ABSTRACT
The study of horizontal gene transfer (HGT) in microbial communities has been revolutionized by significant advances in cultivation-independent methods based on fluorescence reporter gene technologies. Recently, the combination of these novel approaches with flow cytometry has presented itself as one of the most powerful tools to study the spread of mobile genetic elements (MGEs) in the environment. However, the use of fluorescent markers, like green fluorescent protein (GFP) and mCherry, is limited by environmental constraints, such as oxygen availability and pH levels, that affect the correct maturation of their fluorophores. Few studies have characterized the effects of such environmental conditions in a systematic way, and the sheer amount of distinct protein variants requires each system to be examined in an individual fashion. The lack of efficient and reliable markers to monitor HGT in anaerobic environments, coupled to the abundance of ecologically and clinically relevant oxygen-deprived niches in which bacteria thrive, calls for the urgent development of suitable tools that permit its study. In an attempt to devise a process that allows the implementation of the mentioned dual-labeling system to anoxic milieus, the aerobic fluorescence recovery of mCherry and GFPmut3, as well as the effect of pH on their fluorescence intensities, was studied. The findings present a solution to an intrinsic problem that has long hampered the utilization of this system, highlight its pH limitations, and provide experimental tools that will help broaden its horizon of application to other fields.
IMPORTANCE Many anaerobic environments, like the gastrointestinal tract, anaerobic digesters, and the interiors of dense biofilms, have been shown to be hotspots for horizontal gene transfer (HGT). Despite the increasing wealth of reports warning about the alarming spread of antibiotic resistance determinants, to date, HGT studies mainly rely on cultivation-based methods. Unfortunately, the relevance of these studies is often questionable, as only a minor fraction of bacteria can be cultivated. A recently developed approach to monitoring the fate of plasmids in microbial communities is based on a fluorescence dual-labeling system and allows the bypassing of cultivation. However, the fluorescent proteins on which it is founded are constrained by pH levels and by their strict dependence on oxygen for the maturation of their fluorophores. This study focused on the development and validation of an appropriate aerobic fluorescence recovery (AFR) method for this platform, as this embodies the missing technical link impeding its implementation in anoxic environments.
KEYWORDS: aerobic fluorescence recovery, fluorescence, GFPmut3, horizontal gene transfer, plasmid, conjugation, environmental microbiology, mCherry, pKJK5
INTRODUCTION
The knowledge that horizontal gene transfer (HGT) plays a pivotal role in the evolution and adaptation of microbes to environmental changes is far from new (1, 2). This area of research has long attracted much attention and is increasingly becoming of public and clinical concern, mainly due to the accumulating wealth of evidence showcasing how treatments for bacterial infections are gradually losing efficacy and, consequently, ever more often failing (3–5). This alarming setting is mainly attributed to the dissemination of antibiotic resistance genes in bacterial communities, and shedding light on the underlying mechanisms that govern the spread, dynamics and transfer extent of mobile genetic elements (MGEs) has thus become a crucial task (6).
Several different processes by which MGEs can spread among bacteria have been identified, namely, transformation, transduction, and conjugation. However, it is important to highlight that most studies focus primarily on conjugation, as it is generally regarded as the mechanism bearing the greatest impact on the architecture and dynamic shaping of the communal gene pool (7, 8). Although several approaches have been developed for its study, the successfulness of these experimental setups is often questionable, since achieving environmentally relevant conditions yet remains as the greatest challenge. Recently, there has been a shift from the use of the more traditional cultivation-based methods toward the development of novel approaches, aiming to minimize this inherent source of bias (9, 10).
In this context, a technique to study plasmid transfers in microbial communities was recently developed that has been validated though independent transconjugant enumeration methods such as microscopy and CFU counting (11). This platform not only allows the possibility to bypass in vitro cultivation, but also enables in situ visualization of cells. The key to the success of this system is that it is based on a dual-labeling technique in which the donor is chromosomally tagged with a red fluorescence reporter gene (mCherry), a chromosomally encoded lacIq repressor gene, and where the plasmid of interest is labeled with a gfpmut3 gene downstream of a LacI-repressible promoter (12, 13). In this way, plasmid transfers can be elegantly tracked. Donors are constitutively red, for even though they harbor the GFP encoding plasmid, its expression is repressed by LacIq, and transconjugants become green, since the chromosomally encoded repressor is absent (and/or presumably expressed to a lesser extent) in recipients. The principal advantage of this technique is that flow cytometry can be used not only to count cells and assess plasmid transfer frequencies, but also to sort out transconjugants for subsequent 16S rRNA gene analysis, ultimately allowing the study of the extent of HGT in the recipient community. This technique has yielded unprecedented results regarding HGT in complex soil and wastewater microbial communities (12–15) and has the potential to provide highly valuable insights about the spread of MGEs in other environments.
However, many of the key biological systems in which HGT should be assessed are, to some extent, devoid of molecular oxygen, e.g., thick biofilms, the mammalian gastrointestinal tract, deep sediments, anaerobic digesters, etc. Also, some of these systems, like the large intestine of humans, can be subject to pH shifts toward slightly acidic values (5.2 to 7) (16, 17). The facts that the fluorescence of these proteins can be severely affected by their environmental pH, and that the complete maturation of their fluorophores strictly requires oxygen (18, 19), constitute a major drawback in terms of their applicability. Notably, the oxic constraint has motivated many to search for and develop alternative fluorescent genetic markers for imaging under low-oxygen conditions. It is worth mentioning the emergence of flavin-based fluorescent proteins (FbFPs) as a promising new family of small-sized oxygen-independent reporters (20–22). Unfortunately, the fluorescence intensity of these proteins is relatively low compared to that displayed by GFP or DsRed variants (23, 24). Nonetheless, it has been shown how proteins derived from GFP and DsRed can remain in an immature intermediate state until posttranslationally catalyzed to maturation upon exposure to air (25–28). For this reason, the purpose of this study has been the development of a method that successfully couples the aerobic fluorescence recovery (AFR) of both anaerobically expressed fluorescent proteins simultaneously and to characterize the pH limitations associated with the implementation of this system in crucial anaerobic environments, such as the mammalian gut. The implications of the findings presented in this work are paramount for widening the applicability scope of this novel dual-labeling system and provide experimental tools that will usher a deeper understanding of not only HGT, but also of any field in which these fluorescence imaging techniques are applicable.
RESULTS AND DISCUSSION
Characterization of the AFR of mCherry and GFPmut3 expressed in E. coli.
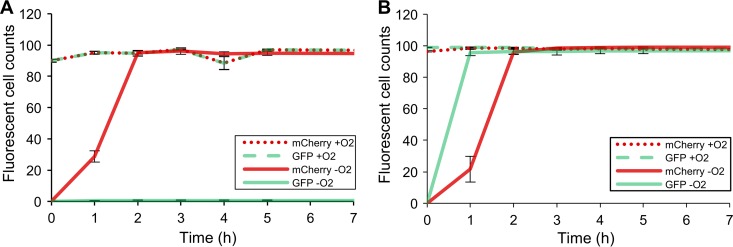
As the purpose of this work is to characterize the AFR constraints and other associated limitations of the proteins integrated in a given platform, the relevant maturation is that taking place in the milieu comprised by intact bacterial cells harboring said system, and not by their protein extracts. This is highly important, since the fluorescence properties of these proteins are greatly affected by their immediate surroundings, and these may be significantly different intra- and extracellularly. A study coupling the AFR of mCherry and GFPmut3 was carried out, using E. coli as the expression host under anaerobic conditions. Upon exposure to air, the AFR of both proteins was measured simultaneously at different time points via flow cytometry, for a total of 50,000 cells per sample (Fig. 1; see also Fig. S2 in the supplemental material). Motivated by the observation that GFPmut3 is highly pH sensitive (29, 30), and taking into account the current growing concern for studying plasmid transfer in the gastrointestinal tract and other environments that may present slightly acidic pH values, the AFR of the system was first assessed at a low extracellular pH. This was achieved by growing the cells overnight in medium lacking the appropriate buffering (pH 5) (Fig. 1A).
FIG 1.
Time course fluorescence recovery of mCherry and GFPmut3 expressed at pH 5 (A) and pH 7 (B). Fluorescent cell counts are illustrated as measured by flow cytometry from triplicates of aerobically (dotted lines) and anaerobically (continuous lines) grown E. coli cells expressing mCherry (red) and GFPmut3 (green) upon subsequent exposure to air. Aerobic incubation was done in phosphate-buffered saline (PBS) supplemented with chloramphenicol (100 μg/ml). Fluorescence is plotted against time as the average % counts captured by the set detection gates (see Fig. S1 in the supplemental material) per 50,000 bacterial counts. The error bars represent standard deviations from the mean. Visualization of the raw flow cytometric data showing the time-wise shift in the population of cells into the set detection gates can be found in Fig. S2 in the supplemental material.
Essentially no recovery was reported for GFPmut3 in the acidified medium (Fig. 1A), an observation that is in accordance with the works of Doherty et al. (29) and Hansen et al. (31), who reported an inhibition of the fluorescence of GFPmut3 when expressed around pH 5. In contrast, mCherry exhibited a complete recovery, an observation that is in agreement with studies reporting a higher resilience of DsRed variants to acidic pH values (29, 32). In order to determine whether the absence of fluorescence recovery of GFPmut3 was indeed a result of its pH lability, the AFR of samples grown in buffered medium (pH 7.4) was monitored (Fig. 1B). Here, GFPmut3 was found to display a complete AFR in less than an hour, a maturation time frame that is consistent with that found by several other studies (29, 33, 34). On the other hand, although some works have described mCherry's maturation as requiring just 30 min (35), we report maturation time to be considerably longer, between 1 to 2 h. Nonetheless, this observation is congruent with other previous works (26, 33).
In vivo pH-dependent emission profiles for mCherry and GFPmut3.
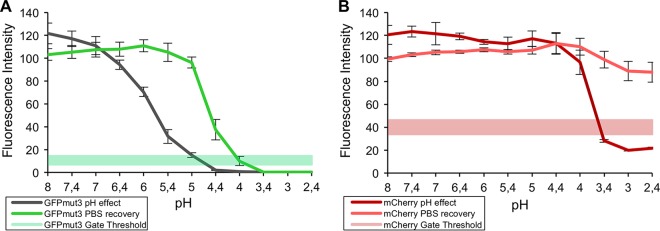
In order to further characterize the effect of the external pH on the fluorescence emission of this system, aerobically grown E. coli cells simultaneously expressing both proteins were subjected to an array of buffers ranging from pH 2.4 to 8. Noteworthy prior studies have characterized the effect of pH on purified extracts of mCherry and GFPmut3 (29). However, as this system is embodied by cells carrying the dual-labeling system, knowledge regarding the impact of the extracellular pH on the fluorescence of these cells is highly desired, and this was thus evaluated via flow cytometry. As shown in Fig. 2A, the fluorescence of GFPmut3 was quenched beyond the flow cytometric gate detection limit at around pH 5, an observation that is consistent with the lack of fluorescence observed in the acidified medium (Fig. 1A). It should be noted that both proteins displayed a higher fluorescence at alkaline pH, suggesting that AFR might be enhanced at higher pH. In line with previous reports (36) and with the data presented in Fig. 2B, mCherry displays a remarkable stability at low pH values, only dropping below the set flow cytometer gate detection threshold at a pH below 3.5.
FIG 2.
Effect of the external pH on the fluorescence emission intensity of intracellular GFPmut3 (A) and mCherry (B). Aerobically grown E. coli cells expressing GFPmut3 and mCherry were subjected overnight, in triplicate, to an array of buffers, ranging from pH 2.4 to 8 (with 100 μg/ml chloramphenicol). The resultant fluorescence emission profiles are shown for GFPmut3, dark green shaded profile (A), and for mCherry, dark red shaded profile (B). Recovery of the acid-induced quenching of fluorescence was studied by incubating the cells in PBS (pH 7.2 to 7.4) overnight, indicated by the light green shaded profile (GFPmut3) (A) and light red shaded profile (mCherry) (B). The set gate thresholds used for the flow cytometric detection of GFPmut3 and mCherry-expressing cells in this system are plotted as a reference, indicated by a continuous green band (A) and a continuous red band (B), respectively. Fluorescence is plotted as the mean fluorescence intensity detected for 50,000 bacterial events (of triplicates), expressed as relative (%) to the intensities the cells exhibited in PBS, before they were subjected to any buffer. The error bars represent the standard deviations from the mean.
The reversion of the observed acid-induced suppression of fluorescence was attempted by incubating the cells in phosphate-buffered saline (PBS) (pH 7.2) overnight, in the presence of chloramphenicol (100 μg/ml) to avoid de novo protein synthesis. As depicted in Fig. 2B, the fluorescence of mCherry was recovered at all of the pH values tested. In contrast, it was only possible to regain the fluorescence of GFPmut3 over a narrow pH range, pH 4.2 to 6.7 (Fig. 2A), where full recovery was only achieved in incubations performed at pH values greater than 5. The reversibility window observed suggests this protein might suffer degradation, and thus irreversible quenching of its fluorescent properties, at low pH values (37). These results seem to contradict the absence of fluorescence recovery observed for GFPmut3 during the AFR experiments performed with the unbuffered cultures (grown at pH 5). This suggests that the reversibility window may only be possible once the proteins are expressed and matured and not during their synthesis, as here it may lead to irreversible improper folding of the polypeptide backbone. However, this finding does not present a drastic impediment to the application potential of this system, as it is rare that one should encounter a biological milieu displaying such a low pH.
To complement these findings, the fluorescence recovery of mCherry and GFPmut3 was additionally recorded over time (see Fig. S3 in the supplemental material). In relation to the ability to recover fluorescence of both proteins, the findings are parallel to those depicted in Fig. 2. However, it can be observed that recovery of fluorescence is extremely quick, demonstrating the suitability of these proteins as sensitive pH indicators (38–40). Practically all of the recovery occurs within the first few minutes, although recovery is slower or absent for GFPmut3 at a pH of <5. It is believed that fluorescence is directly affected by the protonation state of the chromophore, and consequently, that this is responsible for the observed pH sensitivity (37, 41, 42). When exposed to a pH of <5 however, it is likely that GFP undergoes a conformational change that renders it irreversibly inadequate for recovery.
Solid surface filter mating coupled to AFR.
An experiment was performed to demonstrate the suitability of this dual-labeling system for HGT studies in anaerobic environments, such as the gastrointestinal tracts of humans and animals. Since the application of this technology to such complex environments would entail a whole new study in itself, the combination of solid surface filter matings with the controlled anaerobic conditions provided by an anoxic chamber was used as a proxy to validate this system's general anaerobic applicability. E. coli was chosen for this study not only by virtue of being a facultative anaerobe, but also because it has been shown to be a common host for a wide range of plasmids containing antibiotic resistance genes (43), and because it is an indigenous bacterium of the gut and other HGT hotspots. In this regard, E. coli constitutes a relevant candidate donor in future experiments investigating HGT in natural environments.
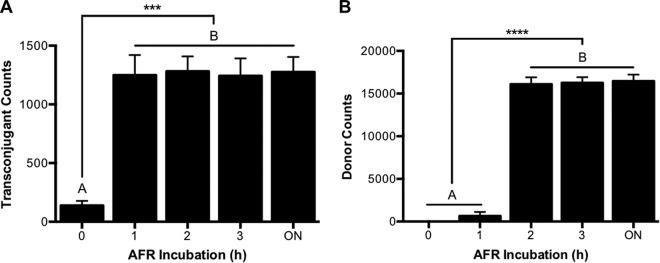
Taking into account the pH and maturation constraints of these proteins, an AFR method was devised to enable the detection of donor and transconjugant cells through flow cytometry. Since a slightly alkaline pH had been shown to enhance the fluorescence intensity of these proteins, AFR was carried out in a phosphate buffer at pH 8. As expected, 1 h of incubation was sufficient to recover the fluorescence of GFPmut3, enabling the detection of transconjugants (Fig. 3A). Congruent with the data displayed in Fig. 1B, no significant increase in transconjugant counts was observed at longer incubation times. Interestingly, a minor fraction of the transconjugant population was already detectable at time zero upon exposure to air. We believe that the preparation of the filter mating cell suspensions for flow cytometric detection introduced a short delay in the measurements, and owing to the remarkably fast maturation times reported for GFPmut3 (33), this resulted in the detection of some transconjugants. Oxic contamination should be discarded, as mCherry did not exhibit detectable fluorescence at this time. In accordance with our previous findings (Fig. 1A), mCherry-expressing cells (donors) present the limiting factor regarding the length of the AFR incubation required for this dual-labeling system, as their possible detection lags behind that of GFPmut3-expressing cells (transconjugants). During the first hour of AFR incubation, very low numbers of donor cells were detected (Fig. 3B). However, consistent with the maturation time we have reported for mCherry (Fig. 1A), within 2 h, practically all donor cells were readily detected, and no increase in the detection of donor cells was observed at longer maturation times.
FIG 3.
Effect of AFR incubation time over the flow cytometric enumeration of transconjugant (A) and donor (B) cell counts originating from anaerobic filter matings. Transconjugant and donor counts were detected via flow cytometry for a total of 50,000 bacterial events. Detection was performed hourly for 3 h and a final measurement was taken after overnight (ON) incubation. Filter matings were carried out overnight inside an anaerobic chamber, using a 1:1 donor/recipient ratio. E. coli MG1655 lacIq::mCherry was used as the donor harboring the pKJK5-gfpmut3 plasmid, and E. coli MG1655 as the recipient strain. Matings were performed in triplicate, and error bars indicate the standard errors of the mean. AFR was performed in phosphate buffer (pH 8) in the presence of chloramphenicol (100 μg/ml). Means with different letters are significantly different (analysis of variance [ANOVA]-Tukey's multiple-comparison test; P values of <0.0005 [***] and <0.00005 [****]).
Altogether, the findings presented in this work serve as proof of concept that the mentioned dual reporter system, when coupled to AFR, is suitable for the in situ detection and quantification of plasmid transfer in anoxic environments. However, the susceptibility of GFPmut3 to acidic pH values was identified as the principal caveat hindering its practical application. It is therefore important to highlight the importance of controlling the pH in the studied environment, especially when in the presence of lactic acid bacteria and other microorganisms that grow fermentatively. Likewise, one should also bear in mind that there might be other limitations which could be species or strain dependent. Our findings set the foundation for future studies investigating the appositeness of our approach to track plasmid conjugative transfers in oxygen-deprived environments and further asses its usage in a broader range of taxa. Furthermore, we believe that AFR is a powerful tool that has been unexploited so far, and its potential is certainly not restricted to the realm of HGT studies; the experimental tools presented here should prompt the use of these proteins in virtually any anaerobic system in which fluorescent imaging techniques are applicable.
MATERIALS AND METHODS
Strain, plasmids, and media.
The strain used in this work was E. coli K-12 MG1655, which was chromosomally tagged with a lacIq repressor gene and a constitutively expressed mCherry gene under the control of the E. coli murein-lipoprotein promoter, pLpp (44). Additionally, this strain harbored a 54-kbp broad-host-range IncP-type plasmid (pKJK5) (45) tagged with a gfpmut3 gene (34) downstream of a synthetic LacI-repressible PA1/04/03 promoter (46) and a kanamycin resistance gene (47). The simultaneous expression of both mCherry and the LacI-repressed GFPmut3 was achieved in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (11). The strain was routinely grown at 37°C for 24 h in lysogeny broth (LB) broth medium (10 g/liter NaCl). Additionally, cultures were supplemented with 50 μg/ml kanamycin to avoid plasmid loss.
Aerobic fluorescence recovery.
Anaerobic medium was prepared as described by Uchino and Ken-Ichiro (48) in anaerobic glass vessels that were closed with rubber stoppers. Media vessels were purged by flushing with nitrogen gas for 5 to 10 min, ensuring that the air in the headspace was completely replaced. Vessels were sealed with aluminum caps and subsequently autoclaved, a process by which the medium undergoes further oxygen purging. The reducing reagent cysteine hydrochloride, in its monohydrate form, is often the preferred reducing agent because of its low toxicity (49). It was introduced via syringe injection, after nitrogen purging and autoclaving, at a final concentration of 0.5 g/liter, to ensure the depletion of any traces of oxygen in the anaerobic medium (49–52). Buffered medium was prepared by adding sodium phosphate buffer at a final concentration of 0.1 M (pH 7.4). The E. coli strain expressing both mCherry and GFPmut3 was grown in triplicate, aerobically and anaerobically, both in buffered and unbuffered media. Cultures were started by inoculating 10 μl of an overnight seeding culture of the E. coli strain into 15 ml of LB medium supplemented with 50 μg/ml kanamycin, corresponding to an initial concentration of about 7 × 105 cells/ml.
The AFR of the proteins upon exposure to air was evaluated by means of flow cytometry. Cultures were diluted in PBS to a concentration corresponding to 1,000 to 3,000 events/second when run at flow rate 1 (10 μl/min). As low temperatures are believed to favor the maturation of these proteins (27, 53), samples were incubated at 4°C during AFR. In order to ensure that the increase in the fluorescence signal was exclusively due to the oxic maturation of the proteins and not as a result of de novo protein synthesis under aerobic conditions, chloramphenicol was used as an efficient translational inhibitor (54). Chloramphenicol was added at a final concentration of 100 μg/ml, greatly surpassing the reported MIC for this species (55). A set of replicates treated without IPTG induction was employed as a negative control for induction of GFPmut3 and to evaluate possible differences when expressing it along mCherry.
Resazurin was included routinely in the media (1 mg/liter) to monitor anaerobiosis (48). This dye was not included when samples were analyzed via flow cytometry since in its pink form it exhibits a strongly red fluorescence that interferes with the fluorescence emission spectrum of mCherry (56). However, samples run at time zero upon exposure to air acted as a consistent control for anaerobiosis, as proteins do not fluoresce in the absence of oxygen. In contrast, aerobically grown cultures served as a positive control for fluorescence.
External pH effect on fluorescence emission of GFPmut3 and mCherry.
To determine the effect of the pH on the fluorescence emission of mCherry and GFPmut3, the E. coli strain was grown aerobically overnight and then subjected overnight to an array of 12 citrate-phosphate buffers (McIlvaine buffers) ranging from pH 2.4 to 8. This was done by mixing 20 μl of the overnight culture with 1 ml of the corresponding buffer solution. The buffer solutions were prepared by mixing stock solutions of 0.2 M disodium hydrogen phosphate (Na2HPO4) and 0.1 M citric acid (C6H8O7) as described previously (57).
Flow cytometry.
Flow cytometric detection of cells was performed by using a FACSAria Illu instrument (Becton Dickinson Biosciences, San Jose, CA) as described by Klümper et al. (12). Samples were adjusted by dilution in PBS to a cell count of 1,000 to 3,000 events per second at flow rate 1 to ensure optimal counting, and the software was set to terminate counts after recording 50,000 bacterial events. The technical settings relevant for this study were as follows: a 70-μm nozzle was used with a sheath fluid pressure of 70 lb/in2; forward scatter (FSC) was set at 505 V; side scatter (SSC) at 308 V; and the detectors for green fluorescence (fluorescein isothiocyanate [FITC]; bandpass filter, 530/30 nm) at 508 V and red fluorescence (PE-Texas Red-A; bandpass filter, 610/20) at 500 V. Thresholds were set at 200 for both FSC and SSC, and BD FACSDiva software v.6.1.3 was used for both operation and analysis of the results.
The total population of bacteria was readily distinguishable from background noise in a bivariate contour plot using the area of FSC (FSC-A) versus area of SSC (SSC-A), and a gate was established around this population. Green-fluorescing cells were gated on a bivariate contour plot using the area of FITC (FITC-A) versus SSC-A. Red-fluorescing cells were gated on a bivariate contour plot using the area of PE-Texas Red (PE-Texas Red-A) versus SSC-A. Construction of the appropriate gating is depicted in Fig. S1 in the supplemental material.
Anaerobic filter mating experiments on solid-surface media coupled to AFR.
Solid-surface filter matings were carried out using E. coli MG1655 lacIq::mCherry/pKJK5-gfpmut3, harboring the conjugative plasmid pKJK5, as the donor strain, and its wild-type counterpart, E. coli MG1655, as the recipient strain. A modified protocol from that developed by Musovic et al. (58) was employed. Briefly, strains were grown anaerobically to an optical density at 600 nm (OD600) of 0.6, equal volumes of plasmid donor and recipient strains were mixed, and 40 μl of the resultant suspension was transferred onto sterile 0.2-μm nitrocellulose filters (Advantec) that were placed over Gifu anaerobic medium (GAM) modified agar (Nissui Pharmaceuticals Co.) plates and incubated at 37°C for 24 h. The area of the filter exposed was estimated to be 54 mm2, resulting in an initial cell count of approximately 3.6 × 105 cells/mm2. The filter mating experiments were carried out anaerobically in triplicate, and controls included plating the donor and recipient strains alone, as well as plating the donor strain on medium supplemented with 1 mM IPTG. Bacteria were harvested from the filters by vortexing in 2 ml of PBS and used immediately for flow cytometric counting. AFR was carried out in McIlvaine buffer solution (pH 8) supplemented with 100 μg/ml chloramphenicol. Note that donor and recipient populations are greatly diluted in the AFR incubation buffer, rendering cell-to-cell contacts between them highly improbable and thus conjugation events negligible. Additionally, even in the unlikely event of transfer, the detection of putatively new transconjugants is not an issue, since chloramphenicol would inhibit the synthesis of newly horizontally acquired GFPmut3.
Anaerobic filter matings and anoxic manipulations were performed in an anaerobic chamber (Concept 400; Thermo Scientific, Waltham, MA) with oxygen levels of <0.02% and where medium was allowed to equilibrate to the anoxic gaseous environment for a minimum of 3 days prior to its use (28). Cells harvested from filters were transferred into 2-ml airtight glass vials (Schuett Biotec, Göttingen, Germany), which were sealed inside the anoxic bench to enable anoxic transportation to the flow cytometer facility. Airtight vials were only opened immediately prior to flow cytometry analysis, thus allowing the necessary controlled exposure to air.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stefan Morberg Milani for sharing his experience with the FACSDiva, Martin Iain Bahl from the Gut Microbiology and Immunology Group at the Technical University of Denmark for his helpful comments during the initial stages of this project, David Mayo Muñoz for his assistance in data handling and generous sharing of his software knowledge, and Mette Kolpen from the Unit of Bacteriology, Department of Immunology and Microbiology, University of Copenhagen for her kind assistance with and provision of an anaerobic chamber.
This research was funded by Innovation Fund Denmark (file 5157-0005B).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02507-17.
REFERENCES
- 1.Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol 19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 2.Aminov RI. 2011. Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huddleston JR. 2014. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 6.Schjørring S, Krogfelt KA. 2011. Assessment of bacterial antibiotic resistance transfer in the gut. Int J Microbiol 2011:312956. doi: 10.1155/2011/312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. 2011. The repertoire of ice in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman A, Hansen LH, Sørensen SJ. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkay T, Smets BF. 2005. Horizontal gene flow in microbial communities. ASM News 71:412–419. [Google Scholar]
- 10.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S, Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol 3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 11.Sørensen SJ, Sørensen AH, Hansen LH, Oregaard G, Veal D. 2003. Direct detection and quantification of horizontal gene transfer by using flow cytometry and gfp as a reporter gene. Curr Microbiol 47:129–133. doi: 10.1007/s00284-002-3978-0. [DOI] [PubMed] [Google Scholar]
- 12.Klümper U, Riber L, Dechesne A, Sannazzarro A, Hansen LH, Sørensen SJ, Smets BF. 2015. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J 9:934–945. doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klümper U, Dechesne A, Riber L, Brandt KK, Gülay A, Sørensen SJ, Smets BF. 2017. Different metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J 11:152–165. doi: 10.1038/ismej.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquiod S, Brejnrod A, Morberg SM, Abu Al-Soud W, Sørensen SJ, Riber L. 2017. Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol Ecol 26:3556–3571. doi: 10.1111/mec.14138. [DOI] [PubMed] [Google Scholar]
- 15.Shintani M, Matsui K, Inoue J, Hosoyama A, Ohji S, Yamazoe A, Nojiri H, Kimbara K, Ohkuma M. 2014. Single-cell analyses revealed transfer ranges of incP-1, incP-7, and incP-9 plasmids in a soil bacterial community. Appl Environ Microbiol 80:138–145. doi: 10.1128/AEM.02571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook MT, Tzortzis G, Charalampopoulos D, Khutoryanskiy VV. 2012. Microencapsulation of probiotics for gastrointestinal delivery. J Control Release 162:56–67. doi: 10.1016/j.jconrel.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Fallingborg J. 1999. Intraluminal pH of the human gastrointestinal tract. Lægeforeningens forlag, Copenhagen, Denmark. [PubMed] [Google Scholar]
- 18.Craggs TD. 2009. Green fluorescent protein: structure, folding and chromophore maturation. Chem Soc Rev 38:2865–2875. doi: 10.1039/b903641p. [DOI] [PubMed] [Google Scholar]
- 19.Strack RL, Strongin DE, Mets L, Glick BS, Keenan RJ. 2010. Chromophore formation in DsRed occurs by a branched pathway. J Am Chem Soc 132:8496–8505. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee A, Schroeder CM. 2015. Flavin-based fluorescent proteins: emerging paradigms in biological imaging. Curr Opin Biotechnol 31:16–23. doi: 10.1016/j.copbio.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Wingen M, Potzkei J, Endres S, Casini G, Rupprecht C, Fahlke C, Krauss U, Jaeger KE, Drepper T, Gensch T. 2014. The photophysics of LOV-based fluorescent proteins—new tools for cell biology. Photochem Photobiol Sci 13:875–883. doi: 10.1039/C3PP50414J. [DOI] [PubMed] [Google Scholar]
- 22.Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl J-K, Wendorff M, Losi A, Gärtner W, Jaeger K-E, Krauß U, Guterl J-K, Wendorff M, Losi A, Gärtner W, Jaeger K-E. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol 25:443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 23.Drepper T, Gensch T, Pohl M. 2013. Advanced in vivo applications of blue light photoreceptors as alternative fluorescent proteins. Photochem Photobiol Sci 12:1125–1134. doi: 10.1039/c3pp50040c. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee A, Walker J, Weyant KB, Schroeder CM. 2013. Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS One 8:e64753. doi: 10.1371/journal.pone.0064753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C, Xing XH, Lou K. 2005. Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol Lett 249:211–218. doi: 10.1016/j.femsle.2005.05.051. [DOI] [PubMed] [Google Scholar]
- 26.Ransom EM, Ellermeier CD, Weiss DS. 2015. Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol 81:1652–1660. doi: 10.1128/AEM.03446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott KP, Mercer DK, Glover LA, Flint HJ. 1998. The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol Ecol 26:219–230. doi: 10.1111/j.1574-6941.1998.tb00507.x. [DOI] [Google Scholar]
- 28.Line L, Alhede M, Kolpen M, Kuhl M, Ciofu O, Bjarnsholt T, Moser C, Toyofuku M, Nomura N, Hoiby N, Jensen PO. 2014. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front Microbiol 5:554. doi: 10.3389/fmicb.2014.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doherty GP, Bailey K, Lewis PJ. 2010. Stage-specific fluorescence intensity of GFP and mCherry during sporulation in Bacillus subtilis. BMC Res Notes 3:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. 2009. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 31.Hansen MC, Palmer RJ, Udsen C, White DC, Molin S, Palmer RJ Jr, Udsen C, White DC, Molin S. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383–1391. doi: 10.1099/00221287-147-5-1383. [DOI] [PubMed] [Google Scholar]
- 32.Shaner NC, Lin MZ, McKeown MR, Steinbach P a, Hazelwood KL, Davidson MW, Tsien RY. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebisch E, Knebel J, Landsberg J, Frey E, Leisner M. 2013. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PLoS One 8:e75991. doi: 10.1371/journal.pone.0075991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 35.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 36.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 37.Campbell TN, Choy FYM. 2001. The effect of pH on green fluorescent protein: a brief review. Mol Biol Today 2:1–4. [Google Scholar]
- 38.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. 1998. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci U S A 95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilks JC, Slonczewski JL. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol 189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitko RD, Wilks JC, Garduque GM, Slonczewski JL. 2010. Osmolytes contribute to pH homeostasis of Escherichia coli. PLoS One 5:e10078. doi: 10.1371/journal.pone.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robey RB, Ruiz O, Santos AVP, Ma J, Kear F, Wang L, Li C, Bernardo AA, Arruda JAL, April RV, Re V, Recei M, June V. 1998. pH-dependent fluorescence of a heterologously expressed aequorea green fluorescent protein mutant: in situ spectral characteristics and applicability to intracellular pH estimation. Biochemistry 37:9894–9901. doi: 10.1021/bi980857x. [DOI] [PubMed] [Google Scholar]
- 42.Kneen M, Farinas J, Li Y, Verkman AS. 1998. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J 74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura K, Inouye M. 1979. DNA sequence of the gene for the outer membrane lipoprotein of E. coli: an extremely AT-rich promoter. Cell 18:1109. doi: 10.1016/0092-8674(79)90224-1. [DOI] [PubMed] [Google Scholar]
- 45.Sengeløv G, Kristensen KJ, Sørensen AH, Kroer N, Sørensen SJ. 2001. Effect of Genomic location on horizontal transfer of a recombinant gene cassette between Pseudomonas strains in the rhizosphere and spermosphere of barley seedlings. Curr Microbiol 42:160–167. doi: 10.1007/s002840010197. [DOI] [PubMed] [Google Scholar]
- 46.Lanzer M, Bujard H. 1988. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A 85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klümper U, Droumpali A, Dechesne A, Smets BF. 2014. Novel assay to measure the plasmid mobilizing potential of mixed microbial communities. Front Microbiol 5:730. doi: 10.3389/fmicb.2014.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uchino Y, Ken-Ichiro S. 2011. A simple preparation of liquid media for the cultivation of strict anaerobes. J Pet Environ Biotechnol S3:001. doi: 10.4172/2157-7463.S3-001. [DOI] [Google Scholar]
- 49.Fukushima RS, Weimer PJ, Kunz DA. 2002. Photocatalytic Interaction of resazurin N-oxide with cysteine optimizes preparation of anaerobic culture media. Anaerobe 8:29–34. doi: 10.1006/anae.2001.0405. [DOI] [Google Scholar]
- 50.Dien BS, Nichols NN, Bothast RJ. 2001. Recombinant Escherichia coli engineered for production of L-lactic acid from hexose and pentose sugars. J Ind Microbiol Biotechnol 27:259–264. doi: 10.1038/sj.jim.7000195. [DOI] [PubMed] [Google Scholar]
- 51.Stieglmeier M, Wirth R, Kminek G, Moissl-Eichinger C. 2009. Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms. Appl Environ Microbiol 75:3484–3491. doi: 10.1128/AEM.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji K, Wang W, Zeng B, Chen S, Zhao Q, Chen Y, Li G, Ma T. 2016. Bacterial cellulose synthesis mechanism of facultative anaerobe Enterobacter sp. FY-07. Sci Rep 6:21863. doi: 10.1038/srep21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siemering KR, Golbik R, Sever R, Haseloff J. 1996. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6:1653–1663. doi: 10.1016/S0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 54.Armentrout SA, Weisberger AS. 1967. Inhibition of directed protein synthesis by chloramphenicol: effect of magnesium concentration. Biochem Biophys Res Commun 26:712–716. doi: 10.1016/S0006-291X(67)80131-1. [DOI] [PubMed] [Google Scholar]
- 55.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 56.Bueno C, Villegas ML, Bertolotti SG, Previtali CM, Neumann MG, Encinas MV. 2002. The excited-state interaction of resazurin and resorufin with amines in aqueous solutions. Photophysics and photochemical reactions. Photochem Photobiol 76:385–390. [DOI] [PubMed] [Google Scholar]
- 57.McIlvaine TC. 1921. A buffer solution for colorimetric comparison. J Biol Chem 49:183–186. [Google Scholar]
- 58.Musovic S, Dechesne A, Sørensen J, Smets BF. 2010. Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl Environ Microbiol 76:4813–4818. doi: 10.1128/AEM.02713-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.