, specifically that miR-34a dysregulation could induce 5-fluorouracil (5-FU) resistance in human CRC DLD-1 cells [16]. This study was designed to establish the association between miR-34a and 5-FU through analysis of the relationship of miR-34a expression to clinical response in patients treated with fluorouracil-based chemotherapy. We conducted an in vitro cell experiment and in vivo animal experiment to assess the influence of miR-34a on the sensitivity to 5-FU in CRC and to determine its underlying mechanism.
Materials and methods
Patients and sample collection
Data concerning patients with stage II/III cases were extracted directly from our previous study [15]. Here, 37 CRC patients with stage IV treated in our department from July 2013 to June 2015 were included in this study. These 37 patients provided serum samples prior to chemotherapy. All serum samples were stored at -80°C for future research. The patients received first-line fluorouracil-based chemotherapy. The clinical data mainly included patients’ clinical responses, which were obtained from their medical records. Clinical response evaluation was performed by computed tomography (CT) scan according to the RECIST 1.1 criteria. Response evaluation included partial response (PR; at least a 30% decrease in the sum of diameters of target lesion), progressive disease (PD; at least a 20% increase in the sum of diameters of target lesions), and stable disease (SD; neither sufficient shrinkage to qualify for PR nor sufficient increase to qualify for PD). All patients provided written informed consent for their samples to be used in research, and this study was approved by the ethics committee of Peking University Cancer Hospital.
Cell lines and antibodies
Seven human CRC cell lines were used here: Caco 2, HT29, HCT116, HCT116 p53-/-, Lovo, SW620, and SW480. Caco 2, HT29, HCT116, Lovo, SW620, and SW480 were purchased from ATCC and cultured in RPMI-1640 medium (Gibco BRL, Carlsbad, CA, U.S.) supplemented with 10% fetal bovine serum (Gibco BRL). HCT116 p53-/- was provided by Professor Bert Vogelstein of Johns Hopkins University and cultured in McCoy’s 5A modified medium (Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL). All cell lines were incubated in a humidified 37°C incubator under 5% CO2. Antibodies of cleaved caspase 9 (#7237), cleaved caspase 8 (#8592), cleaved PARP (9532), and Bcl-2 (#15071) were purchased from Cell Signal Technology (CST, Danvers, MA). Antibody of β-actin (#014M4759) was purchased from Sigma-Aldrich (St. Louis, MO).
Analyses of miR-34a expression
Data of miR-34a expression from tumor tissues was collected from our previous reported study. RNA extraction from serum samples and cell line was conducted using a Trizol LS kit (Invitrogen, Carlsbad, CA, U.S.) and Trizol (Invitrogen) according to the manufacturers’ instructions. After normalization with endogenous control RNU6B, miR-34a expression was evaluated, which was described in previous reports [15].
Cell viability assay
HCT116 and SW480 cells were transfected with miR-34a or negative control miR-Ctrl (GenePharma, Shanghai, China) using lipofectamine 2000 (Invitrogen). After 48 h of transfection, cell viability was analyzed using a Cell Counting Kit-8 (CCK8, Dojindo) according to the manufacturer’s instructions. Absorbance was measured at 450 nm using the microplate spectrophotometer once a day for 5 consecutive days. All reactions were run in triplicate.
Annexin V apoptosis assay
HCT116 and SW480 cells transfected with miR-34a or negative control miR-Ctrl were harvested and stained with phycoerythrin (PE)-annexin V and 7-amino-actinomycin (7-AAD) (BD Biosciences, Erembodegem, Belgium) at room temperature in the dark for 15 min. Cells were then detected using flow cytometry and analyzed using FlowJo 7.6 software (FlowJo, LLC.).
Cell cycle assay
Forty-eight hours after transfection with miR-34a or miR-Ctrl, cells were harvested and fixed in 70% cold ethanol for at least 12 h at 4°C. Fixed cells were treated with DNase-free RNaseA at 37°C for 30 min followed by staining with 50 μg/mL propidium iodide (BD Biosciences) at room temperature in the dark for 30 minute. Cells were then detected by FACS Calibur system (BD Biosciences) and results were analyzed with ModFit 3.0 software (BD Biosciences).
Western blot analysis
Total protein was extracted from cell or tissue samples using RIPA Lysis Buffer (Beyotime, Shanghai, China) on ice. After the determination of protein concentration, about 50 micrograms of protein was separated on 10% SDS-PAGE, after which protein was transferred to nitrocellulose membranes (GE Healthcare, Piscataway, NJ, U.S.). Nitrocellulose membrane was then incubated with primary antibodies overnight at 4°C and secondary antibody at room temperature for 1 h (antibodies are depicted in Supplementary Table 1). Proteins were visualized using ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Animal experiments
SW480 cells with miR-34a and miR-Ctrl expression were established using a lentivirus system (GenePharma, Shanghai, China) and injected subcutaneously into the dorsal flanks of 6-week-old NOD/SCID mice (Beijing HFK Bio-Technology Co, LTD., Beijing, China) to establish SW480-derived xenografts. When tumors reached approximately 150 mm3, mice were randomly divided into two groups (n = 5 per group): 5-FU group (15 mg/kg, intraperitoneal injection, twice a week for 2 weeks) and control group (0.9% saline solution, intraperitoneal injection, twice a week for 2 weeks) for xenografts with miR-34a or miR-Ctrl expressions. Tumor size and body weight were measured with digital calipers and electronic scale every three days, and the tumor volume was calculated using the following formula: Volume = 0.5 × (Length × Width2), where length was the long diameter and width was the short diameter of xenografts. The antitumor activity was depicted by tumor growth inhibition (TGI). TGI was calculated as the following formula: TGI = ΔT/ΔC × 100% (ΔT = tumor volume change of experimental group indicated by tumor volume at the final day of study minus tumor volume at the initial treatment, ΔC = tumor volume change of control group indicated by tumor volume at the final day of study minus tumor volume at the initial treatment). Animal experiments were approved by the independent ethics committee at Peking University Cancer Hospital.
Prediction and validation of targets of miR-34a
In this study, three online target prediction software packages including TargetScan (http://www.targetscan.org/), DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r = microtv4/index), and miRDB (http://microrna.org) were used to predict possible target genes of miR-34a. Moreover, quantitative PCR for assessment of the expression of CREB1, Bcl-2, Notch1, Sirt1, E2F3, HDAC1, and BIRC5 was conducted using SYBR Green master mixture (Applied Biosystems) with the housekeeping gene GAPDH serving as an internal control. The primers for CREB1, Bcl-2, Notch1, Sirt1, E2F3, HDAC1, and BIRC5 are listed in Supplementary Table 1.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5 (La Jolla, CA, U.S.). Crosstable analysis was employed to analyze the difference of 3-year recurrence rate between patients with high or low miR-34a expression. The differences in miR-34a expression between different groups were analyzed using Wilcoxon rank-sum tests. Wilcoxon rank-sum tests were also used to analyze the correlations of miR-34a expression with cell viability, cell apoptosis, and tumor growth. A two-side P value <0.05 was considered statistically significant.
Results
miR-34a expression acts as a predictor for fluorouracil sensitivity in CRC patients
The correlation between miR-34a expression in tumor tissues and 3-year recurrence rate was analyzed in cohort 1 including 91 patients with stage II/III CRC. The characteristics of patients including gender, age, primary tumor sites, differentiation, depth of penetration, and lymph node metastasis were listed in Table 1. According to our previous study, we set the expression level of 0.3072866 as a cutoff value [15], and miR-34a expression higher than the cut-off level was defined as high expression. Among patients with adjuvant chemotherapy, the 3-year recurrence rate in patients with high levels of miR-34a expression was significantly lower than in patients with low levels of miR-34a expression (37.0% vs. 75.0%, P<0.01; Figure 1A). These results suggested that miR-34a expression was closely related to the patient response to fluorouracil-based adjuvant chemotherapy.
Table 1.
Characteristics of patients in cohort I
| Characteristics | Recurrence group | Non-recurrence group | P | ||
|---|---|---|---|---|---|
|
|
|||||
| No. | % | No. | % | ||
| Gender | |||||
| Male (n = 57) | 31 | 34.1 | 26 | 28.6 | 0.136 |
| Female (n = 34) | 13 | 14.3 | 21 | 23.1 | |
| Age (years) | |||||
| <60 (n = 50) | 25 | 27.5 | 25 | 27.5 | 0.728 |
| ≥60 (n = 41) | 19 | 20.9 | 22 | 24.2 | |
| Tumor site | |||||
| Colon (n = 43) | 20 | 22.0 | 23 | 25.3 | 0.740 |
| Rectum (n = 48) | 24 | 26.4 | 24 | 26.4 | |
| Differentiation* | |||||
| Good (n = 71) | 35 | 38.5 | 36 | 39.6 | 0.734 |
| Poor (n = 20) | 9 | 9.9 | 11 | 12.1 | |
| Depth of penetration | |||||
| T3 (n = 27) | 12 | 13.2 | 15 | 16.5 | 0.628 |
| T4 (n = 64) | 32 | 35.2 | 32 | 35.2 | |
| Lymph node | |||||
| No (n = 38) | 14 | 15.4 | 24 | 26.4 | 0.005 |
| N1 (n = 28) | 11 | 12.1 | 17 | 18.7 | |
| N2 (n = 25) | 19 | 20.9 | 6 | 6.6 | |
| Lymph-vascular Invasion | |||||
| Yes (n = 21) | 14 | 15.4 | 7 | 7.7 | 0.056 |
| No (n = 70) | 30 | 33.0 | 40 | 44.0 | |
Good including well-differentiated and moderately differentiated adenocarcinoma; Poor including poor-differentiated adenocarcinoma, mucinous adenocarcinoma, and signet ring cell carcinoma.
Figure 1.
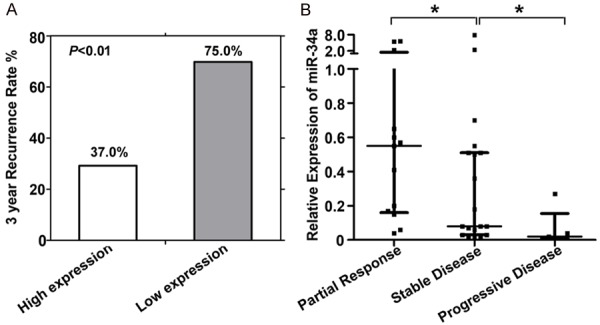
(A) Correlations of miR-34a level with 3-year recurrence rate in stage II/III patients, and (B) correlations of miR-34a level with clinical response evaluation in stage IV patients. Partial response, at least a 30% decrease in the sum of diameters of target lesion; progressive disease, at least a 20% increase in the sum of diameters of target lesions; stable disease, neither sufficient shrinkage to qualify for partial response nor sufficient increase to qualify for progressive disease; *P<0.05.
To further validate the hypothesis given above, we analyzed the association between miR-34a expression and clinical response in 37 stage IV CRC patients treated with first-line fluorouracil-based regimens. The basic characteristics of patients with stage IV CRC and clinical response evaluations were shown in Table 2. The numbers of patients with PR, SD, and PD were 13 (35.1%), 19 (51.4%), and 5 (13.5%), respectively. As expected, patients with partial response had higher levels of miR-34a expression than patients with stable disease or progressive disease (Figure 1B). For another, patients with high levels of miR-34a expression could benefit more from fluorouracil-based chemotherapy than other patients. Results indicated that miR-34a expression could predict the patient response to fluorouracil and that there could be some synergy between miR-34a and fluorouracil.
Table 2.
Characteristics of patients in cohort II
| Partial response (n = 13) | Disease stable (n = 19) | Progressive disease (n = 5) | |
|---|---|---|---|
| Age, years | |||
| Median (range) | 57 (29-70) | 56 (31-72) | 62 (55-79) |
| Gender | |||
| Female/Male | 7/6 | 9/10 | 3/2 |
| Tumor site | |||
| Colon/Rectum | 11/2 | 14/5 | 4/1 |
| Differentiation | |||
| Good/Poor | 11/2 | 16/3 | 4/1 |
Ectopic expression of miR-34a enhanced the ability of cell growth inhibition of 5-fluorouracil
There was less miR-34a expression in CRC tissues than in matched non-tumor tissues in our previous study, which we here confirmed. Compared to three normal colon tissues that served as surrogates due to the absence of normal colon cell lines, miR-34a expression was significantly downregulated in seven CRC cell lines (Figure 2A). As shown in a previous study, both miR-34a and 5-FU alone could inhibit the growth of CRC cells. The current work addresses whether miR-34a has any synergistic effect with 5-FU. HCT116 and SW480 cell lines were used in the following experiments, and ectopic expression of miR-34a was confirmed by quantitative polymerase chain reaction (PCR; Figure 2B). After treatment, cell viability indicated by OD450 was highest in cells treated with miR-Ctrl (the mean OD450 was 6.49 in HCT116 cell and 10.47 in SW480 cell), moderate in cells treated with miR-34a (the mean OD450 was 3.55 in HCT116 cell and 6.20 in SW480 cell) or miR-Ctrl+5-FU (the mean OD450 was 2.13 in HCT116 cell and 6.11 in SW480 cell), and lowest in cells treated with miR-34a+5-FU (the mean OD450 was 0.98 in HCT116 cell and 2.42 in SW480 cell). These results suggested that miR-34a could increase the sensitivity of CRC cells to 5-FU in vitro (Figure 2C and 2D).
Figure 2.
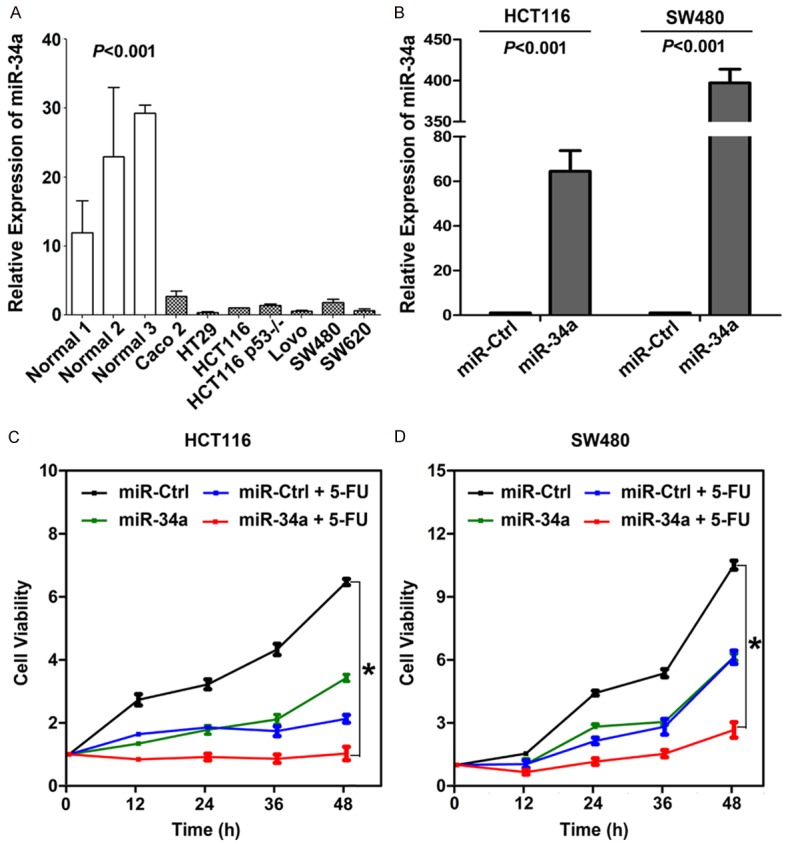
(A) Levels of miR-34a in CRC cells were here found to be higher than in normal colon tissues (P<0.001). (B) miR-34a expression validation in HCT116 and SW480 after ectopic expression (P<0.001). (C, D) miR-34a administered in combination with 5-FU resulted in significantly lower viability in (C) HCT116 cells and (D) SW480 cells than in any other group (*P<0.05).
miR-34a increased 5-FU sensitivity by inducing cell apoptosis
To explore the mechanisms by which miR-34a increased 5-FU sensitivity, we used flow cytometry to analyze the cellular apoptotic rate. As shown in Figure 3A-D, compared to control (8.1% and 13.7% in HCT116 and SW480), miR-34a (16.8% and 17.5% in HCT116 and SW480) or 5-FU (10.5% and 16.1% in HCT116 and SW480) alone could increase the proportion of apoptotic cells in both cells. Moreover, more apoptotic cells in both cells were found when miR-34a combined with 5-FU (20.5% and 19.8% in HCT116 and SW480). Induction of cell apoptosis was further confirmed by western blot to detect apoptosis-related proteins. Accompanied by cell apoptosis, the expression of cleaved caspase-9, caspase-8 and PARP was enhanced, but the anti-apoptosis gene Bcl-2 was inhibited (Figure 3E). These data suggested that induction of cell apoptosis was one of the mechanisms involved in miR-34a expression increasing 5-FU sensitivity.
Figure 3.
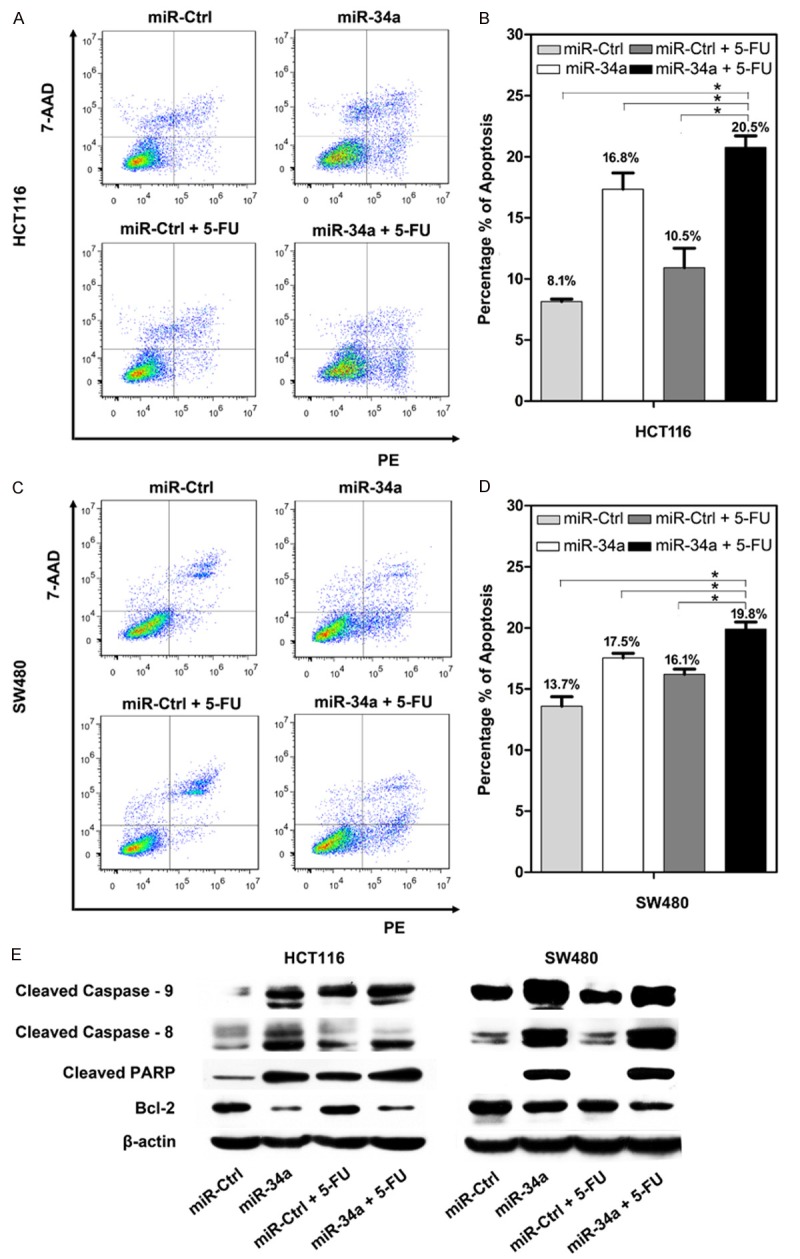
Cell apoptosis were detected in (A and B) HCT116 and (C and D) SW480 cells after treatment with miR-Ctrl, miR-34a, miR-Ctrl+5-FU, and miR-34a+5-FU (*P<0.05). The expression levels of apoptotic proteins including cleaved caspase-9, 8, cleaved PARP, and Bcl-2 were evaluated in HCT116 and SW480 cells (E).
miR-34a increased the inhibitory activity of 5-FU in vivo
SW480 cells transfected with miR-34a or miR-Ctrl were confirmed by quantitative PCR (Figure 4A) and then subcutaneously inoculated into the dorsal flank of NOD/SCID mice to establish xenografts in vivo. After the tumors reached about 150 mm3, mice were treated with 5-FU or 0.9% saline solution as a control. We found that, unlike in cells transfected with miR-Ctrl and treated with 0.9% saline solution, the growth of tumors in other three groups was suppressed to a certain extent. Tumor suppression was most significant in cells transfected with miR-34a and treated with 5-FU, with tumor growth inhibition (TGI) 91% (Figure 4B and 4C), which was consistent with the results shown in Figure 2 and confirmed that miR-34a increased the antitumor activity of 5-FU in CRC in vitro and in vivo.
Figure 4.
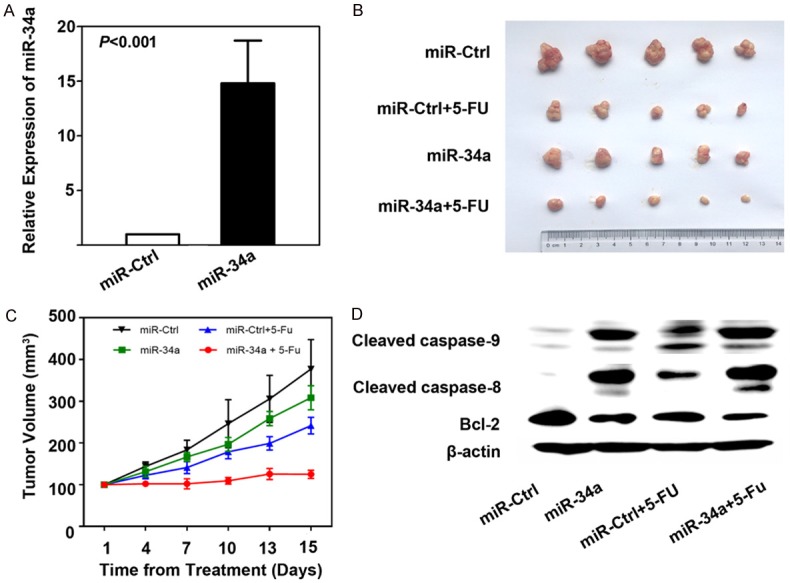
A. After stable expression of miR-34a, the level of miR-34a in SW480 cells was confirmed (P<0.001). B. The gross specimens of xenografts in mice after treated with miR-Ctrl, miR-34a, miR-Ctrl+5-FU, and miR-34a+5-FU. C. The growth curves of xenografts during the treatment with miR-Ctrl, miR-34a, miR-Ctrl+5-FU, and miR-34a+5-FU. Tumor volume was expressed as Mean ± S.D. D. The expression levels of apoptotic proteins including cleaved caspase-9, 8, and Bcl-2 were evaluated in xenografts.
As presented above, cell apoptosis was also analyzed using Western blot analysis to detect the expressions of apoptosis-related proteins in xenografts. As shown in Figure 4D, cell apoptosis was confirmed according to the changes in cleaved caspase-9, caspase-8, and Bcl-2.
Potential target genes of miR-34a
To facilitate further study of the underlying mechanism, we predicted possible targets of miR-34a by searching three reliable databases (MicroRNA-org, DIANA, TargetScan). Figure 5A lists several target genes including CREB1, Bcl-2, Notch 1, Sirt1, E2F3, HDAC1, and BIRC5, which were reported to be involved in the regulation of 5-FU chemosensitivity [15-20], and the binding sites of miR-34a in 3’-UTR were also presented in Figure 5B. Our results demonstrated that the expression of CREB1, Bcl-2, Notch 1, Sirt1, and E2F3 was decreased in HCT116 cells transfected with miR-34a compared to miR-Ctrl (Figure 5C), which suggested these genes might be the targets of miR-34a. However, whether these genes are the direct targets of miR-34a remains unclear and needs to be validated further.
Figure 5.
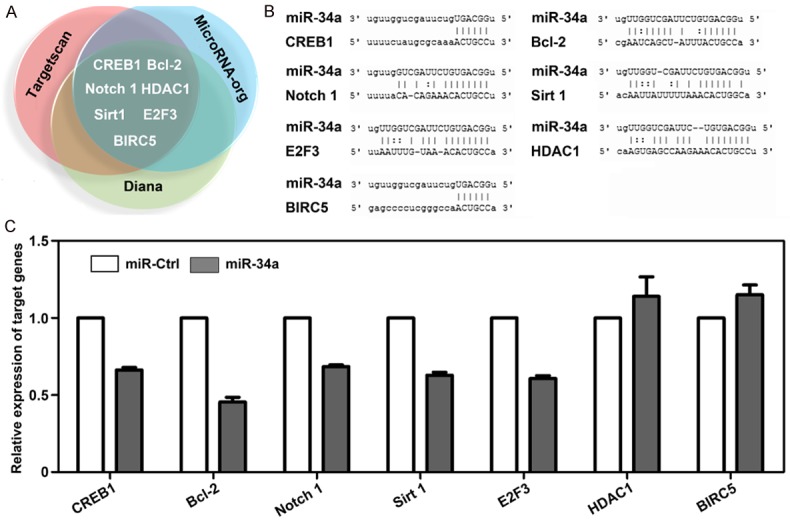
A. CREB1, Bcl-2, Notch 1, Sirt1, E2F3, HDAC1, and BIRC5 were predicted as targets of miR-34a by three prediction software packages. B. The potential binding sites of miR-34a in 3’UTR of each target gene. Alignment in the figure indicated by vertical lines means the direct binding site of miR-34a with each target gene. C. After ectopic expression of miR-34a, the expressions of CREB1, Bcl-2, Notch 1, Sirt1, and E2F3 were decreased, however, the expressions of HDAC1 and BIRC5 were increased.
Discussion
miR-34a has been confirmed to be downregulated in multiple tumors, including CRC, in several studies [15,21-24]. We previously demonstrated that the expression of miR-34a can serve as an independent prognostic factor for recurrence [15]. In this study, we analyzed the 3-year recurrence rate of 91 stage II/III CRC patients who received fluorouracil-based adjuvant chemotherapy stratified by miR-34a expression. We found that patients with high levels of miR-34a had lower 3-year recurrence rate than patients with low levels of miR-34a. It is here suggested that miR-34a has some relationship to fluorouracil.
To validate the above, we evaluated patients with stage IV CRC treated with first-line fluorouracil-based regimens. As expected, patients with high levels of miR-34a expression were found to benefit more from fluorouracil-based chemotherapy than patients with lower levels of expression. Our results indicate for the first time that miR-34a expression could predict the patient response to fluorouracil-based chemotherapy in patients. In clinical practice, almost all patients received fluorouracil as part of their combination regimens. As a consequence, our results suggested that there might be some correlation between miR-34a and fluorouracil, which we confirmed using in vitro cell experiments and in vivo animal experiments.
Our cell viability assay in HCT116 and SW480 cells demonstrated that cell growth was significantly inhibited by miR-34a combined with 5-FU relative to miR-34a or 5-FU alone. This was further confirmed in SW480-cell-derived xenografts. These results suggested that miR-34a could increase 5-FU therapeutic sensitivity in vitro and in vivo. Consistent with our results, other groups reported that miR-34a was downregulated in 5-FU-resistant DLD-1 cells, and that miR-34a could resensitize 5-FU-resistant DLD-1 cells to 5-FU. All of this data showed that miR-34a was promising for use in patients to enhance the sensitivity of 5-FU treatments.
To identify the mechanisms involved in the increased effects of miR-34a and 5-FU, we evaluated two important processes: the cell cycle and cell apoptosis. Consistent with our previous study, significant induction of miR-34a-mediated apoptosis via the caspase apoptosis pathway was verified in this study. There was less overall 5-FU-mediated apoptosis than miR-34a apoptosis, but the most apoptotic cells were observed when miR-34a and 5-FU were administered in combination than miR-34a or 5-FU alone in both cell lines. There were also differences in the make-up of apoptosis-related proteins, which were assessed in both in vitro cells (Figure 3) and in vivo xenografts (Figure 4).
In regard to cell cycle, compared to control, miR-34a was confirmed to induce cell cycle arrest at G1 phase, however, 5-FU was verified to induce cell cycle arrest at S phase. As a result, the progress of the cell cycle in cells exposed to both miR-34a and 5-FU might be complicated and the results may be difficult to interpret, which was shown in Supplementary Figure 1.
MicroRNA acts by regulating target genes, and several target genes have been reported to be regulated by miR-34a, such as Notch1, c-Myc, Sirt1, Bcl-2, LDHA, and KLF4. In this study, three reliable databases (MicroRNA-org, DIANA, TargetScan) were employed to predict the possible target genes of miR-34a. Seven potential targets including CREB1, Bcl-2, Notch 1, Sirt1, E2F3, HDAC1, and BIRC5 were screened and preliminarily validated. We found that ectopic expression of miR-34a could not suppress the expression of HDAC1 or BIRC5 at the mRNA level. Among the remaining five genes downregulated by miR-34a, further studies would be conducted to confirm the genes directly regulated by miR-34a and involved in 5-FU sensitivity. It was reported that Bcl-2 was an integral outer mitochondrial membrane protein that blocked the apoptotic death of some cells such as lymphocytes [27]. Moreover, numerous studies reported that Bcl-2 participated in regulating chemosensitivity in multiple tumors [17]. Wu and his colleagues reported that the phosphorylated Bcl-2 regulated by PXN contributed to 5-FU based chemotherapy resistance in CRC [17]. Similarly, numerous studies indicated that CREB1, Notch 1, Sirt1, and E2F3 were reported to play a role of oncogenes and involved in regulating response to chemotherapy [16,18-22]. Further studies would be conducted to identify the direct genes regulated by miR-34a and involved in 5-FU sensitivity.
Also, there are limitations in our study and the results were needed to be further validated due to the relative small samples of stage IV CRC patients. In addition, more mechanisms underlying cell proliferation and chemosensitivity would be investigated such as autophagy and necrosis, and the identification of target genes of miR-34a would be continued. In summary, we found for the first time that miR-34a might function as a predictor of fluorouracil chemosensitivity in patients with CRC, and a combination strategy of miR-34a with fluorouracil was expected to be more beneficial for CRC patients.
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Benson AB 3rd, Bekaii-Saab T, Chan E, Chen YJ, Choti MA, Cooper HS, Engstrom PF, Enzinger PC, Fakih MG, Fenton MJ, Fuchs CS, Grem JL, Hunt S, Kamel A, Leong LA, Lin E, May KS, Mulcahy MF, Murphy K, Rohren E, Ryan DP, Saltz L, Sharma S, Shibata D, Skibber JM, Small W Jr, Sofocleous CT, Venook AP, Willett CG, Gregory KM, Freedman-Cass DA National Comprehensive Cancer Network. Localized colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11:519–28. doi: 10.6004/jnccn.2013.0069. [DOI] [PubMed] [Google Scholar]
- 3.Benson AB 3rd, Venook AP, Bekaii-Saab T, Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ, Fuchs CS, Grem JL, Grothey A, Hochster HS, Hunt S, Kamel A, kirilcuk N, Leong LA, Lin E, Messersmith WA, Mulcahy MF, Murphy JD, Nurkin S, Rohren E, Ryan DP, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Gregory KM, Freedman-Cass D. Rectal Cancer, Version 2.2015. J Natl Compr Canc Netw. 2015;13:719–28. doi: 10.6004/jnccn.2015.0087. [DOI] [PubMed] [Google Scholar]
- 4.Allegra CJ, Yothers G, O’Connell MJ, Sharif S, Petrelli NJ, Lopa SH, Wolmark N. Bevacizumab in stage II-III colon cancer: 5-year update of the National Surgical Adjuvant Breast and Bowel Project C-08 trial. J. Clin. Oncol. 2013;31:359–64. doi: 10.1200/JCO.2012.44.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassidy J, Tabernero J, Twelves C, Brunet R, Butts C, Conroy T, Debraud F, Figer A, Grossmann J, Sawada N, Schoffski P, Sobrero A, Van Cussem E, Diaz-Rubio E. XELOX (capecitabine plus oxaliplatin): active first-line therapy for patients with metastatic colorectal cancer. J. Clin. Oncol. 2004;22:2084–91. doi: 10.1200/JCO.2004.11.069. [DOI] [PubMed] [Google Scholar]
- 6.Schmoll HJ, Twelves C, Sun W, O’Connell MJ, Cartwright T, McKenna E, Saif M, Lee S, Yothers G, Haller D. Effect of adjuvant capecitabine or fluorouracil, with or without oxaliplatin on survival outcomes in stage III colon cancer and the effect of oxaliplatin on post-relapse survival: a pooled analysis of individual patient data from four randomised controlled trials. Lancet Oncol. 2014;15:1481–92. doi: 10.1016/S1470-2045(14)70486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmoll HJ, Cunningham D, Sobrero A, Karapetis CS, Rougier P, Koski SL, Kocakova I, Bondarenko I, Bodoky G, Mainwaring P, Salazar R, Barker P, Mookerjee B, Robertson J, Van Cutsem E. Cediranib with mFOLFOX6 versus bevacizumab with mFOLFOX6 as first-line treatment for patients with advanced colorectal cancer: a double-blind, randomized phase III study (HORIZON III) J. Clin. Oncol. 2012;30:3588–95. doi: 10.1200/JCO.2012.42.5355. [DOI] [PubMed] [Google Scholar]
- 8.Schwartzberg LS, Rivera F, Karthaus M, Fasola G, Canon JL, Hecht JR, Yu H, Oliner KS, Go WY. PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J. Clin. Oncol. 2014;32:2240–7. doi: 10.1200/JCO.2013.53.2473. [DOI] [PubMed] [Google Scholar]
- 9.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. microRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, Shen L, Yu J. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–52. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 16.Akao Y, Noguchi S, lio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Wu DW, Huang CC, Chang SW, Chen TH, Lee H. Bcl-2 stabilization by paxillin confers 5-fluorouracil resistance in colorectal cancer. Cell Death Differ. 2015;22:779–89. doi: 10.1038/cdd.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, Zhang N, Lyu X, Huang G, Chen B, Zhang Y, He F. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Fan H, Ma Y, Liang D, Huang R, Wang J, Zhou F, Kan Q, Ming L, Li H, Giercksky KE, Nesland JM, Suo Z. Notch1 is a 5-fluorouracil resistant and poor survival marker in human esophagus squamous cell carcinomas. PLoS One. 2013;8:e56141. doi: 10.1371/journal.pone.0056141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Li X, Li X, Hou J, Ding Y, Zhang J, Xu W, Zhang Y. Discovery of multi-target anticancer agents based on HDAC inhibitor MS-275 and 5-FU. Med Chem. 2016;12:30–6. doi: 10.2174/1573406411666150714111045. [DOI] [PubMed] [Google Scholar]
- 21.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–93. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 23.Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, Rimm DL, Gyorffy B, Santarpia L, Pusztai L, Saltzman WM, Slack FJ. miR-34a silences cSRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res. 2016;76:927–39. doi: 10.1158/0008-5472.CAN-15-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–9. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 25.Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288–96. doi: 10.4161/cbt.7.8.6284. [DOI] [PubMed] [Google Scholar]
- 26.Cozzolino AM, Pedace L, Castori M, De Simone P, Preziosi N, Sperduti I, Panetta C, Mogini V, De Bernardo C, Morrone A, Catricala C, Grammatico P. Analysis of the miR-34a locus in 62 patients with familial cutaneous melanoma negative for CDKN2A/CDK4 screening. Fam Cancer. 2012;11:201–8. doi: 10.1007/s10689-011-9502-6. [DOI] [PubMed] [Google Scholar]
- 27.Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem Sci. 2014;39:101–11. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


