Abstract
More than 50 years have passed since the presentation of the Replicon Model which states that a positively acting initiator interacts with a specific site on a circular chromosome molecule to initiate DNA replication. Since then, the origin of chromosome replication, oriC, has been determined as a specific region that carries sequences required for binding of positively acting initiator proteins, DnaA-boxes and DnaA proteins, respectively. In this review we will give a historical overview of significant findings which have led to the very detailed knowledge we now possess about the initiation process in bacteria using Escherichia coli as the model organism, but emphasizing that virtually all bacteria have DnaA proteins that interacts with DnaA boxes to initiate chromosome replication. We will discuss the dnaA gene regulation, the special features of the dnaA gene expression, promoter strength, and translation efficiency, as well as, the DnaA protein, its concentration, its binding to DnaA-boxes, and its binding of ATP or ADP. Furthermore, we will discuss the different models for regulation of initiation which have been proposed over the years, with particular emphasis on the Initiator Titration Model.
Keywords: DnaA protein, DnaA box, dnaA gene, DnaA ADP/ATP, oriC, initiation control models, initiator titration, cell cycle
Introduction
The Replicon Model hypothesizes that to initiate DNA replication a positively acting initiator molecule interacts with a specific site, the replicator, on a circular chromosome (Figure 1; Jacob et al., 1963). The model was probably designed based on two main findings: (1) that the bacterial chromosome was circular (Cairns, 1963); and (2) that arrest of protein and/or RNA synthesis led to inhibition of initiation but allowed termination of ongoing rounds of replication (Maaløe and Hanawalt, 1961).
Figure 1.

The replicon model. A drawing in the sand (provided by M. Mechali to an EMBO meeting report by Skarstad et al. (2003, Copyright, John Wiley and Sons) to celebrate the 40th anniversary of the model.
Since then, several models, to be addressed later, have been designed to explain some of the characteristics of the coupling between the cell cycle and the growth of Escherichia coli and possibly also several other bacteria. Escherichia coli can grow at very different growth rates depending on the carbon source and other nutrients present in the growth medium. The generation time in rich media, such as LB medium, is ~20 min at 37°C whereas generation times in minimal media with poor carbon sources can be several hours. At all these generation times it is characteristic for E. coli (and most other bacteria) that fast growing cells are big and slowly growing cells are small, but the size of a baby cell will always be very close to half the size of the mother cell independent of the growth rate (Schaechter et al., 1958; Maaløe and Kjeldgaard, 1966).
It should be emphasized that the present review is the story about DnaA and oriC. For more detailed reviews see e.g., Messer (2002), Skarstad and Katayama (2013), or Leonard and Grimwade (2015).
The cell cycle
A very important early input to characterization of the bacterial cell cycle was obtained in the work with the baby cell machine; a membrane elution technique (Helmstetter and Cummings, 1963, 1964). In this method the cells of an exponentially growing culture are attached to a membrane filter, the filter holder is inverted and growth medium is passed through the filter. The bacteria grow on the filter and one daughter cell detaches upon cell division and is collected. The main findings were that if cells grew with generation times shorter than 60 min the time it took to replicate the chromosome was close to 40 min, this was termed the C-period, and that the time it took from termination of chromosome replication to cell division was close to 20 min, termed the D-period (Helmstetter, 1967; Helmstetter and Cooper, 1968; Helmstetter et al., 1968). These primary studies were carried out using the Escherichia coli B/r strain that behaved very well in the baby cell machine, i.e., the mother cells were firmly attached to the filter (Helmstetter and Cummings, 1963, 1964). Two different approaches have been used in baby cell experiments. One where DNA was labeled with radioactivity before the cells were attached to the filter and radioactivity was determined in the effluent at different times, the other (and conceptually simpler) where newborn cells were collected on ice to stop growth and an experiment was started with bacterial babies transferred to the desired growth temperature getting synchronized cell division (Clark and Maaløe, 1967). This has been the preferred method for later experiments addressing cell cycle parameters. Cells with generation times longer than 60 min very often have a period with only one non-replicating chromosome, this period was termed the B-period (Helmstetter and Pierucci, 1976). Cells growing with generation times longer than 60 min have longer C- and D-periods than given above, and the B-, C-, and D-period show some variation in different strains of E. coli (Helmstetter, 1996; Michelsen et al., 2003).
The initiation mass
The determination of the cell cycle parameters (see above) and the very careful measurements of DNA and cell mass in a study of macromolecular concentrations at different growth rates of Salmonella typhimurium (Schaechter et al., 1958) led to the suggestion that bacteria like E. coli and to S. typhimurium, initiate replication when a certain mass (or volume) is present per origin of replication, the initiation mass (Donachie, 1968) or the critical volume (Pritchard et al., 1969). There has been some dispute whether the initiation mass is the same at all growth rates (Wold et al., 1994). However, it is generally accepted that the initiation mass does not vary very much in E. coli K-12 growing at different growth rates (Herrick et al., 1996).
Phenomenology of control of chromosome replication
The results obtained with the baby cell machine can only be explained if all chromosomal origins in a cell are initiated once and only once per cell cycle. The length of the C and D periods for fast growing bacteria implies that these cells contain several origins and have several ongoing replication forks. That this is the case was demonstrated clearly by flow cytometry of cultures treated with rifampicin that inhibits initiation of chromosome replication but allows ongoing replication forks to terminate (Skarstad et al., 1986). To determine origins per cell at the time of drug addition it is necessary also to stop cell division (Løbner-Olesen et al., 1989) by simultaneous addition of cephalexin (Begg and Donachie, 1985). In the fast growing cells which carried more than one origin at the time of initiation (and rifampicin addition) flow cytometry showed in addition that all origins were initiated synchronously, i.e., cells had either 2n or 2(n+1) replicated chromosomes (Skarstad et al., 1986) which (with simultaneous cephalexin addition) will be equal to the number of origins per cell (Skarstad et al., 1986; Løbner-Olesen et al., 1989). Thus, a model for control of chromosome replication should satisfy what we know about the cell cycle and the behavior of chromosomes and origins. However, before going into the modeling mode we will give a relatively detailed description of the dnaA gene and its gene product, which is a positively acting initiator, and the origin of replication, which is the replicator. In Francois Jacob's laboratory, Masamichi Kohiyama isolated temperature sensitive DNA replication mutants some of which continued to replicate at a non-permissive growth temperature until they terminated replication, presumably at the terminus of replication (Jacob et al., 1963; Kohiyama et al., 1963). Some of these mutants were dnaA mutants. This historical tale is about the initiator, the DnaA protein and its interaction with the replicator, the origin of replication - oriC, and will focus on details about the dnaA gene encoding the DnaA protein as well as the requirements for the interaction of the DnaA protein with oriC.
The dnaA mutants
The temperature sensitive K-12 strains CRT46 and CRT83, were the first strains carrying mutations in the dnaA gene, dnaA46, and dnaA83, respectively. These mutations were genetically mapped and found to be located in the vicinity of the ilv locus (Hirota et al., 1968). In the following years several strains carrying mutations in the dnaA gene (see Figure 5 below) were isolated in different laboratories (Kuempel, 1969; Carl, 1970; Wechsler and Gross, 1971; Beyersmann et al., 1974; Sevastopoulos et al., 1977). The PC2 strain (Carl, 1970) exhibits a slow stop phenotype similar to that of dnaA mutants, but carries a mutation in the dnaC gene that encodes the loading factor for DnaB, the DNA helicase.
Fine mapping of the dnaA gene
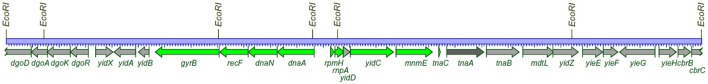
The genetic structure of the dnaA region of the chromosome was characterized using specialized transducing λ bacteriophages, λtna (Hansen and von Meyenburg, 1979; Ream et al., 1980). The chromosomal genes carried on the phages were identified by recombinational rescue of mutations known to be located close to the ilv locus, and mapped relative to an EcoRI restriction enzyme map constructed from cleavage of different transducing phages. In this study the size of gene products, the proteins, were also characterized by introducing the λtna phages into UV-killed bacteria which had degraded all chromosomal DNA (Murialdo and Siminovitch, 1972). The dnaA and dnaN genes were also characterized by a Japanese group (Sakakibara and Mizukami, 1980). Several recombinant plasmids were constructed using restriction enzymes and DNA from different specialized transducing λtna phages. These plasmids were used to determine the precise position of more genes by complementation of mutants and characterization of gene products in the maxicell system (Sancar et al., 1979; Hansen F. G. et al., 1982; Hansen et al., 1985). The dnaA region as characterized by λtna phages and plasmids derived from the phages is shown in Figure 2.
Figure 2.
The dnaA region of the E. coli chromosome. 26,332 bp between the outmost EcoRI sites is the chromosomal DNA present on two λtna phages (λtna 552 and λ tna 530). The drawing is based on the published complete E. coli sequence version (acc.no. U00096.2). The genes (proteins) stained green are those that could be identified on SDS-polyacrylamide gels. The light gray proteins and the dark gray protein encoded by the tnaA gene were not visible (Hansen and von Meyenburg, 1979; Ream et al., 1980).
The origin of replication—oriC
The origin of replication was found to be located close to the ilv genes in several studies during the 1970s (e.g., Bird et al., 1972; Marsh and Worcel, 1977). The oriC symbol was introduced for the origin of replication (Hiraga, 1976). A minichromosome, pSY211, carrying oriC was constructed by combining an ~9.5 kb EcoRI fragment with a non-replicating fragment conferring ampicillin resistance (Yasuda and Hirota, 1977). It was also obtained on specialized transducing λasn phages (von Meyenburg et al., 1979) which could be established as minichromosomes when transduced into a recombination deficient recA strain lysogenic for λ and requiring asparagine. Such strains are heat-sensitive due to the presence of the λcI857 gene on the λ-minichromosome. Small minichromosomes, e.g., pCM959 (von Meyenburg et al., 1979; Buhk and Messer, 1983) carrying exclusively chromosomal DNA, were isolated by selecting heat-resistant Asn+ clones (von Meyenburg et al., 1979). Further characterization of oriC was carried out by deletion analysis of a plasmid carrying a fragment with a selectable marker and the 9.5 kb EcoRI fragment from λasn132 (Messer et al., 1978, 1979). In this way oriC could be located between a BamHI and a XhoI site and this 422 bp region was sequenced (Meijer et al., 1979; Messer et al., 1979; Sugimoto et al., 1979)1. Further deletion analysis limited the oriC region to 245 bp (Tabata et al., 1983).
The DnaA-boxes
The sequence of the E. coli oriC was the first origin of replication to be reported for a cellular organism. The next origin to be sequenced was that of Salmonella typhimurium (Zyskind and Smith, 1980). Similarly to the E. coli oriC the Salmonella sequence carried an unexpected high density of GATC sites (Figure 3), where the position of most were conserved when compared to E. coli.
Figure 3.
The region of the E. coli chromosome carrying oriC, the origin of replication, and neighboring genes. A recent review presents a detailed walkthrough of the various binding sites in oriC and the interactions with the DnaA protein and other factors in forming the orisome that is required to initiate replication (Leonard and Grimwade, 2015). DnaA boxes with high (dark blue boxes) and low (light blue boxes) affinity, respectively; positions of GATC sites (*). However, in an exponentially growing culture containing cells of all ages only the R1, R2, and R4 are bound by DnaA protein (Samitt et al., 1989).
Sequencing other origins of the Enterobacteriacea family (Zyskind et al., 1981; Cleary et al., 1982) revealed a repeated sequence (R-box) which was later renamed the DnaA-box. Since these early days hundreds of bacteria have been sequenced. Virtually all bacteria carry a dnaA gene. The approximate position of the origin of replication can in many cases be determined by the Base-Skew-Method (McLean et al., 1998) and in most cases the precise location of the origin can be identified by the presence of DnaA-boxes which are also present in virtually all bacteria.
DnaA sets the initiation mass—is the master regulator of initiation
The first evidence that DnaA was not just essential for the initiation process but could be the initiator regulating initiation came from the studies of dnaA(Ts) mutants at permissive temperature which showed that initiation mass increased with increasing growth temperature (Hansen and Rasmussen, 1977; Tippe-Schindler et al., 1979). Further support for DnaA as the initiator was provided by the dnaAcos mutant (a pseudorevertant of dnaA46) which has a decreased initiation mass at low temperature (Kellenberger-Gujer et al., 1978). The hypothesis that DnaA was the initiator was disputed after the first studies of strains carrying DnaA overproducing plasmids since they did not show an increase in DNA per mass as expected from a decreased initiation mass (Churchward et al., 1983). An analysis measuring also the actual concentration of oriC DNA per mass (Atlung et al., 1987) revealed that initiation was stimulated in cells overproducing DnaA protein, but the replication speed was decreased so much that there was virtually no increase in DNA per mass. A later analysis where we used plac controlled expression to increase the DnaA protein concentration from 1.2- to 4-fold the normal (Atlung and Hansen, 1993) showed that within a limited range the origins per mass responded to the DnaA protein concentration (see Figure 4) showing that DnaA protein is indeed the limiting factor for initiation. Also in these experiments replication speed is decreased inversely with the higher DnaA protein concentration. Supporting this finding are the following experiments: (1) where wild type DnaA protein was induced to different subnormal levels in a dnaA46 mutant at 42°C which showed low DNA per mass (Løbner-Olesen et al., 1989); (2) lowering the expression in a dnaA+ strain by interfering with the expression of the messenger of the chromosomal dnaA gene using the tCRISPRi system (Si et al., 2017); and 3. using a slowly growing dnaA204 mutant where the DnaA protein concentration was significantly reduced, due to instability of the protein, causing an increase in the initiation mass and thus a decrease in origin to mass ratio (Torheim et al., 2000).
Figure 4.
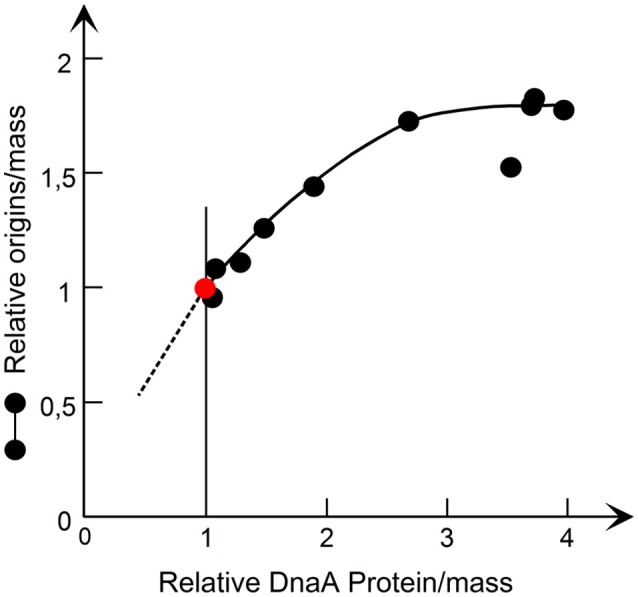
The origin/mass responds to an increase in DnaA protein. The DnaA protein was synthesized from a lacP controlled dnaA gene and origins/mass was determined by flow cytometry. The data are redrawn from Figure 4 in Atlung and Hansen (1993, Copyright, American Society for Microbiology).
The DnaA protein structure
The DnaA protein has four domains (Messer et al., 1999). The domain structure was first suggested by the comparison of the E. coli and B. subtilis proteins (Ogasawara et al., 1985) which showed that there is a moderately conserved N-terminal region and a more highly conserved large C-terminal region separated by a region of highly variable sequence and length. More closely related proteins show a high conservation of the N- and C-terminal domains—e.g., the Enterobacterial proteins have virtually identical sequences in these domains—but many amino acid substitutions, deletions and insertions in the variable region (Skovgaard and Hansen, 1987; Skovgaard, 1990). The C-terminal region contains domains III and IV, with an AAA+ family ATPase motif and the DNA binding domain respectively (Messer et al., 1999). It took a long time after the initial purification of the DnaA protein (Chakraborty et al., 1982; Fuller and Kornberg, 1983) before the first physical structures of DnaA were obtained and as yet no structure of the full length protein has been reported. The structure of domain IV from E. coli was determined by NMR and (Obita et al., 2002) and a year later the crystal structure of domain IV in complex with a DnaA-box was obtained (Fujikawa et al., 2003). This confirmed that the DNA binding is mediated by a combination of a basic loop and a helix-turn-helix motif. The structure of domain III has only been obtained from Aquifex aeolicus (Erzberger et al., 2002). When bound to ATP, but not to ADP, DnaA forms a super-helical structure with four monomers per turn (Erzberger et al., 2006; see Mott and Berger, 2007; for review and for the best figure). The structure of domain I has been determined for E. coli by NMR (Abe et al., 2007) and from three other bacterial species (see Zawilak-Pawlik et al., 2017 for review).
Function of the different domains
The first indication that each of the domains is essential for the activity of the DnaA protein came from the mapping and sequencing of the classical dnaATs mutants (Hansen et al., 1984, 1992), see Figure 5 for position of the mutations and their amino acid changes. Since then many other dnaA mutants have been isolated both by different selections and by screening of in vitro generated mutants (e.g., reviewed in Erzberger et al., 2002; and additional mutants described in the following citations: Felczak and Kaguni, 2004; Asklund and Atlung, 2005; Felczak et al., 2005; Chodavarapu et al., 2013).
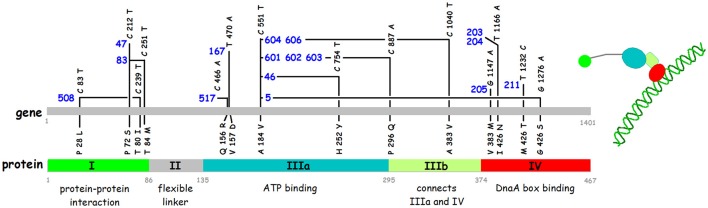
Figure 5.
Nucleotide and amino acid alterations in the dnaA gene of sixteen temperature sensitive mutants. The mutant numbers are in blue. References for the isolation of the different mutants are: dnaA508, dnaA517 (Wechsler and Gross, 1971); dnaA47 (Kuempel, 1969); dnaA46, dnaA83 (Kohiyama et al., 1966); dnaA167 (Abe and Tomizawa, 1971); dnaA601, dnaA602, dnaA603, dnaA604, dnaA606 (Sevastopoulos et al., 1977); dnaA203, dnaA204, dnaA205, dnaA211 (Beyersmann et al., 1974). The sequences of 12 of these mutants are described by Hansen et al. (1992). The four mutations dnaA47, dnaA83, dnaA517, and dnaA603 were sequenced using PCR fragments obtained from the original mutant strains (this work). The domains in the DnaA protein and the borders between them are shown in the lower part of the figure.
The C-terminal domain IV is alone responsible for sequence specific binding to double stranded DNA and the classical dnaATs mutations mapping here reduce the binding (Roth and Messer, 1995).
Domain III contains the AAA+ ATPase motif and is responsible for nucleotide binding (Sekimizu et al., 1987). DnaA binds both ATP and ADP with similar affinities, KD 10–30 nM and 60–100 nM respectively (Sekimizu et al., 1987; Kawakami et al., 2006; Keyamura and Katayama, 2011), and the ATP form is required for the normal control of initiation of replication (Sekimizu et al., 1987). The ATP form facilitates cooperative binding to weak DnaA boxes (Margulies and Kaguni, 1996; Speck et al., 1999; McGarry et al., 2004), is necessary for unwinding of the DUE (Duplex Unwinding Element) in oriC (Sekimizu et al., 1987), and for binding to single stranded (ss) DNA in the DUE (Speck and Messer, 2001; Ozaki and Katayama, 2012). Domain III binds the ss DNA directly through specific amino acids, e.g., V211 and R245 (Ozaki et al., 2008; Duderstadt et al., 2011). The terminal part of Domain III also contains a sequence patch for interaction with the membrane (Garner and Crooke, 1996) which stimulates release of the nucleotide from DnaA (Sekimizu and Kornberg, 1988). Except for dnaA167, and possibly also dnaA517, all the other classical Ts dnaA mutants that map in domain III carry the same mutation, i.e., the A184V change close to the P-loop of the Walker A ATP binding motif and four different secondary mutations (Figure 3), and the DnaA46 protein has been shown to be defective in nucleotide binding (Hwang and Kaguni, 1988; Carr and Kaguni, 1996).
Domain II is thought to be a flexible linker connecting domains I and III. All parts of domain II (between residues 78 and 136) can be deleted without major effects on function as long as the linker is 21–27 amino acids long (Nozaki and Ogawa, 2008) and the entire YFP sequence can be inserted without any effect on function (Nozaki et al., 2009a).
Domain I has several functions in relation to protein-protein interactions (see Zawilak-Pawlik et al., 2017 for review). Domain I is responsible for dimerization of DnaA protein (Weigel et al., 1999). The dimerization is essential for the function of the protein in initiation of replication as deletion of the first 24 amino acids renders the protein inactive in complementation of the dnaA46 mutant (Weigel et al., 1999). Residues 24–86 are essential for interaction with DnaB, but recruitment of DnaB at oriC also requires the first 23 amino acids (Seitz et al., 2000). The DiaA (DnaA initiator associating protein) also interacts with domain I. The diaA gene was identified in a selection for suppressors of the dnaAcos mutant that over initiates (Ishida et al., 2004). The DiaA protein is not essential, but stimulates initiation of replication in vitro, and a diaA null mutant is slightly impaired in initiation and shows moderate asynchrony (Ishida et al., 2004). The DiaA protein is a tetramer (Keyamura et al., 2007, 2009). The amino acids in DnaA involved in dimerization and in DiaA/DnaB interaction are located on opposite faces of the 3 D structure of domain I, therefore the DiaA tetramer protein can act as a connector for more distantly located DnaA dimers and monomers.
Autoregulation of DnaA protein synthesis
The first indication that the dnaA gene is autoregulated came from the studies of dnaA(Ts) mutants. DnaA protein continues to be synthesized at non-permissive temperatures where it is inactive, but in some mutants it can be reactivated when brought back to a permissive growth temperature (Hanna and Carl, 1975; Messer et al., 1975). This reversible initiation capacity, which could be observed in the absence of protein synthesis at the permissive temperature, was higher than expected from the mass increase of the culture and suggested that the DnaA protein synthesis was derepressed at the high growth temperature. These findings led to a detailed study of the dnaA46 mutant at permissive, intermediate, and non-permissive growth temperatures (Hansen and Rasmussen, 1977). The results obtained in this study suggested that the DnaA46 protein became less and less active—the higher the growth temperature—resulting in gradually lower DNA and origin concentrations at the intermediate temperatures. In parallel with the decrease in DnaA protein activity an increase in initiation capacity was observed. Based on these findings Hansen and Rasmussen (1977) proposed that the DnaA protein had a positive role in initiation of replication and a negative role in its own synthesis.
The dnaA promoter region and the dnaA gene was sequenced (Hansen E. B. et al., 1982; Hansen F. G. et al., 1982) and two promoters giving transcripts entering the dnaA gene were identified. The DnaA promoter region carries 9 GATC sites within 225 bp and between the two promoters a sequence homologous to repeats (DnaA-boxes) in the oriC region was identified (box 1 in Figure 6). Both promoters are negatively regulated by DnaA protein (Atlung et al., 1984, 1985; Braun et al., 1985; Kücherer et al., 1986). In these studies the dnaA transcription was found to be increased up to 5-fold in dnaATs mutants at non-permissive temperature and to be repressed 4- to 5-fold by overproduction of DnaA protein. The DnaA-box is essential for the autoregulation of the dnaA gene (Atlung et al., 1984, 1985; Braun et al., 1985). A closer look at the sequence of the dnaA2p promoter region reveals interesting features. Two GATC sites are present in this promoter, one in the−10 sequence and the other in the −35 sequence, and methylation enhances transcription 2-fold from this promoter both in vivo and in vitro (Braun and Wright, 1986). Furthermore, DnaA protein binds with high affinity to sequences upstream of the dnaA2p promoter (Speck et al., 1999). This in vitro study showed that DnaA-ATP as well as DnaA-ADP binds to what they call DnaA box 1 and DnaA box 2, whereas a box they call “DnaA box a” that has two misfits to the consensus sequence (see below) only binds DnaA-ATP. From a comparison of the dnaA2p promoter region with the mioC promoter region (see below) as well as from a closer look at the in vitro binding data (Speck et al., 1999) we suggest that the region act as having four closely spaced DnaA boxes upstream of the dnaA2p promoter region one of which requires DnaA-ATP for binding (Figure 6) and two sites, box b and box c with no homology to DnaA boxes, which also requires DnaA-ATP. The DnaA-ATP form is required for repression of both promoters in vitro (Speck et al., 1999) in accordance with the location of the different kinds of DnaA-boxes.
Figure 6.
The dnaA region with GATC sites (*) and consensus DnaA boxes (box). The position of dnaA promoters p1 and p2 are indicated (Hansen F. G. et al., 1982). The light orange rectangle shows the p2 promoter and potential DnaA boxes upstream of and overlapping the promoter. box1, box2 and the region in between are protected both by DnaA-ADP and DnaA-ATP, the boxes a, b, and c require DnaA-ATP (Speck et al., 1999). The CCGCGG sequence upstream of the +1 position is a Travers box (Travers, 1980).
Other features of DnaA gene expression
Transcription
The dnaA promoter region is fairly strong, giving transcription corresponding to 50% of the tet promoter in pBR322 (Atlung et al., 1984). The dnaA gene proximal promoter p2 is approximately 3-fold stronger than the p1 promoter (Atlung et al., 1985; Kücherer et al., 1986; Chiaramello and Zyskind, 1990). dnaA transcription has been reported to be growth rate regulated decreasing with decreasing growth rate (Chiaramello and Zyskind, 1990; Polaczek and Wright, 1990). The p2 promoter carries a discriminator “Travers box” (Travers, 1980) downstream of the −10 sequence and transcription is, like that of the growth rate regulated rrnP1 promoter, almost completely inhibited by high ppGpp levels.
Translation and mRNA stability
The coding sequence of dnaA has a GUG start codon and a rather poor ribosome binding site (GGAG). The efficiency of translation initiation in mRNA with a SD sequence is 2- to 3-fold lower with a GUG start codon relative to an AUG start codon (Reddy et al., 1985; Donnell and Janssen, 2001). That this is similar for the dnaA mRNA is supported by the finding that the dnaA508cos mutant carrying a GUG to AUG mutation has an increased DnaA protein level sufficient to give auto-suppression of the temperature sensitivity (Eberle et al., 1989). This is similar to suppression of the temperature sensitivity obtained by having a dnaA(Ts) gene on pBR322 derived plasmids and on the chromosome (Hansen et al., 1992; Nyborg et al., 2000).
In addition to the low translation frequency the production of DnaA protein from the mRNA is limited by a short mRNA half-life. The chemical half-life is in the low range, in minimal medium it was amongst the shortest 1% of all mRNAs (1/3 of the mean half-life, Bernstein et al., 2002).
The wild type DnaA protein is stable (Atlung and Hansen, 1999) and is present in K-12 at ~200 monomeric molecules per oriC (Hansen et al., 1991b). Therefore, the cell has to produce 100 new molecules per dnaA gene per generation. The above characteristics of DnaA expression, moderately high transcription rate, low translation initiation frequency and highly unstable mRNA we estimate to give on the average one protein molecule per transcript. These are the features needed to ensure a well-regulated production of protein over the cell cycle that responds smoothly to regulatory changes in transcription initiation (see Figure 8).
DnaA protein at different growth rates
DnaA protein concentration in several different E. coli K-12 strains and in Salmonella typhimurium was determined by immunoblot analysis and found to be very similar in all these strains (Hansen et al., 1991b). This study furthermore showed that the concentration of DnaA protein in all the strains was the same at all growth rates (doubling times between 150 and 22 min). Similar studies using four different primary DnaA antibodies gave the same results (Herrick et al., 1996). The constant DnaA protein concentration might intuitively be in conflict with increasing levels of mRNA with increasing growth rate. But you have to consider two factors: (1) the time available for protein synthesis, i.e., the generation time, (2) the translation efficiency, i.e., the number of proteins produced per mRNA at the different growth rates (see Table 1). If the translation efficiency is the same at 30 and 60 min doubling times, you will need twice as many mRNAs/mass to produce the same number of protein molecules/mass in the faster growing cells. It has been calculated that the average mRNAs are translated more efficiently in faster growing cultures, close to proportionality between number of translations and growth rate (Bremer and Dennis, 1996). The dnaA mRNA is, however, very different from the average mRNA in translation efficiency, we estimated it to be one translation per mRNA (see above) compared to the 30–90 translations for the average mRNA. It is therefore likely that dnaA mRNA efficiency might be more similar at different growth rates.
Table 1.
DnaA protein production at different growth rates.
| Generation time (min) | |||||
|---|---|---|---|---|---|
| Row | Entity | 100 | 60 | 30 | Reference/derivation |
| 1 | Relative total RNA/mass | 0,6 | 0,74 | 1 | Bremer and Dennis, 1996 |
| 2 | % mRNA of total RNA | 2 | 2 | 2 | Bremer and Dennis, 1996 |
| 3 | Relative total mRNA/mass | 0,6 | 0,75 | 1 | From rows 1 and 2 |
| 4 | Average no of translations per mRNA | 27 | 33 | 71 | Bremer and Dennis, 1996 |
| 5 | Relative dnaA mRNA/total RNA | 0,27 | 0,37 | 1 | Chiaramello and Zyskind, 1990 |
| 6 | Relative dnaA mRNA/mass | 0,45 | 0,5 | 1 | from rows 1 and 5 |
| 7 | Relative DnaA per mass | 1 | 1 | 1 | Hansen et al., 1991b |
Initiator titration
The first studies on titration of DnaA protein by DnaA boxes in vivo were conducted looking at derepression of the dnaA gene by DnaA boxes from oriC—mioC and the dnaA promoter region using a single copy dnaA-lacZ fusion (Hansen et al., 1987) and lead to the formulation of the initiator-titration model for control of initiation of replication (see below, Hansen et al., 1991a).
This was followed by two studies aimed at identifying other high affinity DnaA binding sites on the chromosome. In the first only one fragment was obtained and analyzed (Kitagawa et al., 1996). It carried what was later named the datA (DnaA titration) locus. The datA locus contains two consensus DnaA boxes and four 1-misfit boxes (see Figure 14) but was found to titrate much more DnaA protein than the oriC mioC region (Kitagawa et al., 1996). The other study, using a fishing method to isolate high affinity DNA fragments, identified five loci (in addition to oriC) including datA and the mutH promoter (Roth and Messer, 1998) that was later found in the search for DARSs (DnaA Reactivating Sequences, see below; Fujimitsu and Katayama, 2004). The in vivo titration ability—effect on dnaA transcription—of these other four loci has not been studied.
The consensus DnaA box TTWTNCACA was suggested by Schaper and Messer (1995) based on binding of purified DnaA protein to 21 bp oligonucleotides. There are 307 of these consensus DnaA boxes on the E. coli chromosome. The DnaA box quality depends on the base in the fifth position and on the neighboring base sequence. Oligonucleotides which carried a DnaA box of the R1-R4 type (TTATCCACA) and had the neighboring bases as present in oriC were very efficient in binding the protein (KD 10−9 M). Similar experiments were carried out using 17 bp double stranded oligonucleotides with 6 bp upstream and 2 bp downstream of the four consensus boxes starting with TTAT and the mioC (R5) box TTTTCCACA as well as with 104 bp PCR fragments with identical sequences except for the 8 different consensus boxes in the middle of the PCR fragment (Hansen et al., 2006). In the boxes starting with TTAT a C in the fifth position (R1-R4 type) is better than A (R2) or G which is better than T. In the boxes of the mioC type starting with TTTT, C in the fifth position is much better than the other possibilities, which show very weak or unspecific binding. Plasmids carrying the mioC promoter with the 8 different consensus boxes gave varying degrees of titration of DnaA protein that was in good accordance with the in vitro determined relative binding affinities, and the degree of repression of transcription from the mioC promoter also varied as expected from the binding affinities.
As noted above the mioC promoter region carries one consensus DnaA box and the oriC region carries three boxes, which in the gel shift assays were found to be very efficient binders. Nevertheless, the plasmid carrying the mioC promoter region is as efficient in titrating DnaA protein as a plasmid carrying an inactivated oriC region (Hansen et al., 2006). This study also demonstrated that sequences ~100 bp upstream of the promoter contributed to efficient titration and thus derepression of the dnaA-lacZ promoter. An even more detailed study (Hansen et al., 2007) led to definition of DnaA boxes in the mioC region with several misfits (Figure 7). The number of DnaA proteins per origin has been determined to be ~200 monomers in several different E. coli K-12 derivatives growing at different growth rates (Hansen et al., 1991b). The plasmid with the mioC promoter region increases dnaA gene expression 1.4-fold corresponding to 80 DnaA monomers and as the pBR322 derived plasmid carrying the mioC promoter region is present in ~10 copies/oriC (Atlung et al., 1999) it follows that each mioC promoter region titrates approximately eight DnaA monomers. Similar calculations for the titration caused by the dnaA promoter region (Hansen et al., 1987) suggest that the dnaA promoter region titrates 3–4 DnaA proteins.
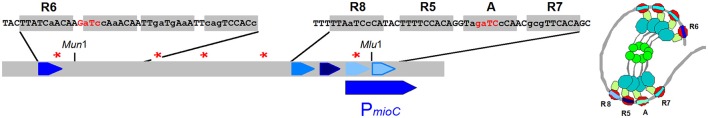
Figure 7.
The five experimentally verified DnaA boxes in the mioC region are named above the sequence boxes (Hansen et al., 2007) but also potential box sequences to the right of the R6 box are indicated. GATC sites are indicated with red stars (or red letters). To the right is a speculative view of DnaA protein bound to the DnaA-boxes in the mioC region (see Figure 5 for domain colors).
The presence of plasmids carrying DnaA boxes affects initiation of replication leading to increased initiation mass in parallel with the increased dnaA gene expression (Christensen et al., 1999; Morigen et al., 2003).
As mentioned above the datA region that carries two consensus DnaA boxes surrounded by closely spaced boxes with up to several misfits (see Figure 14) titrates large amounts of DnaA protein and plasmids carrying these boxes are difficult to handle in wild type cells.
GATC sites, SeqA protein, and the eclipse period
The average density of the GATC sequence in a DNA sequence containing equal amounts of the four bases is one in 256 bp. The density of GATC sites in the oriC sequence and the dnaA promoter sequence is approx. ten times higher (Figures 3, 6). Unmethylated plasmids carrying the oriC or the pBR322 origin cannot transform Dam+ strains, and fully methylated plasmids cannot transform a Dam− strain. In both of these cases the plasmids will become hemimethylated upon replication in the recipient cell, therefore it was concluded that full methylation was essential for the establishment of the plasmids (Russell and Zinder, 1987). The oriC and the dnaA regions become hemimethylated after initiation of replication and stays hemimethylated (are sequestered) for a considerable time period (1/3 and 1/5 of a doubling time, respectively) while regions with few GATC-sites become methylated within 1–2 min (Lark, 1968; Campbell and Kleckner, 1990).
The SeqA protein binds to closely spaced hemi-methylated GATC sites (Brendler et al., 2000) and the oriC and dnaA promoter regions contains several closely spaced GATC sites (see Figures 3, 6). These regions were “sequestered” in much shorter periods when the seqA gene was mutated or absent (Lu et al., 1994; von Freiesleben et al., 1994). A flow cytometric study of the re-initiation kinetics in several dnaA(Ts) mutants which had been initiation aligned at high temperatures showed that after the immediate initiation of one round of replication at the permissive temperature there was a constant number of origins until a second initiation took place (Hansen, 1995). This period, the eclipse, was studied in more detail in strains producing normal, less than normal, and higher than normal Dam-methylase (von Freiesleben et al., 2000a). These experiments showed clearly that the length of the eclipse (sequestration) period was inversely related to the cellular level of Dam-methylase.
The initiation cascade
Flow cytometry studies of wild type and dam mutant cells using synchronous cultures obtained by the baby cell machine (Løbner-Olesen et al., 1994) gave the experimental evidence for the initiation cascade as a model for how E. coli containing multiple origins initiates all of these once and only once per cell cycle (Hansen et al., 1991a). The cascade states that in wild type cells initiation of one origin will lead to release of DnaA protein bound to that origin, these DnaA molecules cannot bind to the two new oriC sequences due to the sequestration of the hemimethylated DNA by the SeqA protein. Therefore, the level of unbound DnaA protein is raised and will promote initiation at another “old” fully methylated origin. Thus, initiation will continue until all origins have initiated. To prevent reinitiation in the same cell cycle the availability of unbound initiation competent DnaA protein must be lowered before the end of the eclipse period.
The initiation cascade also explains the lack of incompatibility between minichromosomes and the chromosome despite the fairly high copy number of the minichromosomes (average 8–10 copies per chromosomal oriC; Løbner-Olesen et al., 1987). Before the isolation and characterization of the first minichromosomes it was expected, based on the observations with plasmids, that there would be incompatibility between the chromosomal and minichromosomal oriCs, i.e., that the presence of minichromosomes would inhibit initiation of chromosomal replication. Baby cell experiments showed that the minichromosomes initiate at the same time in the cell cycle as the chromosome (Leonard and Helmstetter, 1986), and flow cytometric analysis showed that the presence of a minichromosome only has a very small effect on the synchronous initiation of 4 or 8 chromosomal origins in fast growing cells (Løbner-Olesen and von Freiesleben, 1996; Skarstad and Løbner-Olesen, 2003). In contrast DnaA boxes carried on plasmids like pBR322 that replicate randomly during the cell cycle disturbs the initiation cascade leading to asynchrony (Christensen et al., 1999; Morigen et al., 2003).
Synthesis of DnaA protein during the cell cycle
Transcript analysis using an initiation aligned dnaC2 mutant showed that after 30 min at 40°C transcription of the mioC gene was completely shut down, dnaA transcription was halved and gidA transcription was virtually unaffected (Theisen et al., 1993). This suggests that DnaA protein synthesized during the incubation of the culture at 40°C will be filling the DnaA boxes in the mioC promoter region effectively repressing all transcription, while the dnaA promoter will be less affected since it can only be partially repressed. The gidA promoter, that has no DnaA boxes, is not affected at the high temperature. Soon after initiation at the permissive temperature, there was virtually no transcription from the dnaA promoter and only a little from the gidA promoter whereas mioC transcription was restored after a few minutes. One minute after initiation the dnaA gene is replicated and the dnaA promoter region will become hemimethylated and sequestered by SeqA for a considerable period (~10 min; Campbell and Kleckner, 1990; Lu et al., 1994) inactivating the two promoters. The gidA promoter is located close to the sequestered oriC and is probably partially co-sequestered with the oriC region for some time. The mioC promoter is not sequestered and repression will be reduced by decreased DnaA activity due to titration and RIDA as replication proceeds. The findings obtained with the dnaC aligned system was confirmed using synchronous cultures obtained with the baby cell machine (Theisen et al., 1993). A considerable number of non-sequestered genes was also analyzed using these two synchronization techniques (Zhou et al., 1997) but no dramatic changes during the cell cycle were observed.
The dnaA transcription data presented above (Theisen et al., 1993) allow us to speculate about the relative numbers of DnaA protein and DnaA boxes during the cell cycle in the simple situation of a slowly growing culture. Assuming that DnaA protein is synthesized in proportion to the DnaA messenger we can predict how the number of DnaA proteins increases during a cell cycle (Figure 8). There is a period before initiation where dnaA messenger and thus protein is accumulated slowly due to autorepression. Soon after initiation the dnaA promoter region is sequestered and both dnaA promoters are inactivated. When the dnaA promoters are released from sequestration following methylation, the DnaA protein synthesis rate will be high, due to a low concentration of DnaA protein relative to titrating DnaA boxes. The level of DnaA-ATP, that is the active form repressing the dnaA promoter, will also be low, due to RIDA and DDAH (see later).
Figure 8.
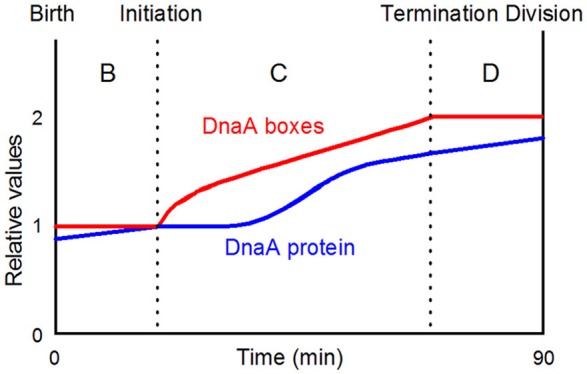
A simplistic view of when initiation takes place in the cell cycle of slowly growing bacteria. The DnaA protein curve is drawn on the basis of the data presented by Theisen et al. (1993). DnaA-boxes with any distribution will be doubled during the C-period; the actual distribution of all consensus DnaA-boxes and selected groups of consensus boxes are presented in Figure 10; the figure is redrawn from Figure 2A in Herrick et al. (1996, Copyright, John Wiley and Sons).
Models for control of initiation of chromosome replication
The Replicon Model (Jacob et al., 1963) states that an initiator structure has to be build to interact with the origin to initiate replication. At initiation new origins are synthesized and the initiator structure is destroyed or at least prevented from reinitiating at newly synthesized origins. New initiator structures have to be constructed/reformed before initiation can take place in the following cell cycle. Basically such a model is valid but at the time it was presented the molecular basis for the initiator structure could not be addressed.
A model which attracted a lot of attention at these early times was the Inhibitor Dilution Model (Pritchard et al., 1969). It postulates that a fixed amount of messengers coding for inhibitor molecules are synthesized in a burst soon after initiation. These inhibitors would be diluted during growth of the cell and when the concentration reaches a threshold allow initiation to take place followed by the burst of new inhibitors. The molecular mechanism for how to synthesize a “fixed” amount of inhibitor after initiation was not discussed. This model inspired formulation of new models for how initiation in E. coli could be controlled. The inhibitor dilution model has later been formally rejected by Margalit and Grover (1987).
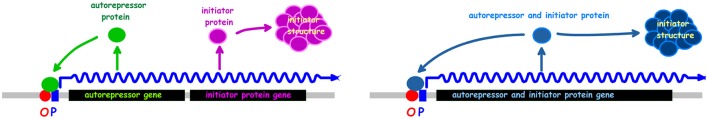
The autorepressor model (Sompayrac and Maaløe, 1973) hypothesizes the existence of an operon with two genes, one coding for a repressor protein, the other for an initiator protein (Figure 9). The repressor interacts with an operator in the promoter and represses its own synthesis to keep the repressor concentration constant and thus the production of initiator molecules constant. The model proposes that the initiator molecules build an initiator structure which will be ready to initiate when the cell reaches its initiation mass and will be destroyed at initiation. After initiation the cell can start building a new initiator structure which can be used to initiate the following replication cycle. An analysis of the model using a stochastic approach suggested that the synthesis of 250 initiator molecules shows the best fit to the experimental observations in a culture synchronized with the baby cell machine (Clark and Maaløe, 1967). This number is surprisingly close to the number of DnaA proteins per oriC found in several E. coli strains (Hansen et al., 1991b). The autorepressor model has been tested in a very detailed theoretical study by Margalit et al. (1984) in which it was concluded that the model could be valid provided that the location of the operon is close to oriC, that the promoter is moderate to strong, translation efficiency is low, and that the repressor binds with a moderate affinity (KD ~10−9 M); the dnaA gene and DnaA protein fulfills all of these requirements.
Figure 9.
The autorepressor model. The left drawing presents the model in its original formulation (Sompayrac and Maaløe, 1973). The right drawing shows an alternative where the autorepressor and initiator is the same protein.
The model published by Mahaffy and Zyskind (1989) was based on data stating that the DnaA protein concentration increases at increasing growth rates (Chiaramello and Zyskind, 1989). This is at variance with the now generally accepted invariance of DnaA protein concentration in K-12. It should be noted that Mahaffy and Zyskind (1989) had to use different nucleotide bound forms of DnaA to get the model to fit with the different DnaA concentrations at different growth rates.
The Initiator Titration Model, which we will cover in some detail, was first presented at the EMBO workshop in Segovia, Spain in 1987 and it did not change before it was published (Hansen et al., 1991a).
The model hypothesizes that:
The initiator—DnaA protein—has a high affinity for specific sites—DnaA boxes on the chromosome.
The initiator—DnaA protein—has a lower affinity for sites involved in the formation of the initiation complex.
DnaA protein bound to DnaA boxes is released (pushed off) by replication.
A newly replicated origin is refractory to initiation for the period it takes to regain a normal topology.
The model was analyzed by computer simulations using a stochastic approach. A main goal was to find ONE set of parameters to put into in silico experiments which could be compared to different in vivo experimental data. To this end the comprehensive theoretical study which concluded that the autorepressor model might be a possible model (Margalit et al., 1984) was very helpful. Some of the parameters we have used are presented in Table 2. When we designed parameters for the model we hypothesized that the active form of DnaA protein was a tetramer and most often we used 75 DnaA-boxes distributed on the chromosome, usually with a higher occurrence of DnaA-boxes close to the origin. The actual consensus DnaA box distribution on the chromosome is presented in Figure 10.
Table 2.
Selected parameters used in the Initiator Titration Model.
| Input variables | Calculated numbers | ||
|---|---|---|---|
| τ | Doubling time | V | Cell volume |
| C | Replication time | T | Age of cells |
| D | Time from termination to cell division | Tn's | Replication fork positions |
| Ai | Number of DnaA molecules required for initiation | n | DnaA boxes per growing cell |
| KD | Dissociation constant for DnaA-DnaA box interaction | G | Genome equivalents per cell |
| kdnaA1p | Promoter constant dnaA1p (3)a | g | dnaA genes per cell |
| kdnaA2p | Promoter constant dnaA2p (15)a | A | DnaA proteins per cell |
| kdnaAcp | Promoter constant dnaA constitutive component (3)a | m | dam genes per cell |
| kdam | Promoter constant damP (0.5)a | δ | Fraction of time the dnaA promoters are open (calculated using KD, A, n, V) |
The numbers in parentheses are relative promoter activities.
Figure 10.
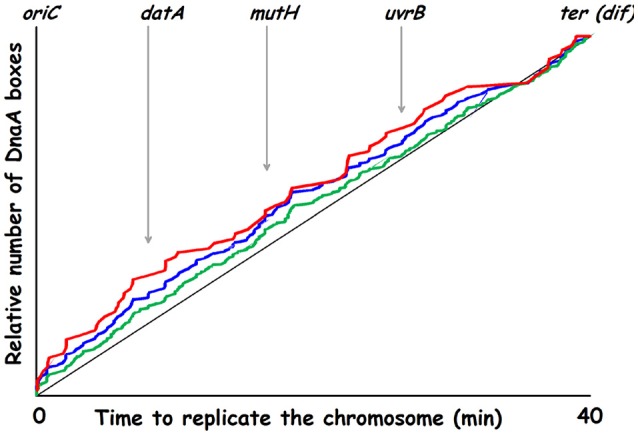
Replication time for DnaA boxes on the K-12 chromosome. Green curve: 307 consensus DnaA boxes (TTWTNCACA); blue curve: 164 DnaA boxes (TTATNCACA + TTTTCCACA); red curve: 78 DnaA boxes (TTATNCACA where N is either C, A, or G). Genes in or lying close to chromosomal regions involved in nucleotide conversions are indicated.
In the computer experiments we started with one predefined unit baby cell that was allowed to run through 50 generations where one randomly chosen daughter was discarded. After 50 generations randomly selected daughters (baby cells) were saved in an array until arriving at a chosen cell number (e.g., 1,000 cells). These computer cells can be used directly as a perfectly synchronized culture, or they can be randomized to obtain an exponential culture by starting the baby cells one by one at different times. The computer culture grows and accumulates cells, until the cell number has doubled, then the computer culture is diluted 2-fold.
The in silico experiment shown in Figure 11B was first carried out with the assumption that the mother cell divided into two equally sized baby cells. To our surprise these simulations resulted in a computer culture behaving incredibly synchronous and that kept the synchrony for a much longer time than should be expected (data not shown). This in silico result was the reason why we changed the cell division mode to have a stochastic positioning of the division septum. To illustrate our stochastic approach we present an example where we use experimental data from the literature to address cell size variation after cell division. The cell length of mothers and daughters of slowly growing E. coli B/r cells were analyzed (Schaechter et al., 1962; Koppes et al., 1978) and found to exhibit some variation (coefficient of variation, CV, of 8–10%) in the positioning of the septum. Figure 12 demonstrates how we handle every single cell at division.
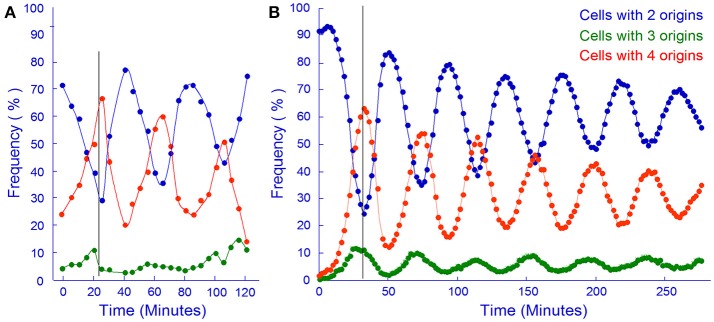
Figure 11.
In vivo and in silico synchronized cultures. (A) Samples from a synchronous culture obtained using the baby cell machine were incubated with rifampicin and the frequency of cells containing different numbers of origins were determined by flow cytometry (redrawn from Figure 4A in Løbner-Olesen et al., 1994, Copyright, John Wiley and Sons). (B) Same as A but using computer simulation of the Initiator Titration Model to obtain baby cells. The cell size variation in this simulation had a CV of 6.3.
Figure 12.
The picture shows a constricted cell of E. coli which will divide asymmetrically. The cell length and the position of the constriction was found using a macro written for the Image-Pro Plus software (Nielsen and Hansen, 2010). To the left is C-code showing how we handle cell division in our simulation. c is the starting cell that increases exponentially in size with a constant generation time. At division a sum of random numbers are generated and divided with the number of random numbers. For this calculation we assume that the division take place in the central 40% of the cell (~the multiplication by 0.4), thus one of new cells will inherit 30% of the size of the old cell plus what came out of the random number considerations. If the No_of_random_no is 8 or 13 it will give CVs of 8.3 or 6.3, respectively.
Consequences of asymmetric divisions
The synthesis of DnaA protein can be described by the formulas:
| (1) |
Ccell is a component describing the general status of the cell at a given time point:
| (2) |
where PSS is the amount of ribosomes, RNA polymerase, and other proteins, as well as nucleotides, amino acids and other small molecules needed to synthesize proteins, RNA, and DNA. The PSS will be proportional to the cell size. The g is the actual number of dnaA genes in the cell at any given time in the cell cycle, and G is the actual number of genome equivalents in the cell which will be proportional to all the other genes. And the promoter component:
| (3) |
that is growth rate dependent (kμ), α is 0 if the promoter region is sequestered or 1 if the region is accessible, the kdnaA1 and kdnaA2 are promoter constants, kdnaAc describes the DnaA protein synthesis that cannot be repressed. For each cell in the computer at each time step as the cell grows the degree of promoter openness (δ) will be calculated from KD, the number of DnaA proteins, and the number of DnaA boxes.
Thus if a cell divides asymmetrically one of the babies will become bigger and will therefore contain more PSS, but each baby will be born with the same number of number of dnaA genes (g) and the same amount of genome equivalents (G). The net result of the asymmetric division will be that the big daughter synthesizes DnaA protein a little faster than the smaller and therefore will be ready for the next initiation a little faster than the other. The overall result of this is that the two cells have about the same size (but not the same age) at the next initiation.
Today we know much more than we did in 1987, and we have to include the different nucleotide bound forms of DnaA protein (see later). However, the model with its original parameter setting can simulate several experiments as published earlier (Hansen et al., 1991a) and the one shown in Figure 11.
Recent additional features of regulation of initiation by DnaA
The initiator titration model was formulated based on the concept that the concentration of DnaA protein determines the initiation mass and the findings of titration of DnaA protein by plasmids carrying DnaA boxes. The model incorporated detailed knowledge about regulation of dnaA gene expression, the eclipse period and the role of Dam, and the initiation synchrony of multiple origins in fast growing cells. In these last sections we will review the newer findings about the role of the datA locus as well as the DARS1 and DARS2 loci and the interconversion of the two nucleotide forms of DnaA (Figure 13) and evaluate their respective contributions to initiation control before concluding remarks about revision of the model for control of initiation to include these aspects.
Figure 13.
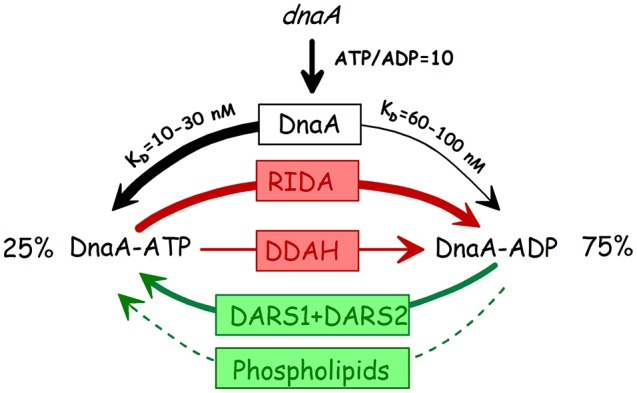
The three states of the DnaA protein. The thickness of the arrows indicate the approximate relative contributions of the different elements to the interconversion of the different forms. Stipled arrow: the relative contribution of phospholipid mediated nucleotide exchange (Sekimizu and Kornberg, 1988; Aranovich et al., 2006, 2015) is unknown. RIDA: Replicative inactivation of DnaA (Katayama et al., 1998; Kato and Katayama, 2001); DDAH: datA dependent ATP hydrolysis (Kasho and Katayama, 2013); DARS: DnaA Reactivating sequence (Fujimitsu et al., 2009).
DnaA-ATP hydrolysis—HDA and datA
Replicative inactivation of DnaA (RIDA) through hydrolysis of DnaA-ATP to DnaA-ADP was first discovered in vitro and shown to be dependent on replication and to require the “IdaB” (now Hda) protein and the β-clamp (DnaN; Katayama et al., 1998). A large increase in the DnaA-ATP form is found at high temperature in the dnaN59(Ts) and several other temperature sensitive replication mutants (Katayama et al., 1998; Kurokawa et al., 1999), showing that RIDA is also working in vivo. The hda gene was found in a search for clones that complemented (new) mutants defective in F plasmid maintenance at permissive temperature and were Ts for growth (Kato and Katayama, 2001). The gene was named hda (homologous to DnaA) and the Hda protein has extensive homology to domain III of DnaA. The Hda protein was shown to be identical to the IdaB protein through in vitro complementation, and an hda null mutant was shown to have a high percentage of DnaA-ATP (Kato and Katayama, 2001). In this study the hda(Ts) strain was also characterized in vivo and found to have increased oriC DNA per mass and increased oriC/terC indicating “abortive replication.”
Until recently, there has been some controversy regarding the essentiality of the hda gene. In the initial identification of hda (Kato and Katayama, 2001) it was found that the gene could not be deleted and only conditional mutants could be obtained. A little later isolation of an hda null mutant was reported (Camara et al., 2003). The hda null mutants are, however, slowly growing under standard conditions (aerobically on rich medium at 37°C) and genetically very unstable and quickly acquire suppressor mutations of many different types (Riber et al., 2006; Charbon et al., 2011). The growth defects are due to replication fork collapse caused by oxidative DNA damage so hda mutants have virtually no growth defects when grown anaerobically (Charbon et al., 2014) and also grow well in minimal media (Charbon et al., 2017). When grown under either of these permissive conditions the hda mutants still exhibit a decreased initiation mass (0.70–0.75) and moderate asynchrony.
The datA locus was originally believed to exert its effect on regulation of initiation through titration of DnaA protein, similar to, but much stronger than, the titration by plasmids with the mioC or dnaA promoters (Kitagawa et al., 1998; Morigen et al., 2005). Later it was found that datA mediates DnaA-ATP hydrolysis of DNA bound protein, a process named DDAH (Kasho and Katayama, 2013). Only two of the five original DnaA-boxes (box 2 and 3) were absolutely required together with the IHF binding site (IBS) located between them. Further studies showed that a 2-misfit box next to box 2 (box 7, see Figure 14) was also essential for the DDAH activity (Kasho et al., 2017). The contribution of DDAH to DnaA-ATP hydrolysis is fairly small, in an otherwise wt strain deletion of datA increased DnaA-ATP a few % (Katayama et al., 2001) and when the datA deletion was introduced into the rnhA ΔoriC- Δhda strain the percentage of DnaA-ATP was increased from 72 to ~90% (Kasho and Katayama, 2013).
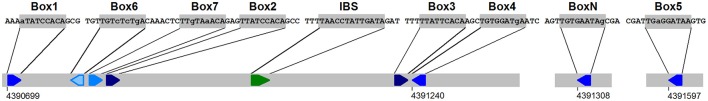
Figure 14.
The datA region responsible for DDAH (datA dependent ATP hydrolysis, Kitagawa et al., 1998; Morigen et al., 2005). The DnaA boxes 2, 7, 3, and IBS are absolutely required for DDAH activity (Kasho et al., 2017). The coordinates are from E. coli K12 (acc.no. U00096.2). IBS: IHF binding site. The color code of the boxes is the same as in Figure 7. An additional one-misfit box (BoxN) is also indicated.
Deletion of the datA locus has no effect on the growth rate of the bacteria but does affect the initiation control, leading to a decrease in the initiation mass (Kitagawa et al., 1998; Morigen et al., 2005; Frimodt-Møller et al., 2016). There is some controversy as to the degree of asynchrony conferred by deletion of datA. The first study which found a rather pronounced asynchrony in ΔdatA cells (Kitagawa et al., 1998) also showed that such cells have close to the same DNA content and the same cell size distribution as wild type cells. Morigen et al. (2005) showed that much of the apparent asynchrony was due to rifampicin resistant initiations. It should be noted (emphasized) that rifampicin resistant initiations lead to overestimation of the effect on initiation mass. Rifampicin resistant initiation of chromosome replication has also been observed in ihf mutants, however, such initiations did not take place in strains carrying ihf and dnaA46 mutations (von Freiesleben et al., 2000b) suggesting that the rifampicin resistant initiation phenotype of ihf mutants might be due to reduced function of datA in DDAH.
The phenotype of the ΔdatA strain must primarily be due to the lack of DDAH activity since different point mutations inactivating the DDHA activity—mutation of the IBS, or inactivation of box 2 or box 7 have the same phenotype (Ogawa et al., 2002; Nozaki et al., 2009b; Kasho et al., 2017). The apparent very high titration activity of plasmids with the intact datA locus can largely be explained by the DDAH activity which lowers the DnaA-ATP level and gives increased expression from promoters which require this form of DnaA for repression. It is, however, clear that the datA locus also directly titrates DnaA protein by binding to the boxes, both from the initial deletion analysis of the datA plasmids where deletions outside the box 1–4 region gave reduced titration (Kitagawa et al., 1996) and from the analysis of plasmids with a box 2 or an IBS point mutation that only reduced titration (Nozaki et al., 2009b).
DnaA-ADP rejuvenation
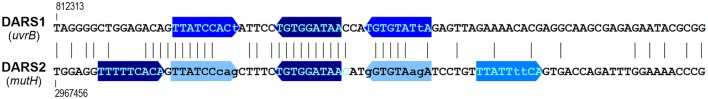
The first DARS DnaA-reactivating sequence, which is present in the ColE1 plasmid origin, was found somewhat fortuitously in a search for factors mediating in vitro DnaA-ADP to ATP exchange (Fujimitsu and Katayama, 2004). The full ColE1 DARS was delimited to a 70 bp sequence with three DnaA boxes, where two of the boxes in correct orientation were essential for activity. Subsequently two chromosomal DARSs were identified in the genome sequence amongst 52 pairs of similarly arranged DnaA boxes with 0 or 1 misfit to the sequence TTATNCACA (Fujimitsu et al., 2009). The DARS1 sequence is located in the uvrB promoter region and DARS2 in the mutH promoter (Figures 10, 15), both previously identified as sequences that interact with DnaA (van den Berg et al., 1985; Roth and Messer, 1998). The presence of extra DARS1 or DARS2 sequences on low copy number plasmids leads to decreased initiation mass (Fujimitsu et al., 2009; Charbon et al., 2011) and introduction of higher copy number plasmids with DARS1 or DARS2 is deleterious to the cells (van den Berg et al., 1985; Fujimitsu et al., 2009). Deletion of either DARS1 or DARS2 from the chromosome increases initiation mass by ~20% (Fujimitsu et al., 2009; Frimodt-Møller et al., 2016) and the two DARS sequences act synergistically since deletion of both increase initiation mass to close to 150% of the normal (Fujimitsu et al., 2009). Deletion of either DARS sequence reduce the DnaA-ATP percentage in the rnhA ΔoriC-Δhda strain, and deletion of both DARS's almost returns the DnaA-ATP level to that of the hda+ strain (Fujimitsu et al., 2009). The effect of the DARS deletions, especially DARS2, is sufficient to give suppression of the lethality associated with lack of Hda mediated ATP hydrolysis (Fujimitsu et al., 2009; Charbon et al., 2011).
Figure 15.
The DARSs (DnaA reactivating sequences). The coordinates given above and below the DARS sequences (Fujimitsu et al., 2009) are from E. coli K12 (acc.no. U00096.2). Vertical lines between the sequences indicate identical bases. See Figure 7 for color code for the boxes.
Intriguingly, it is only plasmids with DARS2, not with DARS1, that give asynchrony (Fujimitsu et al., 2009; Charbon et al., 2011), and only the DARS2 deletion that leads to asynchrony (Fujimitsu et al., 2009; Frimodt-Møller et al., 2016). This might be due to a higher efficiency of DARS2 in DnaA rejuvenation giving a stronger effect on DnaA-ATP % (Fujimitsu et al., 2009). It should be noted that all the data concerning initiation mass and asynchrony referred to above are from cells grown in minimal glucose casamino acids supplemented medium at 37°C. No systematic studies of the effect of DARS have been published for other growth media—i.e., slow growing cells.
We have summarized the effects of the different elements, de novo synthesis, RIDA, DDAH, and DARS on the interconversion of the two nucleotide bound forms of DnaA in Figure 13.
Initiator titration revisited
A fundamental feature of the initiator titration model is that at the end of the eclipse (oriC sequestration) period the replication process has increased the number of DnaA boxes sufficiently to titrate so many DnaA proteins that there is not enough available for the low affinity binding at oriC. The additional feature with the two nucleotide forms of the DnaA protein and their interconversion does not essentially change this concept, in this case the amount of DnaA-ATP protein has to be reduced below a threshold during the eclipse period. Let us first consider a relatively slowly growing cell, e.g., 60 min doubling with a C time of 40 min. As soon as replication starts RIDA begins to work due to the presence of β-clamps remaining on the DNA after passage of the replisome, and when the datA locus is duplicated (8 min after initiation) the DDAH process will increase the hydrolysis rate a little. During these first approx. 10 min there is no DnaA protein synthesis and only one copy of the DARS sequences, and therefore the DnaA-ATP level will fall quickly. When the dnaA gene is no longer sequestered the DnaA-ATP dependent autorepression, will be low, and the DnaA-ATP level will start to increase due to synthesis of new proteins. At 16 min DARS2 is duplicated. In this relatively slow growing cell DARS2 might, however, be inefficient in nucleotide release since it requires Fis for this activity (Kasho et al., 2014) and Fis concentration is low in slow growing cells (Ball et al., 1992). At 26 min also DARS1 is duplicated and this will increase the building up of DnaA-ATP levels. At 40 min replication terminates and soon after RIDA will cease due to unloading of the β-clamps from the DNA which only takes a few minutes (Leu et al., 2000)2. In faster growing cells with overlapping replication cycles RIDA will be active throughout the cell cycle, but will vary with the number of replication forks, e.g., in a cell growing with 25 min doubling time the old forks will terminate 15 min after initiation, reducing the RIDA activity to two thirds, thus accelerating the buildup of DnaA-ATP. Unfortunately the variation in DnaA-ATP levels over the cell cycle has not been investigated using baby cells, it has only been measured in dnaC initiation synchronized cells (Kurokawa et al., 1999), which is an artificial situation probably with an unnaturally high DnaA-ATP starting level.
It will be interesting to incorporate the DnaA-ATP/ADP conversion systems into the initiator titration model and see how that behaves compared to the original initiator titration model, and to a version of the model where we insert the actual high affinity DnaA-box distribution.
It is clear that the original initiator titration model can to a large extent simulate the behavior of E. coli during exponential growth. And the dnaA46 mutant with a protein that does not bind ATP (in vitro) is quite fit at low temperature, growing with an almost normal growth rate and having a reasonably uniform cell size distribution despite the asynchrony of initiation (and increased initiation mass). This raises the question why has E. coli (and other bacteria) evolved an additional level of regulation with the DnaA-ATP/DnaA-ADP interconversion systems? It will be interesting to see if these systems are necessary and under which growth conditions—how does an E. coli strain that is deleted for hda, datA, and the two DARSs behave? It will probably be necessary to simultaneously reduce the dnaA gene expression to get a cell with a normal initiation mass avoiding the side effects of over initiation.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are particularly grateful to Knud V. Rasmussen who as a Master project supervisor started our scientific entry into the field of initiation of chromosome replication and to Kaspar von Meyenburg who took part in much of the early work on the subject. We are also grateful to deceased Walter Messer who has participated in a number of open discussions of what his group did at the Max Planck Institute in Berlin and what we did in Copenhagen. At different periods we have also had close collaboration with Anders Løbner-Olesen, Ole Skovgaard, and Bjarke Bak Christensen.
Footnotes
1At the Cold Spring Harbor meeting in 1978 where the 422 bp sequence carrying oriC was presented (Messer et al., 1979), Hirota's group presented the complete sequence of a 2.1 kb BamHI fragment. After the meeting K. von Meyenburg and F. G. Hansen went to Arthur Kornberg's laboratory at Stanford University where we after comparing the sequences could see that the small 91 bp BamHI fragment was missing from the original presentation of the Japanese group. Therefore minichromosome pSY211, which was available at Stanford University, was restricted with BamHI and analyzed on non-denaturing poly-acrylamide gels that allowed the separation of very small fragments. F.G.Hansen found that minichromosome pSY211 also contained 2 two small BamHI fragment. The sequence of the 91 bp BamHI fragments was added to the final report from the Japanese group (Hirota et al., 1979) and referred to as “Hausen and Schaller, personal communication.”
2The calculated average residence time for the β-clamps (3.5–7 min) is based on a very careful determination of the number of molecules of different polymerase subunits per cell, giving a number of 350 β-clamp dimers/cell in an exponentially growing culture of C600 in LB medium. However, the final calculation uses only 2–4,000 Okazaki fragments produced per cell in 40 min, corresponding to only one replicating chromosome per cell, but C600 growing in LB medium has mostly 8 origins per cell, and thus 4 pairs of replisomes, so the average residence time therefore has to be 3- to 4-fold shorter.
References
- Abe M., Tomizawa J. (1971). Chromosome replication in E. coli K12 mutant affected in DNA initiation. Genetics 69, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. (2007). Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282, 17816–17827. 10.1074/jbc.M701841200 [DOI] [PubMed] [Google Scholar]
- Aranovich A., Braier-Marcovitz S., Ansbacher E., Granek R., Parola A. H., Fishov I. (2015). N-terminal-mediated oligomerization of DnaA drives the occupancy-dependent rejuvenation of the protein on the membrane. Biosci. Rep. 35:e00250. 10.1042/BSR20150175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranovich A., Gdalevsky G. Y., Cohen-Luria R., Fishov I., Parola A. H. (2006). Membrane-catalyzed nucleotide exchange on DnaA. Effect of surface molecular crowding. J. Biol. Chem. 281, 12526–12534. 10.1074/jbc.M510266200 [DOI] [PubMed] [Google Scholar]
- Asklund M., Atlung T. (2005). New non-detrimental DNA-binding mutants of the Escherichia coli initiator protein DnaA. J. Mol. Biol. 345, 717–730. 10.1016/j.jmb.2004.10.092 [DOI] [PubMed] [Google Scholar]
- Atlung T., Christensen B. B., Hansen F. G. (1999). Role of the Rom protein in copy number control of plasmid pBR322 at different growth rates in Escherichia coli K-12. Plasmid 41, 110–119. 10.1006/plas.1998.1386 [DOI] [PubMed] [Google Scholar]
- Atlung T., Clausen E., Hansen F. G. (1984). Autorepression of the dnaA gene of Escherichia coli, in Proteins Involved in DNA Replication, eds Huebscher U., Spadari S. (New York, NY; London: Plenum Press; ), 199–207. [DOI] [PubMed] [Google Scholar]
- Atlung T., Clausen E. S., Hansen F. G. (1985). Autoregulation of the dnaA gene of Escherichia coli. Mol. Gen. Genet. 200, 442–450. 10.1007/BF00425729 [DOI] [PubMed] [Google Scholar]
- Atlung T., Hansen F. G. (1993). Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J. Bacteriol. 175, 6537–6545. 10.1128/jb.175.20.6537-6545.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Hansen F. G. (1999). Low-temperature-induced dnaa protein synthesis does not change initiation mass in Escherichia coli K-12 low-temperature-induced dnaa protein synthesis does not change initiation mass in Escherichia coli K-12. J. Bacteriol. 181, 5557–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T., Løbner-Olesen A., Hansen F. G. (1987). Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in E. coli. Mol. Gen. Genet. 206, 51–59. 10.1007/BF00326535 [DOI] [PubMed] [Google Scholar]
- Ball C. A., Osuna R., Ferguson K. C., Johnson R. C. (1992). Dramatic change of fis level upon nutrient upshift in Escherichia coli. J. Bacteriol. 174, 8043–8056. 10.1128/jb.174.24.8043-8056.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. (1985). Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J. A., Khodursky A. B., Lin P. H., Lin-Chao S., Cohen S. N. (2002). Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 99, 9697–9702. 10.1073/pnas.112318199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyersmann D., Messer W., Schlicht M. (1974). Mutants of Escherichia coli B/r defective in DNA initiation: dnaI, a new gene for replication. J. Bacteriol. 118, 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird R. E., Louarn J., Maruscelli J., Caro L. (1972). Origin and sequence of chromosome replication in Escherichia coli. J.Mol.Biol. 70, 549–566. 10.1016/0022-2836(72)90559-1 [DOI] [PubMed] [Google Scholar]
- Braun R. E., O'Day K., Wright A. (1985). Autoregulation of the DNA replication gene dnaA in E. coli. Cell 40, 159–169. [DOI] [PubMed] [Google Scholar]
- Braun R. E., Wright A. (1986). DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol. Gen. Genet. 202, 246–250. 10.1007/BF00331644 [DOI] [PubMed] [Google Scholar]
- Bremer H., Dennis P. P. (1996). Modulation of chemical composition and other parameters of the cell by growth rate, in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds Neidhardt F. C., Curtiss R. I., Ingraham J. L., Lin E. C. C., Brooks Low K., Magasanik B., et al. (Washington, DC: American Society for Microbiology; ), 1553–1569. [Google Scholar]
- Brendler T., Sawitzke J., Sergueev K., Austin S. (2000). A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 19, 6249–6258. 10.1093/emboj/19.22.6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhk H.-J., Messer W. (1983). The replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene 24, 265–279. 10.1016/0378-1119(83)90087-2 [DOI] [PubMed] [Google Scholar]
- Cairns J. (1963). The chromosome of Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 28, 43–46. 10.1101/SQB.1963.028.01.011 [DOI] [PubMed] [Google Scholar]
- Camara J. E., Skarstad K., Crooke E. (2003). Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 185, 3244–3248. 10.1128/JB.185.10.3244-3248.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. (1990). E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62, 967–979. 10.1016/0092-8674(90)90271-F [DOI] [PubMed] [Google Scholar]
- Carl P. L. (1970). E. coli mutants with temperature sensitive synthesis of DNA. Mol. Gen. Genet. 109, 107–122. 10.1007/BF00269647 [DOI] [PubMed] [Google Scholar]
- Carr K. M., Kaguni J. M. (1996). The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 20, 1307–1318. 10.1111/j.1365-2958.1996.tb02649.x [DOI] [PubMed] [Google Scholar]
- Chakraborty T., Yoshinaga K., Lother H., Messer W. (1982). Purification of the E. coli dnaA gene product. EMBO J. 1, 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon G., Bjørn L., Mendoza-Chamizo B., Frimodt-Møller J., Løbner-Olesen A. (2014). Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res. 42, 13228–13241. 10.1093/nar/gku1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon G., Campion C., Chan S. H. J., Bjørn L., Weimann A., da Silva L., et al. (2017). Re-wiring of energy metabolism promotes viability during hyperreplication stress in E. coli. PLoS Genet. 13:e1006590. 10.1371/journal.pgen.1006590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbon G., Riber L., Cohen M., Skovgaard O., Fujimitsu K., Katayama T., et al. (2011). Suppressors of DnaA(ATP) imposed overinitiation in Escherichia coli. Mol. Microbiol. 79, 914–928. 10.1111/j.1365-2958.2010.07493.x [DOI] [PubMed] [Google Scholar]
- Chiaramello A. E., Zyskind J. W. (1989). Expression of Escherichia coli dnaA and mioC genes as function of growth rate. J. Bacteriol. 171, 4272–4280. 10.1128/jb.171.8.4272-4280.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaramello A. E., Zyskind J. W. (1990). Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J. Bacteriol. 172, 2013–2019. 10.1128/jb.172.4.2013-2019.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu S., Felczak M. M., Simmons L. A., Murillo A., Kaguni J. M. (2013). Mutant DnaAs of Escherichia coli that are refractory to negative control. Nucleic Acids Res. 41, 10254–10267. 10.1093/nar/gkt774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B. B., Atlung T., Hansen F. G. (1999). DnaA boxes are important elements in setting the initiation mass of Escherichia coli. J. Bacteriol. 181, 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Holmans P., Bremer H. (1983). Increased expression of the dnaA gene has no effect on DNA replication in a dnaA+ strain of Escherichia coli. Mol. Gen. Genet. 192, 506–508. 10.1007/BF00392197 [DOI] [PubMed] [Google Scholar]
- Clark D. J., Maaløe O. (1967). DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23, 99–112. 10.1016/S0022-2836(67)80070-6 [DOI] [Google Scholar]
- Cleary J. M., Smith D. W., Harding N. E., Zyskind J. W. (1982). Primary structure of the chromosomal origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae: comparisons and evolutionary relationships. J. Bacteriol. 150, 1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W. D. (1968). Relationship between cell size and time of initiation of DNA replication. Nature 219, 1077–1079. 10.1038/2191077a0 [DOI] [PubMed] [Google Scholar]
- Donnell S. M. O., Janssen G. R. (2001). The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5 ′ untranslated leader. J. Bacteriol. 183, 1277–1283. 10.1128/JB.183.4.1277-1283.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt K. E., Chuang K., Berger J. M. (2011). DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213. 10.1038/nature10455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle H., van de Merwe W., Madden K., Kampo G., Wright L., Donlon K. (1989). The nature of an intragenic suppressor of the Escherichia coli dnaA508 temperature-sensitive mutation. Gene 84, 237–245. 10.1016/0378-1119(89)90497-6 [DOI] [PubMed] [Google Scholar]
- Erzberger J. P., Mott M. L., Berger J. M. (2006). Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13, 676–683. 10.1038/nsmb1115 [DOI] [PubMed] [Google Scholar]
- Erzberger J. P., Pirruccello M. M., Berger J. M. (2002). The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21, 4763–4773. 10.1093/emboj/cdf496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felczak M. M., Kaguni J. M. (2004). The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 279, 51156–51162. 10.1074/jbc.M409695200 [DOI] [PubMed] [Google Scholar]
- Felczak M. M., Simmons L. A., Kaguni J. M. (2005). An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 280, 24627–24633. 10.1074/jbc.M503684200 [DOI] [PubMed] [Google Scholar]
- Frimodt-Møller J., Charbon G., Krogfelt K. A., Løbner-Olesen A. (2016). DNA replication control is linked to genomic positioning of control regions in Escherichia coli. PLoS Genet. 12:e1006286. 10.1371/journal.pgen.1006286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., et al. (2003). Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 31, 2077–2086. 10.1093/nar/gkg309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K., Katayama T. (2004). Reactivation of DnaA by DNA sequence-specific nucleotide exchange in vitro. Biochem. Biophys. Res. Commun. 322, 411–419. 10.1016/j.bbrc.2004.07.141 [DOI] [PubMed] [Google Scholar]
- Fujimitsu K., Senriuchi T., Katayama T. (2009). Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 23, 1221–1233. 10.1101/gad.1775809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Kornberg A. (1983). Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. U.S.A. 80, 5817–5821. 10.1073/pnas.80.19.5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner J., Crooke E. (1996). Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 15, 3477–3485. [PMC free article] [PubMed] [Google Scholar]
- Hanna M. H., Carl P. L. (1975). Reinitiation of deoxyribonucleic acid synthesis by deoxyribonucleic acid initiation mutants of E. coli: role of ribonucleic acid synthesis, protein synthesis, and cell division. J. Bacteriol. 121, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. B., Atlung T., Hansen F. G., Skovgaard O., von Meyenburg K. (1984). Fine structure genetic map and complementation analysis of mutations in the dnaA gene of Escherichia coli. Mol. Gen. Genet. 196, 387–396. 10.1007/BF00436184 [DOI] [PubMed] [Google Scholar]
- Hansen E. B., Hansen F. G., von Meyenburg K. (1982). The nucleotide sequence of the dnaA gene and the first part of the dnaN gene of Escherichia coli K12. Nucleic Acids Res. 10, 7373–7385. 10.1093/nar/10.22.7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G. (1995). Reinitiation kinetics in eight dnaA(Ts) mutants of Escherichia coli: rifampicin resistant initiation of chromosome replication. Mol. Microbiol. 15, 133–140. 10.1111/j.1365-2958.1995.tb02227.x [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Atlung T., Braun R. E., Wright A., Hughes P., Kohiyama M. (1991b). Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 173, 5194–5199. 10.1128/jb.173.16.5194-5199.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G., Christensen B. B., Atlung T. (1991a). The initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol. 142, 161–167. 10.1016/0923-2508(91)90025-6 [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Christensen B. B., Atlung T. (2007). Sequence characteristics required for cooperative binding and efficient in vivo titration of the replication initiator protein DnaA in E. coli. J. Mol. Biol. 367, 942–952. 10.1016/j.jmb.2007.01.056 [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Christensen B. B., Nielsen C. B., Atlung T. (2006). Insights into the quality of DnaA boxes and their cooperativity. J. Mol. Biol. 355, 85–95. 10.1016/j.jmb.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Hansen E. B., Atlung T. (1982). The nucleotide sequence of the dnaA gene promoter and of the adjacent rpmH gene, coding for the ribosomal protein L34, of Escherichia coli. EMBO J. 1, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G., Hansen E. B., Atlung T. (1985). Physical mapping and DNA sequence of the gene rnpA, that encodes the protein component of ribonuclease P in Escherichia coli. Gene 38, 85–93. 10.1016/0378-1119(85)90206-9 [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Koefoed S., Atlung T. (1992). Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol. Gen. Genet. 234, 14–21. [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Koefoed S., Sørensen L., Atlung T. (1987). Titration of DnaA protein by oriC DnaA-boxes increases dnaA gene expression in Escherichia coli. EMBO J. 6, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G., Rasmussen K. V. (1977). Regulation of the dnaA product in E. coli. Mol. Gen. Genet. 155, 219–225. 10.1007/BF00393163 [DOI] [PubMed] [Google Scholar]
- Hansen F. G., von Meyenburg K. (1979). Characterization of the dnaA, gyrB and other genes in the dnaA region of the Escherichia coli chromosome on specialized transducing phages lambda-tna. Mol. Gen. Genet. 175, 135–144. 10.1007/BF00425529 [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E. (1967). Rate of DNA synthesis during the division cycle of E. coli B/r. J. Mol. Biol. 24, 417–427. 10.1016/0022-2836(67)90228-8 [DOI] [Google Scholar]
- Helmstetter C. E. (1996). Timing of synthetic activities in the cell cycle, in Escherichia coli and Salmonella Cellular and Molecular Biology, eds Neidhardt F. C., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., et al. (Washington, DC: ASM Press; ), 1627–1639. [Google Scholar]
- Helmstetter C. E., Cooper S. (1968). DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J. Mol. Biol. 31, 507–518. 10.1016/0022-2836(68)90424-5 [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E., Cooper S., Pierucci D., Revelas E. (1968). The bacterial life cycle. Cold Spring Harb. Symp. Quant. Biol. 33, 809–822. 10.1101/SQB.1968.033.01.093 [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E., Cummings D. J. (1963). Bacterial synchronization by selection of cells at division. Proc. Natl. Acad. Sci. U.S.A. 50, 767–774. 10.1073/pnas.50.4.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter C. E., Cummings D. J. (1964). An improved method for the selection of bacterial cells at division. Biochim. Biophys. Acta 82, 608–610. 10.1016/0304-4165(64)90453-2 [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E., Pierucci O. (1976). DNA synthesis during the division cycle of three substrains of Escherichia coli B/r. J. Mol. Biol. 102, 477–486. 10.1016/0022-2836(76)90329-6 [DOI] [PubMed] [Google Scholar]
- Herrick J., Kohiyama M., Atlung T., Hansen F. G. (1996). The initiation mess? Mol. Microbiol. 19, 659–666. 10.1046/j.1365-2958.1996.432956.x [DOI] [PubMed] [Google Scholar]
- Hiraga S. (1976). Novel F prime factors able to replicate in E. coli Hfr strains. Proc. Natl. Acad. Sci. U.S.A. 73, 198–202. 10.1073/pnas.73.1.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Ryter A., Jacob F. (1968). Thermosensitive mutants of E. coli affected in the process of DNA synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 33, 677–693. 10.1101/SQB.1968.033.01.077 [DOI] [PubMed] [Google Scholar]
- Hirota Y., Yasuda S., Yamada M., Nishimura A., Sugimoto K., Sugisaki H., et al. (1979). Structural and functional properties of the Escherichia coli origin of DNA replication. Cold Spring Harb. Symp. Quant. Biol. 43, 129–138. 10.1101/SQB.1979.043.01.019 [DOI] [PubMed] [Google Scholar]
- Hwang D. S., Kaguni J. M. (1988). Interaction of dnaA46 protein with a stimulatory protein in replication from the Escherichia coli chromosomal origin. J. Biol. Chem. 263, 10633–10640. [PubMed] [Google Scholar]
- Ishida T., Akimitsu N., Kashioka T., Hatano M., Kubota T., Ogata Y., et al. (2004). DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 279, 45546–45555. 10.1074/jbc.M402762200 [DOI] [PubMed] [Google Scholar]
- Jacob F., Brenner S., Cuzin F. (1963). On the regulation of DNA replication in bacteria. Cold Spring Harb. Symp. Quant. Biol. 28, 329–348. 10.1101/SQB.1963.028.01.048 [DOI] [Google Scholar]
- Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T. (2014). Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 42, 13134–13149. 10.1093/nar/gku1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasho K., Katayama T. (2013). DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 110, 936–941. 10.1073/pnas.1212070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasho K., Tanaka H., Sakai R., Katayama T. (2017). Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA-ATP hydrolysis. J. Biol. Chem. 292, 1251–1266. 10.1074/jbc.M116.762815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T., Fujimitsu K., Ogawa T. (2001). Multiple pathways regulating DnaA function in Escherichia coli: distinct roles for DnaA titration by the datA locus and the regulatory inactivation of DnaA. Biochimie 83, 13–17. 10.1016/S0300-9084(00)01206-2 [DOI] [PubMed] [Google Scholar]
- Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. (1998). The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli Chromosomal replicase. Cell 94, 61–71. 10.1016/S0092-8674(00)81222-2 [DOI] [PubMed] [Google Scholar]
- Kato J., Katayama T. (2001). Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20, 4253–4262. 10.1093/emboj/2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H., Ozaki S., Suzuki S., Nakamura K., Senriuchi T., Su'etsugu M., et al. (2006). The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 62, 1310–1324. 10.1111/j.1365-2958.2006.05450.x [DOI] [PubMed] [Google Scholar]
- Kellenberger-Gujer G., Podhajska A. J., Caro L. (1978). A cold sensitive dnaA mutant of E. coli which overinitiates chromosome replication at low temperature. Mol. Gen. Genet. 162, 9–16. 10.1007/BF00333845 [DOI] [PubMed] [Google Scholar]
- Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. (2009). DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 284, 25038–25050. 10.1074/jbc.M109.002717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., et al. (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP-DnaA-specific initiation complexes. Genes Dev. 21, 2083–2099. 10.1101/gad.1561207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura K., Katayama T. (2011). DnaA protein DNA-binding domain binds to Hda protein to promote inter-AAA+ domain interaction involved in regulatory inactivation of DnaA. J. Biol. Chem. 286, 29336–29346. 10.1074/jbc.M111.233403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R., Mitsuki H., Okazaki T., Ogawa T. (1996). A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19, 1137–1147. 10.1046/j.1365-2958.1996.453983.x [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Ozaki T., Moriya S., Ogawa T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043. 10.1101/gad.12.19.3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohiyama M., Cousin D., Ryter A., Jacob F. (1966). Mutants thermosensibles d'E. coli K-12. I. Isolement et characterisation rapide. Ann. Inst. Pasteur 110, 465–486. [PubMed] [Google Scholar]
- Kohiyama M., Lanfrom H., Brenner S., Jacob F. (1963). Modifications de fonctions indispensables chez des mutants thermosensibles D'escherichia Coli. sur une mutation emp echant la r'eplication du chromosome bact'erien. C. R. Hebd. Seances Acad. Sci. 257, 1979–1981. [PubMed] [Google Scholar]
- Koppes L. J. H., Woldringh C. L., Nanninga N. (1978). Size variation and correlation of different cell cycle events in slow-growing Escherichia coli. J. Bacteriol. 134, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kücherer C., Lother H., Kölling R., Schauzu M. A., Messer W. (1986). Regulation of transcription of the chromosomal dnaA gene of Escherichia coli. Mol. Gen. Genet. 205, 115–121. 10.1007/BF02428040 [DOI] [PubMed] [Google Scholar]
- Kuempel P. L. (1969). Temperature-sensitive initiation of chromosome replication in a mutant of Escherichia coli. J. Bacteriol. 100, 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999). Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 18, 6642–6652. 10.1093/emboj/18.23.6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark C. (1968). Studies on the in vivo methylation of DNA in Escherichia coli 15T-. J. Mol. Biol. 31, 389–399. 10.1016/0022-2836(68)90416-6 [DOI] [PubMed] [Google Scholar]
- Leonard A. C., Grimwade J. E. (2015). The orisome: structure and function. Front. Microbiol. 6:545. 10.3389/fmicb.2015.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A. C., Helmstetter C. E. (1986). Cell cycle-specific replication of E. coli minichromosomes. Proc. Natl Acad. Sci. U.S.A. 83, 5101–5105. 10.1073/pnas.83.14.5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu F. P., Hingorani M. M., Turner J., O'donnell M. (2000). The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem. 275, 34609–34618. 10.1074/jbc.M005495200 [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., Atlung T., Rasmussen K. V. (1987). Stability and replication control of E. coli minichromosomes. J. Bacteriol. 169, 2835–2842. 10.1128/jb.169.6.2835-2842.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Hansen F. G., Rasmussen K. V., Martin B., Kuempel P. L. (1994). The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 13, 1856–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A., Skarstad K., Hansen F. G., von Meyenburg K., Boye E. (1989). The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57, 881–889. 10.1016/0092-8674(89)90802-7 [DOI] [PubMed] [Google Scholar]
- Løbner-Olesen A., von Freiesleben U. (1996). Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 15, 5999–6008. [PMC free article] [PubMed] [Google Scholar]
- Lu M., Campbell J. L., Boye E., Kleckner N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77, 413–426. 10.1016/0092-8674(94)90156-2 [DOI] [PubMed] [Google Scholar]
- Maaløe O., Hanawalt P. C. (1961). Thymine deficiency and the normal DNA replication cycle I. J. Mol. Biol. 3, 144–155. 10.1016/S0022-2836(61)80041-7 [DOI] [PubMed] [Google Scholar]
- Maaløe O., Kjeldgaard N. O. (1966). Control of Macromolecular Synthesis. New York, NY: Amsterdam;Benjamin. [Google Scholar]
- Mahaffy J. M., Zyskind J. W. (1989). A model for the initiation of replication in Escherichia coli. J. Theor. Biol. 140, 453–477. 10.1016/S0022-5193(89)80109-2 [DOI] [PubMed] [Google Scholar]
- Margalit H., Grover N. B. (1987). Initiation of chromosome replication in bacteria: analysis of an inhibitor control model. J. Bacteriol. 169, 5231–5240. 10.1128/jb.169.11.5231-5240.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit H., Rosenberger R. F., Grover N. B. (1984). Initiation of DNA replication in bacteria: analysis of an autorepressor model. J. Theor. Biol. 111, 183–199. 10.1016/S0022-5193(84)80204-0 [DOI] [PubMed] [Google Scholar]
- Margulies C., Kaguni J. M. (1996). Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 271, 17035–17040. 10.1074/jbc.271.29.17035 [DOI] [PubMed] [Google Scholar]
- Marsh R. C., Worcel A. (1977). A DNA fragment containing the origin of replication of the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 74, 2720–2724. 10.1073/pnas.74.7.2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry K. C., Ryan V. T., Grimwade J. E., Leonard A. C. (2004). Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816. 10.1073/pnas.0400340101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M. J., Wolfe K. H., Devine K. M. (1998). Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol. 47, 691–696. 10.1007/PL00006428 [DOI] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., et al. (1979). Nucleotide sequence of the origin of replication of the E. coli K-12 chromosome. Proc. Natl. Acad. Sci. U.S.A. 76, 580–584. 10.1073/pnas.76.2.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. (2002). The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374. 10.1016/S0168-6445(02)00127-4 [DOI] [PubMed] [Google Scholar]
- Messer W., Bergmans H. E., Meijer M., Womack J. E., Hansen F. G., von Meyenburg K. (1978). Minichromosomes: plasmids which carry the E. coli replication origin. Mol. Gen. Genet. 162, 269–275. 10.1007/BF00268852 [DOI] [PubMed] [Google Scholar]
- Messer W., Blaesing F., Majka J., Nardmann J., Schaper S., Schmidt A., et al. (1999). Functional domains of DnaA proteins. Biochimie 81, 819–825. 10.1016/S0300-9084(99)00215-1 [DOI] [PubMed] [Google Scholar]
- Messer W., Dankwarth L., Tippe-Schindler R., Womack J. E., Zahn G. (1975). Regulation of the initiation of DNA replication in E. coli. Isolation of iRNA and the control of iRNA synthesis. ICN-UCLA Symp. Mol. Cell. Biol. 3, 602–617. [Google Scholar]
- Messer W., Meijer M., Bergmans H. E., Hansen F. G., von Meyenburg K., Beck E., et al. (1979). Origin of replication, oriC, of the Escherichia coli K- 12 chromosome: nucleotide sequence. Cold Spring Harb. Symp. Quant. Biol. 43, 139–145. 10.1101/SQB.1979.043.01.020 [DOI] [PubMed] [Google Scholar]
- Michelsen O., Teixeira de Mattos M. J., Jensen P. R., Hansen F. G. (2003). Precise determinations of C and D periods by flow cytometry in Escherichia coli K-12 and B/r. Microbiology 149, 1001–1010. 10.1099/mic.0.26058-0 [DOI] [PubMed] [Google Scholar]
- Morigen Løbner-Olesen A., Skarstad K. (2003). Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50, 349–362. 10.1046/j.1365-2958.2003.03695.x [DOI] [PubMed] [Google Scholar]
- Morigen Molina F., Skarstad K. (2005). Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187, 3913–3920. 10.1128/JB.187.12.3913-3920.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott M. L., Berger J. M. (2007). DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354. 10.1038/nrmicro1640 [DOI] [PubMed] [Google Scholar]
- Murialdo H., Siminovitch L. (1972). The morphogenesis of bacteriophage lambda. IV. Identification of gene products and control of the expression of the morphogenetic information. Virology 48, 785–823. 10.1016/0042-6822(72)90162-6 [DOI] [PubMed] [Google Scholar]
- Nielsen H. J., Hansen F. G. (2010). An automated and highly efficient method for counting and measuring fluorescent foci in rod-shaped bacteria. J. Microsc. 239, 194–199. 10.1111/j.1365-2818.2010.03374.x [DOI] [PubMed] [Google Scholar]
- Nozaki S., Niki H., Ogawa T. (2009a). Replication initiator DnaA of Escherichia coli changes its assembly form on the replication origin during the cell cycle. J. Bacteriol. 191, 4807–4814. 10.1128/JB.00435-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S., Ogawa T. (2008). Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154, 3379–3384. 10.1099/mic.0.2008/019745-0 [DOI] [PubMed] [Google Scholar]
- Nozaki S., Yamada Y., Ogawa T. (2009b). Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells 14, 329–341. 10.1111/j.1365-2443.2008.01269.x [DOI] [PubMed] [Google Scholar]
- Nyborg M., Atlung T., Skovgaard O., Hansen F. G. (2000). Two types of cold sensitivity associated with the A184V change in the DnaA protein. Mol. Microbiol. 35, 1202–1210. 10.1046/j.1365-2958.2000.01790.x [DOI] [PubMed] [Google Scholar]
- Obita T., Iwura T., Su'etsugu M., Yoshida Y., Tanaka Y., Katayama T., et al. (2002). Determination of the secondary structure in solution of the Escherichia coli DnaA DNA-binding domain. Biochem. Biophys. Res. Commun. 299, 42–48. 10.1016/S0006-291X(02)02590-1 [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. (1985). Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 4, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Yamada Y., Kuroda T., Kishi T., Moriya S. (2002). The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44, 1367–1375. 10.1046/j.1365-2958.2002.02969.x [DOI] [PubMed] [Google Scholar]
- Ozaki S., Katayama T. (2012). Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 40, 1648–1665. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S. Y., et al. (2008). A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283, 8351–8362. 10.1074/jbc.M708684200 [DOI] [PubMed] [Google Scholar]
- Polaczek P., Wright A. (1990). Regulation of expression of the dnaA gene in Escherichia coli: role of the two promoters and the DnaA box. New Biol. 2, 564–582. [PubMed] [Google Scholar]
- Pritchard R. H., Barth P. T., Collins J. (1969). Control of DNA synthesis in bacteria. Symp. Soc. Gen. Microbiol. 19, 263–297. [Google Scholar]
- Ream L. W., Margossian L., Clark A. J., Hansen F. G., von Meyenburg K. (1980). Genetic and physical mapping of recF in Escherichia coli K- 12. Mol. Gen. Genet. 180, 115–121. 10.1007/BF00267359 [DOI] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky A., McKenney K. (1985). Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc. Natl. Acad. Sci. U.S.A. 82, 5656–5660. 10.1073/pnas.82.17.5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber L., Olsson J. A., Jensen R. B., Skovgaard O., Dasgupta S., Marinus M. G. (2006). Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 20, 2121–2134. 10.1101/gad.379506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Messer W. (1995). The DNA binding domain of the initiator protein DnaA. EMBO J. 14, 2106–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Messer W. (1998). High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol. 28, 395–401. 10.1046/j.1365-2958.1998.00813.x [DOI] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. (1987). Hemimethylation prevents DNA replication in E. coli. Cell 50, 1071–1079. 10.1016/0092-8674(87)90173-5 [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Mizukami T. (1980). A temperature sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol. Gen. Genet. 178, 541–553. 10.1007/BF00337859 [DOI] [PubMed] [Google Scholar]
- Samitt C. E., Hansen F. G., Miller J. F., Schaechter M. (1989). In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 8, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. (1979). Simple method for identification of plasmid-coded proteins. J. Bacteriol. 137, 692–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M., Maaløe O., Kjeldgaard N. O. (1958). Dependency on medium and temperature of cell size chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19, 592–606. 10.1099/00221287-19-3-592 [DOI] [PubMed] [Google Scholar]
- Schaechter M., Williamson J. P., Hood J. R., Koch A. L. (1962). Growth, cell and nuclear divisions in some bacteria. J. Gen. Microbiol. 29, 421–434. 10.1099/00221287-29-3-421 [DOI] [PubMed] [Google Scholar]
- Schaper S., Messer W. (1995). Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270, 17622–17626. 10.1074/jbc.270.29.17622 [DOI] [PubMed] [Google Scholar]
- Seitz H., Weigel C., Messer W. (2000). The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37, 1270–1279. 10.1046/j.1365-2958.2000.02096.x [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Bramhill D., Kornberg A. (1987). ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50, 259–265. 10.1016/0092-8674(87)90221-2 [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Kornberg A. (1988). Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 263, 7131–7135. [PubMed] [Google Scholar]
- Sevastopoulos C. G., Wehr C. T., Glaser D. A. (1977). Largescale automated isolation of Escherichia coli mutants with thermosensitive DNA replication. Proc. Natl. Acad. Sci. U.S.A. 74, 3485–3489. 10.1073/pnas.74.8.3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F., Li D., Cox S. E., Sauls J. T., Azizi O., Sou C., et al. (2017). Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr. Biol. 27, 1278–1287. 10.1016/j.cub.2017.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye E., Fanning E. (2003). Circles in the sand. EMBO Rep. 4, 661–665. 10.1038/sj.embor.embor888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye E., Steen H. B. (1986). Timing of initiation of chromosome replication in individual E. coli cells. EMBO J. 5, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Katayama T. (2013). Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 5, 1–17. 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Løbner-Olesen A. (2003). Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 22, 140–150. 10.1093/emboj/cdg003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard O. (1990). Nucleotide sequence of a Proteus mirabilis fragment homologous to the 60K-rnpA-rpmH-dnaA-dnaN-recF-gyrB region of Escherichia coli. Gene 93, 27–34. 10.1016/0378-1119(90)90131-A [DOI] [PubMed] [Google Scholar]
- Skovgaard O., Hansen F. G. (1987). Comparison of dnaA nucleotide sequences of Escherichia coli, Salmonella typhimurium, and Serratia Marcescens. J. Bacteriol. 169, 3976–3981. 10.1128/jb.169.9.3976-3981.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L., Maaloe O. (1973). Autorepressor model for control of DNA replication. Nat. New Biol. 241, 133–135. 10.1038/newbio241133a0 [DOI] [PubMed] [Google Scholar]
- Speck C., Messer W. (2001). Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20, 1469–1476. 10.1093/emboj/20.6.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C., Weigel C., Messer W. (1999). ATP- and ADP-DnaA protein, a molecular switch in gene regulation. EMBO J. 18, 6169–6176. 10.1093/emboj/18.21.6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Oka A., Sugisaki H., Takanami M., Nishimura A., Yasuda Y., et al. (1979). Nucleotide sequence of Escherichia coli replication origin. Proc. Natl. Acad. Sci. U.S.A. 76, 575–579. 10.1073/pnas.76.2.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Oka A., Sugimoto K., Takanami M., Yasuda S., Hirota Y. (1983). The 245 bp oriC sequence of the E. coli chromosome directs bidirectional replication at an adjacent region. Nucleic Acids Res. 11, 2617–2626. 10.1093/nar/11.9.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen P. W., Grimwade J. E., Leonard A. C., Bogan J. A., Helmstetter C. E. (1993). Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10, 575–584. 10.1111/j.1365-2958.1993.tb00929.x [DOI] [PubMed] [Google Scholar]
- Tippe-Schindler R., Zahn G., Messer W. (1979). Control of the initiation of DNA replication in E. coli. I. Negative control of initiation. Mol. Gen. Genet. 168, 185–195. 10.1007/BF00431444 [DOI] [PubMed] [Google Scholar]
- Torheim N. K., Boye E., Løbner-Olesen A., Stokke T., Skarstad K. (2000). The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol. Microbiol. 37, 629–638. 10.1046/j.1365-2958.2000.02031.x [DOI] [PubMed] [Google Scholar]
- Travers A. A. (1980). Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J. Bacteriol. 141, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg E. A., Geerse R. H., Memelink J., Bovenberg R. A. L., Magnée F. A., van de Putte P. (1985). Analysis of regulatory sequences upstream of the E. coli uvrB gene; involvement of the DnaA protein. Nucleic Acids Res. 13, 1829–1840. 10.1093/nar/13.6.1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Krekling M. A., Hansen F. G., Løbner-Olesen A. (2000a). The eclipse period of Escherichia coli. EMBO J. 19, 6240–6248. 10.1093/emboj/19.22.6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V., Atlung T., Hansen F. G. (2000b). Rifampicin resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 37, 1087–1093. 10.1046/j.1365-2958.2000.02060.x [DOI] [PubMed] [Google Scholar]
- von Freiesleben U., Rasmussen K. V., Schaechter M. (1994). SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14, 763–772. 10.1111/j.1365-2958.1994.tb01313.x [DOI] [PubMed] [Google Scholar]
- von Meyenburg K., Hansen F. G., Riise E., Bergmans H. E., Meijer M., Messer W. (1979). Origin of replication, oriC, of the E. coli K-12 chromosome: genetic mapping and minichromosome replication. Cold Spring Harb. Symp. Quant. Biol. 43, 121–128. 10.1101/SQB.1979.043.01.018 [DOI] [PubMed] [Google Scholar]
- Wechsler J. A., Gross J. D. (1971). Escherichia coli mutants temperature sensitive for DNA synthesis. Mol. Gen. Genet. 113, 273–284. 10.1007/BF00339547 [DOI] [PubMed] [Google Scholar]
- Weigel C., Schmidt A., Seitz H., Tüngler D., Welzeck M., Messer W. (1999). The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA. Mol. Microbiol. 34, 53–66. 10.1046/j.1365-2958.1999.01568.x [DOI] [PubMed] [Google Scholar]
- Wold S., Skarstad K., Steen H. B., Stokke T., Boye E. (1994). The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 13, 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Hirota Y. (1977). Cloning and mapping of the replication origin of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 74, 5458–5462. 10.1073/pnas.74.12.5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilak-Pawlik A., Nowaczyk M., Zakrzewska-Czerwinska J. (2017). The role of the N-terminal domains of bacterial initiator DnaA in the assembly and regulation of the bacterial replication initiation complex. Genes 8, 1–18. 10.3390/genes8050136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Bogan J. A., Welch K., Pickett S. R., Wang H. J., Zaritsky A., et al. (1997). Gene transcription and chromosome replication in Escherichia coli. J. Bacteriol. 179, 163–169. 10.1128/jb.179.1.163-169.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Harding N. E., Takeda Y., Cleary J. M., Smith D. W. (1981). The DNA replication origin region of the Enterobacteriaceae. ICN-UCLA Symp. Mol. Cell. Biol. 22, 13–25. 10.1016/B978-0-12-583580-0.50007-8 [DOI] [Google Scholar]
- Zyskind J. W., Smith D. W. (1980). Nucleotide sequence of the Salmonella typhimurium_ origin of DNA replication. Proc. Natl. Acad. Sci. U.S.A. 77, 2460–2464. 10.1073/pnas.77.5.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]