Abstract
Drug repositioning is a highly studied alternative strategy to discover and develop anticancer drugs. This drug development approach identifies new indications for existing compounds. Ivermectin belongs to the group of avermectins (AVM), a series of 16-membered macrocyclic lactone compounds discovered in 1967, and FDA-approved for human use in 1987. It has been used by millions of people around the world exhibiting a wide margin of clinical safety. In this review, we summarize the in vitro and in vivo evidences demonstrating that ivermectin exerts antitumor effects in different types of cancer. Ivermectin interacts with several targets including the multidrug resistance protein (MDR), the Akt/mTOR and WNT-TCF pathways, the purinergic receptors, PAK-1 protein, certain cancer-related epigenetic deregulators such as SIN3A and SIN3B, RNA helicase, chloride channel receptors and preferentially target cancer stem-cell like population. Importantly, the in vitro and in vivo antitumor activities of ivermectin are achieved at concentrations that can be clinically reachable based on the human pharmacokinetic studies done in healthy and parasited patients. Thus, existing information on ivermectin could allow its rapid move into clinical trials for cancer patients.
Keywords: Ivermectin, cancer, drug repurposing
Introduction
The antiparasitic drug ivermectin was initially approved in humans in 1987 to orally treat onchocerciasis, also known as river blindness, caused by the blackfly-transmitted parasite Onchocerca volvulus in poor populations around the tropics, mostly in West and Central Africa [1,2]. In humans, its use has improved the nutrition, general health and well-being of billions of people worldwide since it was first used to treat onchocerciasis. Not only that, but in veterinary medicine, ivermectin is used to treat billions of livestock and pets around the world, helping to boost production of food and leather products, as well as to keep billions of companion animals, particularly dogs and horses, healthy. Nowadays, ivermectin by its own has produced sales greater than US$1 billion/annum during the past two decades [3] and is annually taken by close to 250 million people [1].
Ivermectin belongs to the group of avermectins (AVM), which is a group of 16-membered macrocyclic lactone compounds discovered in 1967 in the Japanese Kitasato Institute [1,3] in fermentation broths of actinomycetes cultures with the fungus Streptomyces avermitilis [4-6]. AVM family members include, among others, selamectin, abamectin, monoxidectin and ivermectin (Figure 1), all of which differ from the antibacterial and antifungal 16-membered macrocyclic lactones by owning a bisoleandrosyloxy substituent at the C13 [3]. Ivermectin is the most commonly employed compound from the AVM group, being a more potent and safer semi-synthetic mixture of the two AVMs 22,23-dihydroavermectin-B1a and dihydroavermectin-B1b, at a reason of 4:1, respectively [1]. Since 1981 ivermectin has been employed for agriculture, veterinary and aquaculture purposes [1], and is recognized as nematocidal, acaracidal and insecticidal [3,7]. The antiparasitic efficacy of ivermectin is not limited to onchocerciasis, since it is also effective for filarial infections such as those caused by Wuchereria bancrofti, Brugia malayi, Loa loa, Mansonella perstans, and Mansonella ozzardi [7]. It also eradicates gastrointestinal parasites, including Ascaris lumbricoides, Strongyloides stercoralis, Enterobius vermicularis, Trichuris trichiuria, and Ancylostoma duodenale [7]. Besides, it is employed to treat trypanosomiasis, malaria, leishmaniasis, scabies and head lice [1]. In parasites and helminths, ivermectin, as well as the rest of AVMs, increases the activity of γ-aminobutyric acid (GABA) receptors or glutamate-gated chloride ion channels (Glu-Cl) [3,4,6], which blockades the signal between neuron and muscle [3].
Figure 1.
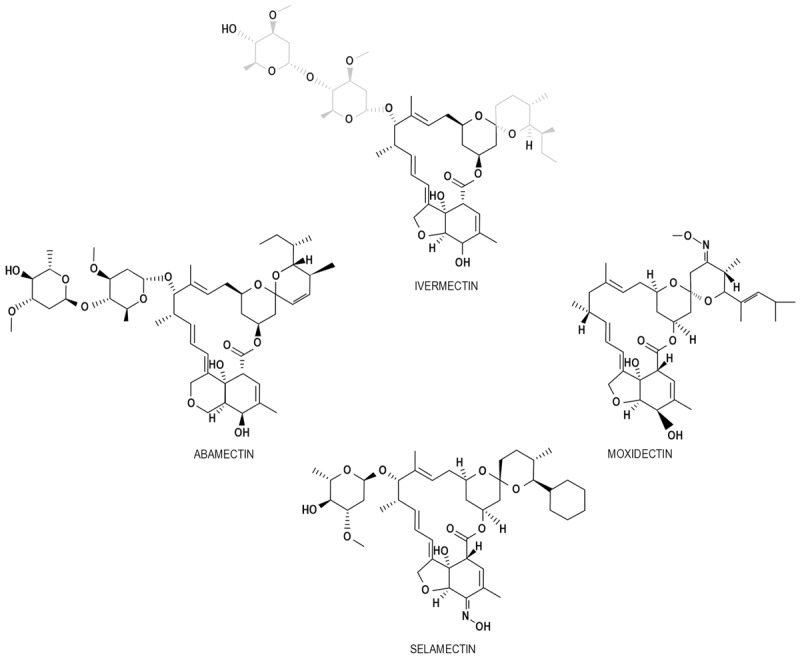
Compounds belonging to the avermectin family. Some AVM family members are showed in the picture. All of them are recognized by a 16-carbon macrocyclic lactone core with the bisoleandrosyloxy substituent at C13. The characteristics substituents of ivermectin are marked in gray.
In mammals, GABA-sensitive neurons are secured by the blood-brain barrier (BBB) within the central nervous system (CNS), protecting vertebrates against potential harmful effects of AVMs [3,6]. In support of this statement, subpopulation of collie dogs that have defective function of the multidrug resistance (MDR) protein (commonly a 4 base-pair deletion of the mdr-1 gene which produces a stop codon), which is an integral part of the BBB and functions as a drug-transport pump in the BBB, have increased neurotoxicity to ivermectin [8]. On the other hand, invertebrates are dose-dependent susceptible due to the widespread allocation of Glu-Cl channels, in whom ivermectin induces the opening of GABA-regulated Cl- channels that generates an influx of Cl- [1,7]. The resulting hyperpolarization impedes the phosphorylation of the regulatory light chain of myosin II by PAK1 [9], promoting paralysis of somatic muscles with concomitant uncoordinated movement, starvation due to inhibition of pharyngeal pumping, and death [1,3,5,7]. Certainly, the affinity of ivermectin for the parasite is 100 times greater than for the mammalian brain, but at least in onchocerciasis, ivermectin action is mostly restricted to the microfilariae stage of Onchocerca volvulus [7] as the macrofilariae form does not require pharyngeal pumping to survive [1]. The rate of reduction in microfilarial worms is close to 98% with only two weeks of ivermectin administration, an effect maintained within the next 12 months [1].
Current use and dosage as an antiparasitic drug
In humans, the most used dose of ivermectin for onchocerciasis, strongyloidiasis and enterobiasis ranges between 150 to 200 µg/kg [10-12], while it is used at higher doses of 400 µg/kg for lymphatic filariasis [13]. It is noteworthy the report of a clinical trial on the use of ivermectin for patients with spinal damage and muscle spasms where the drug was administered up to 1.6 mg/kg subcutaneously twice a week for 12 weeks [14].
Toxicity
This compound has a wide margin of safety in ruminants, pigs and equine, as well as in most of the dog breeds [15,16]. The acute toxicity of ivermectin has been investigated in various species of animals. The signs of toxicity were similar after oral and intraperitoneal administration in rats and mice, and the effects consisted in ataxia, tremors, and reduced activity [17]. In early stages of development, ivermectin at doses of 0.4-0.8 mg/kg in mice, 10 mg/kg in rats, and 3-6 mg/kg in rabbits, increased the incidence of cleft palate, but it was not considered as embryotoxic since the frequency of anomalies was very low [18]. The toxic effects have been related to its interaction with the P-glycoprotein, which limits its access to the CNS. The absence of this protein determines the accumulation of ivermectin in the brain of transgenic mice that do not express it. Finally, in adult Rhesus monkeys that ingested it daily for 16 days at 1.2 mg/kg, no undesirable effects were detected [18].
There are several toxicological reports of ivermectin in different species. The lethal dose 50 (LD50) reported in mice [19] is 25 mg/kg administered orally, whose human equivalent dose (HED) is 2.02 mg/kg. The LD50 increases up to 30 mg/kg when this compound is administered intraperitoneally in mice (HED 2.43 mg/kg). For rats the average lethal dose is 50 mg/kg orally (HED 8.01 mg/kg) and 55 mg/kg intraperitoneally (HED 8.91 mg/kg). In rabbits it is 406 mg/kg in topical application, while in dogs it is 80 mg/kg administered orally (HED 43.24 mg/kg) [20]. Clearly, it seems that the higher the phylogenetic scale the lower toxicity by ivermectin. These data are in accord with the findings in a review paper on avermectins poisoning (14 on suicidal attempt). In this retrospective review, among 18 patients exposed to abamectin and one to ivermectin, 15 were poisoned by oral ingestion. Four were asymptomatic and 8 had minor symptoms with a mean ingestion of 23 mg/kg (range in 4.2-67 mg/kg). Seven patients manifested severe symptoms, such as coma (seven), aspiration with respiratory failure (four), and hypotension (three), after a mean ingestion of 100.7 mg/kg avermectin (15.4 mg/kg for ivermectin and 114.9 mg/kg for abamectin). All 7 seven patients received intensive supportive care; 1 patient died 18 days later as a result of multiple organ failure [21].
In humans it is considered that ivermectin generates low levels of toxicity because its targets are confined within the CNS. Indeed, most patients treated with ivermectin have no side-effects other than those caused by the immune and inflammatory responses against the parasite, such as fever, pruritus, skin rashes and malaise [7,22], and when present, they appear within 24-48 h after treatment [23]. Certainly, moderate symptoms such as arthralgia, dizziness, fever, skin edema, dyspnea and hypotension may be more related with the microfilarial load in the patient rather than with the intrinsic toxicity of ivermectin [24]. Reports on cases of encephalopathy in patients co-infected with onchocerciasis and lymphatic filariasis after 48 h of treatment with ivermectin can be found in the literature [25], but it is believed that this adverse reaction is due to the obstruction of the microcirculation of the brain by the accumulation of dead or paralyzed parasites, which leads to brain embolism [26].
In conclusion, the immense number of patients who have been treated with ivermectin shows that it is a safe and a well-tolerated drug. Beyond the side effects attributable to the immunological and inflammatory reaction elicited by dying or death parasites, there are sympathetic signs related to ivermectin intoxication, including tremors, mydriasis, sialorrhea, motor incoordination and coma [27].
Pharmacokinetics
The pharmacokinetics of ivermectin have been widely studied in various mammals, including humans; is a fat-soluble compound, with a distribution volumen of 46.9 L; it has a mean peak plasma level of ~4 h after oral administration with a second peak at 6-12 h because of enterohepatic recycling [1], and possesses an oral clearance of 1.2 L/h [7]. With a plasma protein binding of 93% [7], this drug experiences low biotransformation within the organism [4-6]. The maximum concentration in plasma is reached 4-5 h after its oral administration; its half-life is approximately 19 h and is metabolized in the liver by the cytochrome CYP1A and CYP3A4 complexes, generating 10 metabolites, mostly demethylated and hydroxylated. Its excretion is mainly by feces and only 1% is excreted in the urine [28]. Table 1 shows the pharmacokinetic data of ivermectin in humans infected with parasites, as well as in healthy humans treated with various doses of ivermectin [28,29]. According with Table 1, the molar concentrations achieved taking into account the total exposure of the drug measured by the area under the curve (systemic exposure) in healthy individuals or patients treated for parasitic diseases are:
Table 1.
Pharmacokinetics of ivermectin in healthy and parasited subjects
| Condition | Doses (mg/kg) | Route | Cmax (ng/mL) | Tmax (h) | AUC (µg/h/mL) |
|---|---|---|---|---|---|
| Parasitic infection | 0.1-0.2 | Oral | 52.0 | 5.2 | 2.852 |
| Healthy | 0.35-0.6 | Oral | 87.0 | 4.2 | 1.444 |
| Healthy | 0.7-1.1 | Oral | 165.2 | 3.6 | 2.099 |
| Healthy | 1.4-2.0 | Oral | 247.8 | 4.2 | 4.547 |
3.25 μM/h in parasitized patients with a dose of 0.1-0.2 mg/kg, 1.64 μM in healthy subjects with a dose of 0.35-0.6 mg/kg, 2.4 μM/h in healthy subjects with a dose of 0.7-1.1 mg/kg, and 5.2 μM/h in healthy subjects with a dose of 2 mg/kg.
Drug repurposing in cancer
Drug repurposing, drug redirecting or drug reprofiling is defined as the identification of novel usages for existing drugs. Both development risks and costs, as well as safety-related failure, are reduced with this approach because such drugs have well-known formulation development, in vitro and in vivo screening, as well as pharmacokinetic and pharmacodynamic profiles. Also, the first clinical phases of many drugs had been completed and can be bypassed to reduce several years of development. Therefore, drug repurposing has the potential to reduce the whole process up to 3-12 years, and consequently, the potential recycle of compounds towards a new indication is an attractive opportunity for patients on need [30]. The relevance of drug repositioning in research is demonstrated with the fact that since the first publication of the subject back to 2004, there are more than 500 papers about it until 2013, at least in PubMed [31]. However, the majority of repositioned agents were discovered before starting with systematic efforts on 2006 to identify drugs with potential additional use, suggesting a serendipitous detection [30,32,33]. Currently, other ways to identify compounds with repositioning potential are informed insights and platforms established to identify in silico repositioning opportunities [32,34]. Furthermore, with the recently assembly of the Drug Repurposing Hub, an online repurposing library that systematically classifies a collection of clinically tested compounds from existing databases, now it is possible to easily search and view drugs according to their clinical status, drug indications, or mechanism of action, allowing to rapidly find agents for further evaluation [35].
Important efforts have been made for drug repositioning in cancer. Pantziarka et al. have recently summarized on this topic. They report at least 235 non-cancer drugs with proven antitumor activity either in vitro or in vivo, and among these, 67 (29%) are in the World Health Organization (WHO) list of essential medicines, and 176 (75%) are off-patent [36]. 133 (57%) had human data in cancer patients [36]. Four were listed in clinical guidelines, namely thalidomide, all-trans retinoic acid, zoledronic acid and non-steroidal anti-inflammatory drugs (NSAID) [36]. Of note, at least 3 drugs have shown a survival benefit in randomized trials: cimetidine (colorectal cancer), progesterone (breast cancer) and itraconazole (lung cancer) [36]. Few examples of drug-target networks analyses show that both simvastatin and ketoconazole are anti-proliferative compounds in breast cancer [32], while gene expression profiles suggest that topiramate can be used to treat small-cell lung cancer and that sirolimus can be useful for glucocorticoid-resistant acute lymphocytic leukemia [35]. On the other hand, drugs that were first formulated to treat cancer might also be useful to treat non-malignant diseases. That is the case of inhibitors of histone deacetylase enzymes that are approved for T-cell lymphoma, but are prospective targets for malaria, leishmaniasis and trypanosomiasis [37].
Molecular mechanisms of the antitumor effects of ivermectin
Despite the relatively short time on which ivermectin has been investigated as a drug for cancer repositioning, a number of molecular mechanisms of action have been discovered (Figure 2 and Table 2). Among these are the following:
Figure 2.
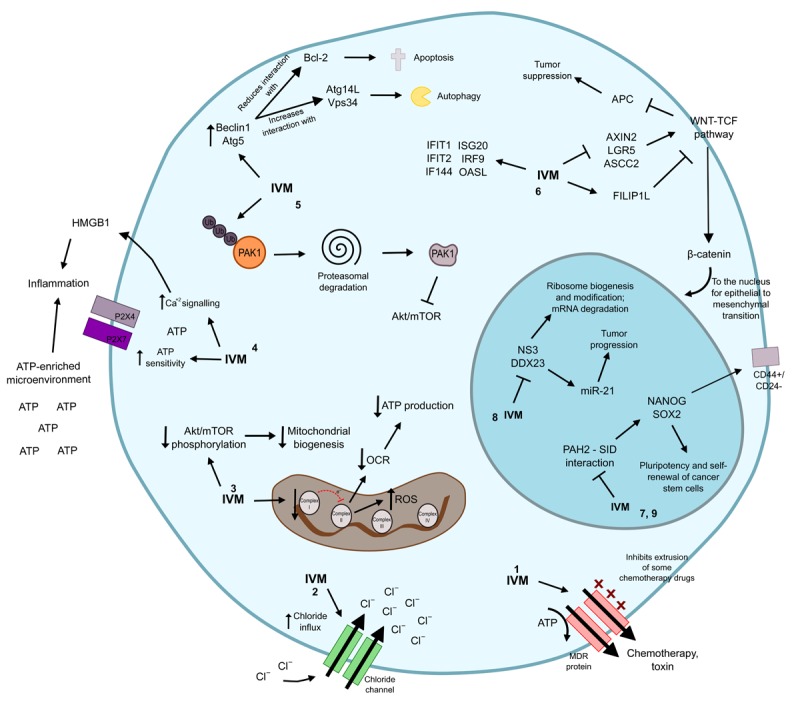
Antitumor mechanisms of ivermectin. 1. Ivermectin inhibits the P-glycoprotein pump, that induces a multidrug phenotype in the cancer cell. 2. Ivermectin acts as an ionophore and up-regulates chloride channels to generate apoptosis and osmotic cell death. 3. By decreasing the function of the mitochondrial complex I, ivermectin limits the electronic movement in the oxidative phosphorylation pathway that stimulates oxygen consumption rate to generate ATP for the cell. Concomitantly there is a reduction in the phosphorylation levels of Akt, impacting in the mitochondrial biogenesis process. Furthermore, alterations in the mitochondrial machinery are related with increased levels of reactive oxygen species that damage DNA. 4. Ivermectin induces ICD through the stimulation of an ATP- and HMGB1-enriched microenvironment, which promotes inflammation. This drug also increases ATP sensitivity and calcium signals in P2X membranal receptors, particularly P2X4 and P2X7, to induce ATP-dependent immune responses. 5. Ivermectin promotes the poly-ubiquitination of the kinase PAK1, which directs it to degradation in the proteasome. Defective PAK1, in turn, inhibits the Akt/mTOR pathway. At the same time, ivermectin stimulates the expression of Beclin1 and Atg5, both related with induction of autophagy and reduces the function of the negative regulator of apoptosis Bcl-2. Together, this generates autophagy and apoptosis. 6. Ivermectin represses AXIN2, LGR5 and ASCL2, all of them positive regulators of WNT-TCF while promotes the repressor of the WNT signaling FILIP1L. Concomitantly, ivermectin promotes the expression of several IFN-related genes, such as IFIT1, IFIT2, IF144, ISG20, IRF9 and OASL. 7. Ivermectin modifies the epigenetic signature and the self-renewal activity in the malignant cell due to its ability to mimic the SIN3-interaction that binds to the PAH2 motif of the cancer-associated deregulators SIN3A and SIN3B. SIN3A naturally induces NANOG and SOX2, which are stimulants of stem cell pluripotency. 8. Ivermectin limits the function of the RNA helicases NS3 and DDX23, both of which are related with ribosome biogenesis and post-transcriptional modifications, as well as with mRNA degradation. DDX23 acts as a promoter of miR-21, which is a well-recognized stimulator of tumor progression. 9. Ivermectin inhibitis preferentially the CSC population and up-regulates pluripotency and self-renewal genes NANOG, SOX2 and OCT4. IVM: ivermectin; ATP: adenosine triphosphate; OCR: oxygen consumption rate; ROS: reactive oxygen species.
Table 2.
Summary of the antitumor targets of ivermectin
| Target | Effect | References |
|---|---|---|
| MDR protein | Inhibition | [39] |
| Chloride channel | Increase of activity | [44] |
| Akt/mTOR pathway | Inhibition | [47] |
| P2X7/P2X7 receptors | Activation | [50,51] |
| PAK1 protein | Inhibition | [9,54] |
| WNT-TCF pathway | Inhibition | [57,58] |
| SIN3 domain | Inhibition | [59] |
| NS3 DDX23 helicase | Inhibition | [64] |
| Nanog/Sox2/Oct4 genes | Downregulation | [67] |
Ivermectin as an inhibitor of the multi-drug resistance (MDR) phenotype
The firsts reports on the potential antitumor activity of ivermectin appeared almost 20 years ago, and these were somehow linked with the recently discovered basis for the MDR phenotype at that time. Gros P et al. in 1986 reported the isolation of DNA clones complementary to the cellular messenger RNA transcripts of MDR genes, and showed that high-level expression of a full-length complementary DNA clone in an otherwise drug-sensitive cell confers a complete MDR phenotype [38]. Later on, Didier and Loor proved by a short-term assay the P-glycoprotein function inhibition, which measures the restoration of the retention of two P-glycoprotein probes in MDR cells to their parental cells, concluding that ivermectin is also a substrate and an inhibitor of P-glycoprotein [39]. Though not named as ivermectin, avermectin IB1 showed the ability to reduce tumor growth in vivo by 50% at day 5 at dose of 1 mg/kg (HED 81 μg/kg) in SHK male mice bearing a solid Ehrlich carcinoma. In addition, at the same dose it inhibits the growth of the carcinoma cell line 755 (C57/BL6 male mice), and the tumor growth inhibition value reached a maximum when avermectin B1 was injected on day 3 after tumor inoculation. Based on the fact that ivermectin inhibits multidrug resistance in tumors, avermectin IB1 was tested with vincristine in the Ehrlich carcinoma, with results indicating that the antitumor effect of vincristine is greatly increased when avermectin 1B1 is administered after vincristine [40]. No further studies have exploited the anti-MDR effect of ivermectin. Nevertheless, the search for novel strategies and/or schedule optimization of MDR inhibitors continues [41], which suggests that there is still room for investigating ivermectin roles in reversing or preventing the development of the MDR phenotype.
Ivermectin as an ionophore drug
The term ‘ionophore’ was first used in 1967 in reference of the ability of organic molecules to bind metal cations and to form lipid soluble complexes that facilitate their transport across cellular membranes. Thus, ionophores can diffuse back and forth between the extracellular and intracellular spaces, or may remain in the plasma membrane as their transport metal ions between intracellular and extracellular spaces [42]. Ionophore antibiotics act by generating pores in biological membranes that dramatically alter the ionic household of cells. Salinomycin is an example of an ionophore antibiotic which generates ion channel-like structures that exhibit strong selectivity for K+, but other monovalent cations are also conducted (e.g., Na+ and H+) [43]. Traditionally, the cell-killing activity of ionophore antibiotics is thought to originate from profoundly deregulating osmosis, as well as from direct cytotoxic effects of the altered biochemical landscape. It is known that malignant cells tend to upregulate chloride channels, which potentially could mark them as more sensitive to alterations in chloride flux, and that an unbalance in intracellular chloride concentrations affects intracellular Ca2+ levels, as well as pH and cell volume, which can lead to apoptosis in the affected cell [44].
In line with these statements, in a screen of a small chemical library of antibiotics and metabolic regulators to identify anti-leukemia compounds, Sharmeen et al. found that ivermectin induces cell death at low micromolar concentrations (IC50 of 10 μM) in HL60, KG1a, and OCI-AML2 acute myeloid leukemia cell lines, as well as in primary patient samples. Cell death was caspase-dependent and interestingly, normal hematopoietic cells were much less sensitive to ivermectin as it did not induce apoptosis at concentrations up to 20 μM. Similar effects were seen when leukemia and normal cells were tested for clonogenicity. They also showed that increased chloride influx correlated with cell death and changes in both cell size and cell hyperpolarization, as these effects were much more marked in sensitive leukemia cells as compared to normal cells. Interestingly, there was a synergistic or additive interaction in OCI-AML2 and U937, but not in normal cells, when they treated with ivermectin plus cytarabine or daunorubicin [44].
Ivermectin as an inductor of mitochondrial dysfunction and oxidative damage
On the basis that anthelmintics or antibiotics may target mitochondria in mammalian cells [45,46], ivermectin was tested in glioblastoma cell lines to identify whether its antitumor effect occurs via inhibition of mitochondrial biogenesis or function. As expected, ivermectin inhibits in a dose-dependent manner the basal and maximum oxygen consumption rate (OCR), most likely by decreasing the enzyme activity of respiratory complex I but not II, IV or V, and consistent with that, both the membrane potential and electrochemical proton gradient decrease while a significant increase in mitochondrial superoxide and decreased ATP are observed. Furthermore, by establishing a subline of the U87 cell line deficient in mitochondrial respiration it was proved that under these conditions ivermectin was unable to induce cell death, as well as when these cells were co-treated with the antioxidants alfa-tocopherol or mannitol. These effects were tracked down by studying the Akt/mTOR pathway, which at least in part controls mitochondria biogenesis and function. Results showed that ivermectin decreases phosphorylation of Akt (S473), mTOR (S2481) and the ribosomal S6 protein (rS6) in U87, T98G and HBMEC cells, indicating that ivermectin inhibits the Akt/mTOR pathway [47]. The effect of ivermectin upon mitochondria and oxidative damage has been recently corroborated in a study on renal cancer cell lines, where it was demonstrated that renal cancer cells do have higher mitochondrial mass and basal and maximal OCR [48]. As in the Liu et al. study, ivermectin in this model also decreased mitochondrial membrane potential as well as basal and maximal respiratory capacities. Of note, ivermectin significantly increased intracellular ROS and 8-OHdG levels, suggesting that the antitumoral effects of ivermectin are related to oxidative stress and DNA damage. This was confirmed by the abolition of the inhibitory effect of ivermectin in these renal cancer cell lines when co-treated with acetyl-L-carnitine (ALCAR) or N-acetyl-L-cysteine (NAC), a stimulant of mitochondrial biogenesis and an antioxidant, respectively [48].
Ivermectin as an inductor of immunogenic cell death (ICD)
Immunogenic cell death (ICD) is characterized by the presence of damage-associated molecular patterns (DAMPs) such as the membranal exposure of calreticulin and the release of both ATP and high-mobility group box 1 (HMGB1) into the extracellular space, which are then recognized by immune cells to elicit antineoplastic activities [49]. Exogenous ATP regulates defense through P1, P2X and P2Y purinergic receptors [50]. However, recently P2X7 overexpression has been correlated with promotion of both tumor growth and metastases [50]. Although within the tumor microenvironment ATPases such CD39 and CD73 degrade ATP to its immunosuppressive form, adenosine, ivermectin can surpass their effect by a potent induction of both HMGB1 and ATP extrusion which, in turn, induces inflammation [50]. Certainly, it has been reported in triple-negative breast cancer cells (TNCB) that ivermectin allosterically potentiates P2X4/P2X7- and caspase-1-mediated ICD due to the stimulation of an ATP-enriched tumor microenvironment, disrupting the balance between the survival and cytotoxic roles of purinergic signaling in malignant cells, which also induces autophagy [50]. One additional work done with human monocyte-derived macrophages corroborated the ivermectin association with PX24 and P2X7 receptors. Such research demonstrated that ivermectin increases ATP sensitivity and delays current deactivation after ATP wash-out in PX24 receptors, which together with the augmentation of ATP-induced currents and Ca2+ signals in P2X7 receptors, suggests that ivermectin may stimulate ATP-dependent immune responses [51]. Altogether, literature indicates that ivermectin could promote and potentiate ICD at the tumor microenvironment.
Ivermectin as an inductor of autophagy
Autophagy, a self-degradative catabolic pathway is characterized by formation of doublemembrane autophagosomes, which sequester excess or defective organelles and fuse with lysosomes for degradation of enclosed materials in the lysosome to mobilize energy and nutrients under certain cellular stimuli such as starvation, developmental transitions, hypoxia and/or oxidative stress [52,53]. Ivermectin in ovarian and glioblastoma cancer cell lines promotes ubiquitination-mediated degradation of the oncogenic kinase PAK1 [1,47], a key protein in cytoskeletal reorganization and nuclear signaling for tumor growing in more than 70% of all human cancers [1]. PAK1 downregulation in turn blockades the repressor of autophagy Akt/mTOR as evidenced by decreased phosphorylation of Akt, mTOR, p70S6K and 4EBP1 via direct interaction of PAK1 with Akt [54]. In fact, one work with multiple breast cancer cell lines treated with ivermectin showed its autophagy-promoter role by the formation of acidic vesicular organelles, with double-membraned autophagosomes by transmission electronic microscopy, and with the promotion of the expression levels of the autophagy-related proteins Beclin 1 and Atg5 in a dose-dependent fashion. Ivermectin, in turn, increases the interaction of Beclin 1 with positive regulators of autophagy, specifically Atg14L and Vps34, while diminishes its interaction with negative regulators such as Bcl-2 [54]. All together it is demonstrated that the ivermectin autophagic effects in this model results from inhibition of the PAK1/Akt/mTOR pathway.
Ivermectin as an inhibitor of the WNT-TCF pathway
The antiproliferative function of ivermectin has been widely documented. A study aimed to find out drug candidates for repositioning with the ability to block the WNT-TCF signaling, which inactivates the tumor suppressor APC in several sporadic human cancers and stimulates the constitutive activation and translocation into the nucleus of β-catenin during neoplastic transformation [55,56], revealed that ivermectin at low micromolar concentrations performs anti-WNT-TCF response in cancer cells [57]. The authors showed the efficacy of ivermectin to inhibit BrdU incorporation in colon cancer, glioblastoma and melanoma cell lines, indicating repression in cell proliferation [57]. Besides, they demonstrated upregulation of activated caspase 3 and repression of the positive WNT-TCF targets AXIN2, LGR5, and ASCL2 in DLD1 and Ls174T colon cancer cells [57], opening the possibility to use ivermectin to block WNT-TCF-dependent cancers, such as those from breast, skin, lung and intestine [1,57]. Moreover, after investigating the ivermectin anti-clonogenic activity by analyzing spheroid formation, they showed that pre-treatment of such cell lines with ivermectin diminishes clonal floating spheroids by up to 73%, which together with the repression of the positive cell cycle regulator cyclin D1, suggests a limiting cancer stem cell formation driven by ivermectin [57]. In fact, ivermectin role as an WNT-TCF inhibitor led to use it as a positive control of such pathway in one research that screened a library of plant and microorganism natural compounds, which confirmed ivermectin suppressor activity by transcriptomic analysis that showed an upregulation of the repressor of WNT signaling FILIP1L up to 10-fold by the use of this compound [58]. Such study also revealed the increase of the interferon-responsive genes ISG20, IFIT1, OASL, IRF9, IF144, and IFIT2, in colon cancer cells [58]. Interestingly, it has been suggested the employment of interferon to inhibit WNT-TCF pathway [58].
Ivermectin as an epigenetic modulator
Other recognized field of action of ivermectin in cancer involves epigenetic regulation. One work done with the breast cancer cell line MDA-MB-231 reported the functional effect of ivermectin after an in silico screen of 2,000 FDA-approved small molecule drugs. The authors evaluated the ability of ivermectin to mimic the SIN3-interaction domain (SID), which naturally binds to the PAH2 motif belonging to the breast cancer-related epigenetic deregulators SIN3A and SIN3B [59]. By nuclear magnetic resonance they demonstrated that ivermectin indeed blocks the PAH2-SID interaction [59]. Furthermore, since SIN3A is part of a complex that positively regulates the stem cell and self-renewal markers NANOG and SOX2, the authors analyzed the role of ivermectin to inhibit such genes and proved that in D3H2LN cells at doses of 0.5 µM it reduces NANOG and SOX2 gene expression by 80%, with a decrease between 90-100% in clonogenic tumorsphere growth [59].
Ivermectin as a helicase inhibitor
RNA helicases represent a large family of proteins implicated in many biological processes, including ribosome biogenesis, splicing, translation and mRNA degradation [60]. Members of the DEAD-box family of RNA helicases play important roles in various aspects of RNA processing, including transcription, spliceosome biogenesis, ribosome biogenesis, splicing, nucleocytoplasmic transport, translation and decay [61,62]. These family members share a conserved core that includes the amino acid sequence D-E-A-D (aspartate-glutamate-alanine-aspartate). They use energy received from ATP hydrolysis to unwind double-stranded RNA, generally act as components of multi-protein complexes, and have diverse functions that depend on their interacting partners. Several DEAD-box RNA helicases are aberrantly expressed in various types of cancer, where they may play important roles in cancer development and/or progression [61,62]. In 2012, Mastrangelo et al. uncovered that ivermectin was an effective inhibitor of the NS3 helicase activity from the Kunjin virus (an Australian variant of the yellow fever virus) by an in silico test, and confirmed it by in vitro helicase enzymatic assays [63]. On the other hand, a recent study in glioma cell lines has found that the RNA helicase DDX23 regulates the oncogenic miR-21 biogenesis at post-transcriptional level [64], and it is over-expressed in glioma tissues as compared to normal brain, which is associated with poor survival of glioma patients. After the authors assayed a number of drugs known to inhibit viral helicases, they found that ivermectin inhibits the DDX23-mediated potentiation of pri-miR-21 processing that, in turn, decreases the levels of both precursor and mature miR-21, a well-recognized poor prognostic upregulated marker in cancer [65]. The treatment with ivermectin in glioma cell lines both in vitro and in vivo was able to induce antitumoral effects, which suggests that the antihelicase role of ivermectin can be considered as another mechanism of its anticancer effect.
Ivermectin as a stem-cell cancer inhibitor
In 2009, Gupta et al. performed a high-throughput screening to discover selective inhibitors of cancer stem cells (CSCs), and found that salinomycin reduces the proportion of CSCs by >100-fold relative to paclitaxel. Besides, they showed that salinomycin inhibits mammary tumor growth in vivo, and that it induces increased epithelial differentiation of tumor cells accompanied by the loss of expression of breast CSC genes [66]. As salinomycin is an antiparasitic drug for veterinary use only, our group searched similar compounds for human use that could also act as selective inhibitors of CSCs. Our results showed that ivermectin has high similarity with salinomycin (similarity of 0.78), and therefore we hypothesized that the antiparasitic drug ivermectin could also have similar biological properties as salinomycin [67]. The results of this study showed that ivermectin has growth inhibitory effects upon MDA-MB-231 cells in the range of 0.2-8 µM, and as predicted, ivermectin preferentially inhibits the viability of CSCs-enriched populations (CD44+/CD24-) and cells growing in spheroids, as compared to bulk cell population. The opposite pattern was observed with paclitaxel where the non-CSCs (CD44+/CD24+) were sensitive to paclitaxel at nanomolar concentrations, while the inhibition of the stem cell subpopulation was only observed at higher drug concentrations. According with this, ivermectin reduces the expression of maintenance of the pluripotency and self-renewal markers Nanog, Oct4 and Sox2 at both mRNA and protein levels [67]. A summary of the molecular mechanisms of the antitumor effects of ivermectin are shown in Table 2 and Figure 2.
Antitumor effects of ivermectin, in vitro and in vivo
There are a number of in vitro and in vivo preclinical studies where ivermectin demonstrates its efficacy against a wide range of malignant conditions, including solid and hematological malignancies. In breast cancer, ivermectin has been studied in MDA-MB-435, MDA-MB-231, MDA-MB-468, MDA-MB-361, MCF-7, HS578T and SKBR3 cell lines where it demonstrates its ability to inhibit cell proliferation, induction of apoptosis, autophagy and reversion of tamoxifen resistance among other effects [51,55,60,67]. These effects are also reported in cancer cell lines from ovarian, prostate, head and neck, colon, and pancreas, as well as in melanoma [9,45,51]. Similar results are also observed in a number of murine cancer cell lines including breast, melanoma and colon [51,60]. Two studies in glioblastoma cell lines show that ivermectin not only induces growth arrest and apoptosis, but induces anti-angiogenic effects [48,58], whereas two more studies extend these observation upon myeloblastic acute leukemia cell lines [41,51]. Of interest, the median concentration used for the in vitro treatment from all studies referred in Table 3 was 5 µM (0.01-100 µM) which is clinically achievable according with the pharmacokinetic data in humans shown in Table 1. Regarding the in vivo evaluation of ivermectin (Table 4), this has been done in immune deficient mice using human acute myeloblastic leukemia, glioblastoma, breast and colon carcinoma, as well as in the murine lymphosarcoma cell line MDAY-D2. These studies show more than a 50% reduction in tumor volumes after ivermectin treatment, which varied from 10 to 42 days of treatment by either oral, intraperitoneal or intratumoral routes (more commonly intraperitoneal). The median dose employed was 5 mg/kg (2.4-40 mg/kg), which is equivalent to 0.40 mg/kg in humans, a dose below to the highest dose safely used in human subjects evaluated so far (2 mg/kg) (Table 1). Thus, the in vitro and in vivo results with ivermectin strongly suggest that its antitumor effects in cancer patients can be achieved at feasible doses.
Table 3.
Antitumor effects of ivermectin in vitro
| Tumor type | Cell lines | [µM] | Effects | Reference |
|---|---|---|---|---|
| Human leukemia | OCI-AML2 | 5, | Induces cell death through upregulation of ROS. | [44] |
| HL60 | 10, | |||
| U937 | 15, | |||
| K61a | 20 | |||
| Prostate cancer | DU145 | |||
| PPC-1 | ||||
| Human glioblastoma | U87 | 1, | Induces growth inhibition, apoptosis and anti-angiogenesis. | [47] |
| T98G | 5, | |||
| 10 | ||||
| Ovarian cancer | TYK-nu | 0.1, | Inhibition of cell proliferation. | [9] |
| KOC7C | 1, | |||
| SKOV3 | 10, | |||
| RMUG-S | 100 | |||
| HEI-193 | ||||
| Breast cancer | MDA-MB-435, | 5, | Stimulates autophagy and inhibition of cell proliferation. | [54] |
| MDA-MB-231 | 10, | |||
| MDA-MB-468 | 15, | |||
| MDA-MB-361 | 20 | |||
| MCF-7 | ||||
| HS578T | ||||
| Murine breast cancer | 4T1.2 | 1, | Induces apoptosis and necrosis. Induces autophagy. | [50] |
| DDHer2 | 4, | |||
| 8, | ||||
| Murine melanoma | C57BL/6 | 16 | Inhibition of cell proliferation and clonogenic capacity. | |
| Murine colon adenocarcinoma | CT.26 | Increases the amount of ROS. | ||
| Human breast cancer | MDA-MB-231 | |||
| MCF7 | ||||
| SKBR3 | ||||
| Human melanoma | A2058 | |||
| A375 | ||||
| Human pancreatic cancer | PANC1 | |||
| MiaPaca2 | ||||
| Human prostate cancer | DU145 | |||
| Human head and neck cancer | A253 | |||
| Human leukemia | MV411 | |||
| Human colon cancer | CC14 | 0.1, | Inhibits cell proliferation and induces apoptosis. | [57] |
| CC36 | 1, | |||
| Ls174T | 5, | |||
| HT29 | 10 | |||
| Human glioblastoma | DLD1 | |||
| U251 | ||||
| Human melanoma | SKMe12 | |||
| Murine breast cancer | 4T1 | 0.01, | Inhibition of invasiveness and restoration of tamoxifen sensitivity. | [59] |
| MMTV-Myc | 0.1, | |||
| Human breast cancer | MDA-MB-231 | 1 | Inhibition of cell growth and metastases. | |
| D3H2LN | ||||
| Human breast cancer | MDA-MB-231 | 0.2, | Preferentially inhibits cell viability and clonogenicity of the stem cell population. | [67] |
| 0.4, | ||||
| 0.8, | ||||
| 1, | ||||
| 5, | ||||
| 8 |
Table 4.
Antitumor effects of ivermectin in vivo
| Tumor type | Tumor cell line | Days of treat. | Dose mg/kg | Mice | Effects of Ivermectin | Ref. |
|---|---|---|---|---|---|---|
| Murine leukemia | MDAY-D2 | 10 | 3, | NOD/SCID mice | Reduces tumor volume up to 70% in all models | [44] |
| 5, | ||||||
| 6 | ||||||
| i.p. | ||||||
| Human leukemia | K562 | 3 oral | ||||
| OCI-AML2 | ||||||
| Human glioblastoma | U87 | 21 | 40 | SCID mice | Reduces tumor volume up to 50% | [47] |
| T98G | i.p. | |||||
| Breast cancer | MDA-MB-231-GFP | 10 | 2.4 | NOD/SCID mice | Reduces tumor volume up to 60% | [54] |
| i.p. | ||||||
| Human glioma | U87MG | 42 | 3, | Balb/c nude mice | Reduces tumor volume up to 50% at 3 mg/kg. | [64] |
| 10 | At 10 mg/kg tumors were not detectable | |||||
| i.t. | ||||||
| Human colon cancer | LDL1 | 21 | 10 | NMRI nude mice | Reduces tumor volume up to 85% (LDL1 cell line). No effect is observed in the tumor TCF-independent cell line (CC14) | [57] |
| CC14 | i.p. | |||||
| HT29 |
i.t.: intratumoral. i.p.: intraperitoneal.
Conclusions
The recognition that drug repositioning is a clever opportunity to accelerate the development of cancer drugs is increasing. So far, at least 235 clinically-approved, non-cancer drugs have proven antitumor activity either in vitro, in vivo, or even clinically. Among these, ivermectin, an antiparasitic compound of wide use in veterinary and human medicine, is clearly a strong candidate for repositioning, based on the fact that i) it is very safe, causing almost no side-effects other than those caused by the immune and inflammatory responses against the parasite in infected patients, and ii) it has proven antitumor activity in preclinical studies. On the other hand, it is now evident that the use of very selective “unitargeted” drugs is commonly associated to early development of resistance by cancer cells, hence the use of “dirty” or “multitargeted” drugs is important to explore. In this sense, ivermectin has this potential as it modulates several targets such as the multidrug resistance protein (MDR), the Akt/mTOR and WNT-TCF pathways, the purinergic receptors, the PAK-1 protein, certain cancer-related epigenetic deregulators such as SIN3A and SIN3B, RNA helicase activity, while stimulates chloride channel receptors leading to cell hyperpolarization, and down-regulates stemness genes to preferentially target cancer stem-cell like population, at least in breast cancer. Importantly, the in vitro and in vivo antitumor activities of ivermectin are achieved at concentrations that can be clinically reachable based on the human pharmacokinetic studies done in healthy and parasited patients. Thus, existing information on ivermectin could allow its rapid move into clinical trials for cancer patients.
Acknowledgements
Mandy Juarez is a student belonging to the Programa de Ciencias Bioquímicas, UNAM. Alejandro Schcolnik-Cabrera is a student belonging to the Plan de Estudios Combinados en Medicina (PECEM), UNAM.
Disclosure of conflict of interest
None.
References
- 1.Crump A. Ivermectin: enigmatic multifaceted ‘wonder’ drug continues to surprise and exceed expectations. J Antibiot (Tokyo) 2017;70:495–505. doi: 10.1038/ja.2017.11. [DOI] [PubMed] [Google Scholar]
- 2.Gloeckner C, Garner AL, Mersha F, Oksov Y, Tricoche N, Eubanks LM, Lustigman S, Kaufmann GF, Janda KD. Repositioning of an existing drug for the neglected tropical disease Onchocerciasis. Proc Natl Acad Sci U S A. 2010;107:3424–9. doi: 10.1073/pnas.0915125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai SH, Ogbourne S. Eco-toxicological effects of the avermectin family with a focus on abamectin and ivermectin. Chemosphere. 2016;154:204–214. doi: 10.1016/j.chemosphere.2016.03.113. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Luo M, Xu W, Yang M, Wang B, Gao J, Li Y, Tao L. Avermectin confers its cytotoxic effects by inducing DNA damage and mitochondria-associated apoptosis. J Agric Food Chem. 2016;64:6895–902. doi: 10.1021/acs.jafc.6b02812. [DOI] [PubMed] [Google Scholar]
- 5.Alberich M, Ménez C, Sutra JF, Lespine A. Ivermectin exposure leads to up-regulation of detoxification genes in vitro and in vivo in mice. Eur J Pharmacol. 2014;740:428–35. doi: 10.1016/j.ejphar.2014.06.052. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Wu J, Xu W, Gao J, Cao H, Yang M, Wang B, Hao Y, Tao L. Cytotoxic effects of Avermectin on human HepG2 cells in vitro bioassays. Environ Pollut. 2017;220:1127–1137. doi: 10.1016/j.envpol.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Ottesen EA, Campbell WC. Ivermectin in human medicine. J Antimicrob Chemother. 1994;34:195–203. doi: 10.1093/jac/34.2.195. [DOI] [PubMed] [Google Scholar]
- 8.Mealey KL, Bentjen SA, Gay JM, Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11:727–33. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto H, Messerli SM, Sudo T, Maruta H. Ivermectin inactivates the kinase PAK1 and blocks the PAK1-dependent growth of human ovarian cancer and NF2 tumor cell lines. Drug Discov Ther. 2009;3:243–6. [PubMed] [Google Scholar]
- 10.Goa KL, McTavish D, Clissold SP. Ivermectin. A review of its antifilarial activity, pharmacokinetic properties and clinical efficacy in onchocerciasis. Drugs. 1991;42:640–58. doi: 10.2165/00003495-199142040-00007. [DOI] [PubMed] [Google Scholar]
- 11.Marti H, Haji HJ, Savioli L, Chwaya HM, Mgeni AF, Ameir JS, Hatz C. A comparative trial of a single-dose ivermectin versus three days of albendazole for treatment of Strongyloides stercoralis and other soil-transmitted helminth infections in children. Am J Trop Med Hyg. 1996;55:477–81. doi: 10.4269/ajtmh.1996.55.477. [DOI] [PubMed] [Google Scholar]
- 12.Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. JAMA. 2008;299:1937–48. doi: 10.1001/jama.299.16.1937. [DOI] [PubMed] [Google Scholar]
- 13.Ottesen EA, Ramachandran CP. Lymphatic filariasis infection and diseases: control strategies. Parasitology Today. 1995;11:129–31. [Google Scholar]
- 14.Costa JL, Diazgranados JA. Ivermectin for spasticity in spinal-cord injury. Lancet. 1994;343:739. doi: 10.1016/s0140-6736(94)91625-x. [DOI] [PubMed] [Google Scholar]
- 15.McKellar QA, Benchaoui HA. Avermectins and milbemycins. J Vet Pharmacol Ther. 1996;19:331–51. doi: 10.1111/j.1365-2885.1996.tb00062.x. [DOI] [PubMed] [Google Scholar]
- 16.Burkhart CN. Ivermectin: an assessment of its pharmacology, microbiology and safety. Vet Hum Toxicol. 2000;42:30–5. [PubMed] [Google Scholar]
- 17.Umbenhauer DR, Lankas GR, Pippert TR, Wise LD, Cartwright ME, Hall SJ, Beare CM. Identification of a P-glycoprotein-deficient subpopulation in the CF-1 mouse strain using a restriction fragment length polymorphism. Toxicol Appl Pharmacol. 1997;146:88–94. doi: 10.1006/taap.1997.8225. [DOI] [PubMed] [Google Scholar]
- 18.Lankas GR, Minsker DH, Robertson RT. Robertson, effects of ivermectin on reproduction and neonatal toxicity in rats. Food Chem Toxicol. 1989;27:523–9. doi: 10.1016/0278-6915(89)90048-3. [DOI] [PubMed] [Google Scholar]
- 19.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–61. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, editor. Pharmacology/Toxicology NDA review and evaluation. 2011 20/12/2017] ; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/202736Orig1s000PharmR.pdf.
- 21.Chung K, Yang CC, Wu ML, Deng JF, Tsai WJ. Agricultural avermectins: an uncommon but potentially fatal cause of pesticide poisoning. Ann Emerg Med. 1999;34:51–7. doi: 10.1016/s0196-0644(99)70271-4. [DOI] [PubMed] [Google Scholar]
- 22.Dourmishev AL, Dourmishev LA, Schwartz RA. Ivermectin: pharmacology and application in dermatology. Int J Dermatol. 2005;44:981–8. doi: 10.1111/j.1365-4632.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- 23.De Sole G, Awadzi K, Remme J, Dadzie KY, Ba O, Giese J, Karam M, Keita FM, Opoku NO. A community trial of ivermectin in the onchocerciasis focus of Asubende, Ghana. II. Adverse reactions. Trop Med Parasitol. 1989;40:375–82. [PubMed] [Google Scholar]
- 24.De Sole G, Awadzi K, Remme J, Dadzie KY, Ba O, Giese J, Karam M, Keita FM, Opoku NO. Lack of adverse reactions in ivermectin treatment of onchocerciasis. Lancet. 1990;335:1106–7. doi: 10.1016/0140-6736(90)92687-d. [DOI] [PubMed] [Google Scholar]
- 25.Boussinesq M, Gardon J, Gardon-Wendel N, Chippaux JP. Clinical picture, epidemiology and outcome of Loa-associated serious adverse events related to mass ivermectin treatment of onchocerciasis in Cameroon. Filaria J. 2003;2(Suppl 1):S4. doi: 10.1186/1475-2883-2-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boussinesq M, Kamgno J, Pion SD, Gardon J. What are the mechanisms associated with post-ivermectin serious adverse events? Trends Parasitol. 2006;22:244–6. doi: 10.1016/j.pt.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Qu J, Li M, Zhao F, Liu C, Zhang Z, Xu S, Li S. Autophagy is upregulated in brain tissues of pigeons exposed to avermectin. Ecotoxicol Environ Saf. 2015;113:159–68. doi: 10.1016/j.ecoenv.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Baraka OZ, Mahmoud BM, Marschke CK, Geary TG, Homeida MM, Williams JF. Ivermectin distribution in the plasma and tissues of patients infected with Onchocerca volvulus. Eur J Clin Pharmacol. 1996;50:407–10. doi: 10.1007/s002280050131. [DOI] [PubMed] [Google Scholar]
- 29.Guzzo CA, Furtek CI, Porras AG, Chen C, Tipping R, Clineschmidt CM, Sciberras DG, Hsieh JY, Lasseter KC. Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. J Clin Pharmacol. 2002;42:1122–33. doi: 10.1177/009127002401382731. [DOI] [PubMed] [Google Scholar]
- 30.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34:267–72. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Langedijk J, Mantel-Teeuwisse AK, Slijkerman DS, Schutjens MH. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today. 2015;20:1027–34. doi: 10.1016/j.drudis.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Kang S, Kim W. Drug repositioning for cancer therapy based on large-scale drug-induced transcriptional signatures. PLoS One. 2016;11:e0150460. doi: 10.1371/journal.pone.0150460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med. 2012;4:27. doi: 10.1186/gm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 35.Corsello SM, Bittker JA, Liu Z, Gould J, McCarren P, Hirschman JE, Johnston SE, Vrcic A, Wong B, Khan M, Asiedu J, Narayan R, Mader CC, Subramanian A, Golub TR. The drug repurposing hub: a next-generation drug library and information resource. Nat Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantziarka PS, Meheus L, Sukhatme VP, Bouche G. Repurposing non-cancer drugs in oncology-How many drugs are out there? bioRxiv. 2017:197434. [Google Scholar]
- 37.Andrews KT, Fisher G, Skinner-Adams TS. Drug repurposing and human parasitic protozoan diseases. Int J Parasitol Drugs Drug Resist. 2014;4:95–111. doi: 10.1016/j.ijpddr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gros P, Ben Neriah YB, Croop JM, Housman DE. Isolation and expression of a complementary DNA that confers multidrug resistance. Nature. 1986;323:728–31. doi: 10.1038/323728a0. [DOI] [PubMed] [Google Scholar]
- 39.Didier A, Loor F. The abamectin derivative ivermectin is a potent P-glycoprotein inhibitor. Anticancer Drugs. 1996;7:745–51. doi: 10.1097/00001813-199609000-00005. [DOI] [PubMed] [Google Scholar]
- 40.Drinyaev VA, Mosin VA, Kruglyak EB, Novik TS, Sterlina TS, Ermakova NV, Kublik LN, Levitman MKh, Shaposhnikova VV, Korystov YN. Antitumor effect of avermectins. Eur J Pharmacol. 2004;501:19–23. doi: 10.1016/j.ejphar.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Seebacher N, Shi H, Kan Q, Duan Z. Novel strategies to prevent the development of multidrug resistance (MDR) in cancer. Oncotarget. 2017;8:84559–84571. doi: 10.18632/oncotarget.19187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pressman BC. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–30. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- 43.Mitani M, Yamanishi T, Miyazaki Y. Salinomycin: a new monovalent cation ionophore. Biochem Biophys Res Commun. 1975;66:1231–6. doi: 10.1016/0006-291x(75)90490-8. [DOI] [PubMed] [Google Scholar]
- 44.Sharmeen S, Skrtic M, Sukhai MA, Hurren R, Gronda M, Wang X, Fonseca SB, Sun H, Wood TE, Ward R, Minden MD, Batey RA, Datti A, Wrana J, Kelley SO, Schimmer AD. The antiparasitic agent ivermectin induces chloride-dependent membrane hyperpolarization and cell death in leukemia cells. Blood. 2010;116:3593–603. doi: 10.1182/blood-2010-01-262675. [DOI] [PubMed] [Google Scholar]
- 45.Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6:4569–84. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, Molina A, Shirihai OS, Collins JJ. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in Mammalian cells. Sci Transl Med. 2013;5:192ra85. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Fang S, Sun Q, Liu B. Anthelmintic drug ivermectin inhibits angiogenesis, growth and survival of glioblastoma through inducing mitochondrial dysfunction and oxidative stress. Biochem Biophys Res Commun. 2016;480:415–421. doi: 10.1016/j.bbrc.2016.10.064. [DOI] [PubMed] [Google Scholar]
- 48.Zhu M, Li Y, Zhou Z. Antibiotic ivermectin preferentially targets renal cancer through inducing mitochondrial dysfunction and oxidative damage. Biochem Biophys Res Commun. 2017;492:373–378. doi: 10.1016/j.bbrc.2017.08.097. [DOI] [PubMed] [Google Scholar]
- 49.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 50.Draganov D, Gopalakrishna-Pillai S, Chen YR, Zuckerman N, Moeller S, Wang C, Ann D, Lee PP. Modulation of P2X4/P2X7/Pannexin-1 sensitivity to extracellular ATP via Ivermectin induces a non-apoptotic and inflammatory form of cancer cell death. Sci Rep. 2015;5:16222. doi: 10.1038/srep16222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norenberg W, Sobottka H, Hempel C, Plötz T, Fischer W, Schmalzing G, Schaefer M. Positive allosteric modulation by ivermectin of human but not murine P2X7 receptors. Br J Pharmacol. 2012;167:48–66. doi: 10.1111/j.1476-5381.2012.01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–72. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 54.Dou Q, Chen HN, Wang K, Yuan K, Lei Y, Li K, Lan J, Chen Y, Huang Z, Xie N, Zhang L, Xiang R, Nice EC, Wei Y, Huang C. Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer. Cancer Res. 2016;76:4457–69. doi: 10.1158/0008-5472.CAN-15-2887. [DOI] [PubMed] [Google Scholar]
- 55.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- 56.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melotti A, Mas C, Kuciak M, Lorente-Trigos A, Borges I, Ruiz i Altaba A. The river blindness drug Ivermectin and related macrocyclic lactones inhibit WNT-TCF pathway responses in human cancer. EMBO Mol Med. 2014;6:1263–78. doi: 10.15252/emmm.201404084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seth C, Mas C, Conod A, Mueller J, Siems K, Kuciak M, Borges I, Ruiz I Altaba A. Long-Lasting WNT-TCF response blocking and epigenetic modifying activities of withanolide f in human cancer cells. PLoS One. 2016;11:e0168170. doi: 10.1371/journal.pone.0168170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon YJ, Petrie K, Leibovitch BA, Zeng L, Mezei M, Howell L, Gil V, Christova R, Bansal N, Yang S, Sharma R, Ariztia EV, Frankum J, Brough R, Sbirkov Y, Ashworth A, Lord CJ, Zelent A, Farias E, Zhou MM, Waxman S. Selective inhibition of SIN3 corepressor with avermectins as a novel therapeutic strategy in triple-negative breast cancer. Mol Cancer Ther. 2015;14:1824–36. doi: 10.1158/1535-7163.MCT-14-0980-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol. 2016;17:426–38. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 61.Abdelhaleem M. Over-expression of RNA helicases in cancer. Anticancer Res. 2004;24:3951–3. [PubMed] [Google Scholar]
- 62.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121–32. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mastrangelo E, Pezzullo M, De Burghgraeve T, Kaptein S, Pastorino B, Dallmeier K, de Lamballerie X, Neyts J, Hanson AM, Frick DN, Bolognesi M, Milani M. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J Antimicrob Chemother. 2012;67:1884–94. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin J, Park G, Lee JE, Choi EY, Park JY, Kim TH, Park N, Jin X, Jung JE, Shin D, Hong JH, Kim H, Yoo H, Lee SH, Kim YJ, Park JB, Kim JH. DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain. 2015;138:2553–70. doi: 10.1093/brain/awv167. [DOI] [PubMed] [Google Scholar]
- 65.Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ, Jeong JA, Jo SH, Kim TH, Min HS, Chae JS, Choi EJ, Kim G, Shin SH, Gwak HS, Kim SK, Hong EK, Lee GK, Choi KH, Kim JH, Yoo H, Park JB, Lee SH. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011;30:2433–42. doi: 10.1038/onc.2010.620. [DOI] [PubMed] [Google Scholar]
- 66.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dominguez-Gomez G, Chavez-Blanco A, Medina-Franco JL, Saldivar-Gonzalez F, Flores-Torrontegui Y, Juarez M, Díaz-Chávez J, Gonzalez-Fierro A, Dueñas-González A. Ivermectin as an inhibitor of cancer stem-like cells. Mol Med Rep. 2018;17:3397–3403. doi: 10.3892/mmr.2017.8231. [DOI] [PubMed] [Google Scholar]


