ABSTRACT
Many Pseudomonas aeruginosa infections are derived from residential, recreational, or surface water sources; thus, these environments represent an important preinfection niche. To better understand P. aeruginosa biology in these environments, we quantified transcriptional changes by microarray after exposure to diluted LB, diluted R2B, potable tap water, and freshwater from a eutrophic pond. Quantitative reverse transcription-PCR (qRT-PCR) confirmed the conservation of these responses in other water sources, and competition experiments were used to test the importance of three implicated metabolic pathways. The global transcriptional responses in potable water and freshwater showed strong induction of genes involved in metabolism of the head groups and acyl tails of phospholipids, as well as nucleotide metabolism, with commensurate decreased transcript expression of genes encoding their synthetic pathways. These data suggest that phospholipids and nucleotides are part of the nutritional milieu of these two environments. A unique response in municipal-delivered potable water was to the metals in the piping system, particularly copper. To identify potential nutrient sources used by P. aeruginosa in these environments, we used competition assays between the wild-type and deletion mutant strains in three pathways induced under these conditions. For phospholipid head-group metabolism, ethanolamine utilization (eutB) was important for competition in potable water, while choline oxidation (betBA) was important for competition in freshwater. Nucleotide utilization, particularly pyrimidine metabolism (dht), showed a trend toward importance in freshwater but was not statistically significant. These findings provide new insights into the P. aeruginosa response to potable water and freshwater and led to the identification of potentially important nutrient sources in these environments.
IMPORTANCE Much of our knowledge about Pseudomonas aeruginosa comes from the infection niche, and much less is known about its lifestyle in the environment. P. aeruginosa is an adaptable bacterium capable of growing in many environments but is particularly common in potable water systems and freshwater. We used the transcriptional responses of P. aeruginosa to these environments to identify important nutrient sources specific to either of these two environments. Additionally, these environments could provide experimental situations to understand gene function for the large number of transcripts with unknown functions induced under these conditions.
KEYWORDS: nutrient limitation, preinfection niche, ethanolamine, choline, copper, water system, water systems
INTRODUCTION
Pseudomonas aeruginosa is a common environmental bacterium particularly abundant in freshwater environments and potable water (PW) systems (1, 2). P. aeruginosa can be a serious opportunistic pathogen causing a variety of different infections, including life-threatening bacteremia and pneumonia in neutropenic patients, chronic infections in cystic fibrosis and chronic obstructive pulmonary disease, serious eye and ear infections, and generally non-life-threatening but common and costly folliculitis and nail infections. P. aeruginosa lives in the PW system (3–6) and can cause infection directly from this environment; this is best studied in folliculitis and ear infections, for which >80% of the cases are PW derived (7–10). For eye infections and cystic fibrosis, evidence suggests some infections are derived from the PW system (11–15). Numerous studies of nosocomial pneumonia and bacteremia in intensive care units (ICUs) (including references 16–27) support a proportion of PW-derived P. aeruginosa infections being between 8 and 50% (28–31), and when patient PW was subject to filtration and sterilization, the contribution of PW-derived infections in the units dropped (16–18, 32–34). Given the evidence for PW impact on P. aeruginosa infection, surprisingly little is known about P. aeruginosa biology in this environment.
PW contains a variety of dissolved organic carbon species of different sizes and chemical classes, some of which are accessible to bacteria for growth, termed assimilable organic carbon (AOC) (35–37). While many studies have characterized bacterial growth on AOC and hydrological and treatment processes that alter AOC (38–43), relatively little is known about the specific carbon sources utilized by bacteria during growth in PW. Our goal in this study was to use the P. aeruginosa transcriptional response to PW and freshwater to better understand these environments from the bacterial perspective and identify potential carbon and nitrogen sources used under these conditions. Here, we describe that P. aeruginosa responds to PW and freshwater by inducing transcripts involved with phospholipid and nucleotide metabolism and downregulating genes involved in their synthesis. The PW transcriptional response includes the signature of the metals present in the plumbing system, with induction of transcripts involved in copper, iron, and zinc stress and downregulation of transcripts involved with uptake of these metals. Competition experiments with mutants in specific metabolic pathways support ethanolamine as an important nutrient source in PW, while choline is important in freshwater. This study provides foundational data on the P. aeruginosa responses to PW and freshwater and lays the groundwork for a better understanding of these environments as important preinfection niches.
RESULTS
P. aeruginosa transcriptomes in diluted R2B and diluted LB are remarkably similar.
Our primary goal in this study was to identify P. aeruginosa genes induced in response to potable water. One of the more common media for the growth of potable water bacteria is full-strength or diluted R2B, and this medium was to serve as our control for the remaining experiments. Because many previous P. aeruginosa transcriptomes were done using LB, we sought to compare our half-strength R2B (1/2R2B) to half-strength LB (1/2LB) to examine the transcriptional differences.
Surprisingly, there were only 34 transcripts that differed in expression more than 3-fold between these two conditions (see Data Set S1 in the supplemental material). Of the 20 transcripts at levels higher in 1/2LB than 1/2R2B, eight were involved in nitric oxide and nitrate reduction (nirJ, nirG, PA0515, nirC, norC, norB, ppyR, and fhp; altered 3.1- to 10.6-fold), three with choline metabolism (encoded by the betIBA operon; altered 6.5- to 29.1-fold), two involved with polyamine response (agtA [6.1-fold] and gbuA [3.8-fold]), two involved with membrane responses (oprH [10.7-fold] and esrC [8.5-fold]), and five transcripts of uncharacterized genes (3.5- to 9.1-fold changes).
Fourteen transcripts were present at higher levels in 1/2R2B than in 1/2LB, with nine of them directly related to iron acquisition and transport (pvdE, fpvA, pvdS, bfd, PA3690, PA4514, PA4515, PA4517, and phuR; altered 3.1- to 10-fold) and three related to altered flux through metabolic pathways (gcvH2 [3.8-fold], antB [4.5-fold], and ureG [3.7-fold]).
These findings suggest that the major differences that P. aeruginosa detects between these two media are iron availability, oxidized nitrogen content or oxygen availability, and altered organic nitrogen species, particularly the abundance of choline in LB from the 10-fold greater concentration of yeast extract compared to R2B.
Response of P. aeruginosa to tap water.
Our primary goal was to determine the transcriptome of P. aeruginosa in the presence of potable (tap) water. We compared the transcriptomes between 1/2R2B and undiluted tap water, as described in Materials and Methods. In total, the expression of 485 transcripts were significantly changed more than 3-fold between 1/2R2B and tap water (Data Set S2), which represents approximately 8.6% of the genome.
Differences between the conditions can be described based on the functions of transcripts altered and their direction (Fig. 1 and Data Set S2). One of the most notable and obvious differences between 1/2R2B and tap water is the concentration of transition metals, particularly copper, resulting in the induction of 22 transcripts involved in transition metal resistance and efflux, and the decrease in the expression of 11 transcripts involved in coproporphyrinogen, hemerythrin, and bacterioferritin production.
FIG 1.
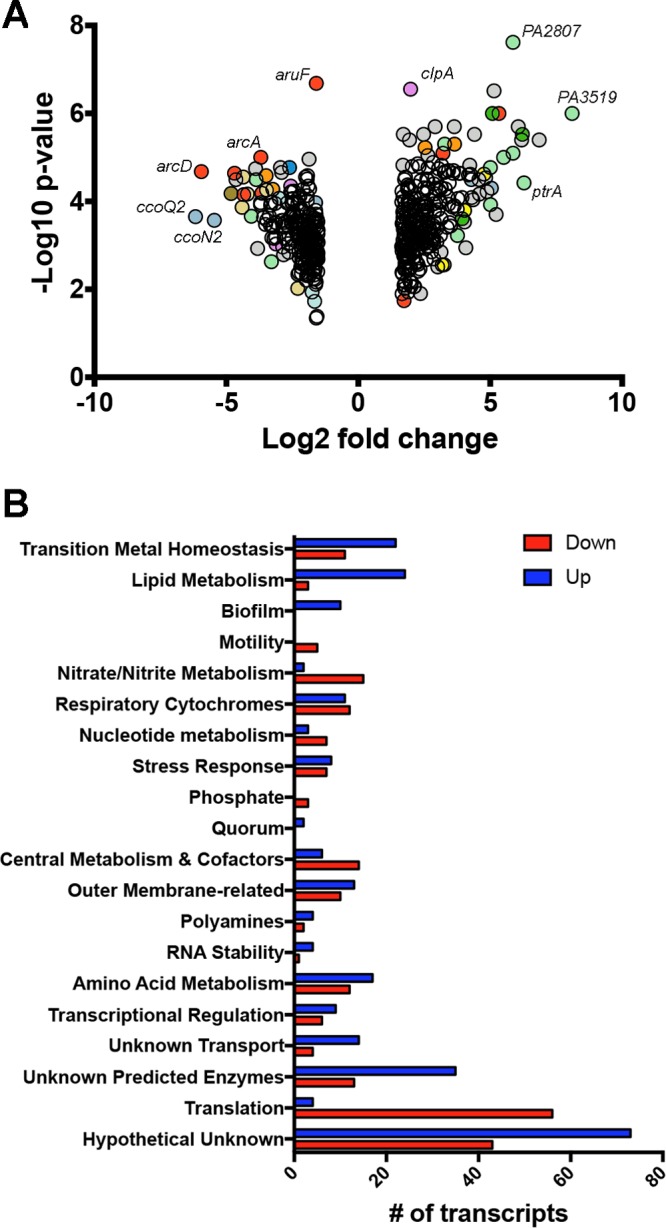
Pseudomonas aeruginosa transcriptome changes in response to tap water. (A) Volcano plot showing the transcripts that met both fold and P value cutoffs. Colors denote functional classes of transcripts, and only transcripts with two or fewer symbol overlaps are colored: light green, transition metal homeostasis; yellow, lipid metabolism; mauve, stress responses; red, amino acid metabolism; light blue, respiratory cytochromes; gray, hypothetical unknowns. (B) Transcripts plotted in panel A clustered by function showing number of transcripts that were present at higher levels in tap water (blue bars) or lower in tap water (red bars).
In addition to the transition metals, transcripts related to lipids, polyamines, biofilm formation, stress, and nucleotide utilization were substantially altered (Fig. 1 and Data Set S2). Tap water led to the induction of 18 transcripts related to lipid acquisition and metabolism with concomitant reduction in the expression of three lipid synthesis transcripts, suggesting a lipid source in tap water. These included the induction of transcripts for the metabolism of choline, ethanolamine, and glycerol, suggesting a source of glycerophospholipids. A similar pattern was seen for transcripts related to polyamines and nucleotides, where the expression of transcripts encoding utilization and transport components were increased and those encoding biosynthetic pathways were decreased. Ten transcripts involved in biofilm formation, particularly the Fap-based pathway, were induced, while the expression of five transcripts related to flagellar synthesis or regulation were decreased. Eight transcripts encoding heat shock chaperones or parts of the misfolded protein response system were induced, while the expression of a number of transcripts coding for members of the universal stress protein family were decreased.
It was also evident that the differential availability of amino acids and carbohydrates between the two conditions altered both amino acid-related pathways and central metabolism (Fig. 1 and Data Set S2). In particular, the expression of transcripts for enzymes of the glyoxylate shunt were increased, while those of some related to glycolysis were decreased. Also increased are the expression of transcripts related to amino acid uptake, while the expression of most of the arc transcripts, involved in anaerobic arginine utilization, are decreased.
There were also reductions in the expression of transcripts involved in microaerophilic and anaerobic metabolism in PW compared to 1/2R2B, including the nir and nor genes and the cbb3-2 cytochrome oxidase complex genes. This is likely a direct consequence of more rapid growth and oxygen depletion under the 1/2R2B condition, leading to increased expression of these microaerobic and anaerobic respiratory transcripts (44, 45) than an actual repression under the PW condition.
Finally, a notable pattern in the transcripts involves those genes related to translation and growth. In total, the expression of 56 transcripts involved in translation and growth are lower in PW than in 1/2R2B, while the expression of only four transcripts are increased. One of those transcripts increased in PW is rmf, a gene whose expression is greatly dependent on growth rate, being strongly induced during slower growth. Together, these patterns suggest slower growth in tap water than in 1/2R2B, which is not surprising given the difference in nutrient concentrations between these two conditions.
The response of P. aeruginosa to pond water.
The P. aeruginosa response to surface water from a eutrophic pond was somewhat similar to the tap water response compared to 1/2R2B (Fig. 2A and Data Set S3). In total, the expression of 600 transcripts were differentially expressed more than 3-fold, and many followed the same pattern as for tap water: decreased growth signaled via altered expression of translation-related genes, changes to central metabolism particularly glyoxylate shunt induction, induction of transcripts for branched-chain amino acid utilization, and increases in the expression of nucleotide utilization transcripts with decreases in nucleotide synthesis-related gene expression. Given the large number of changes between 1/2R2B and pond water, we decided to focus on those genes differentially expressed between tap and pond water.
FIG 2.
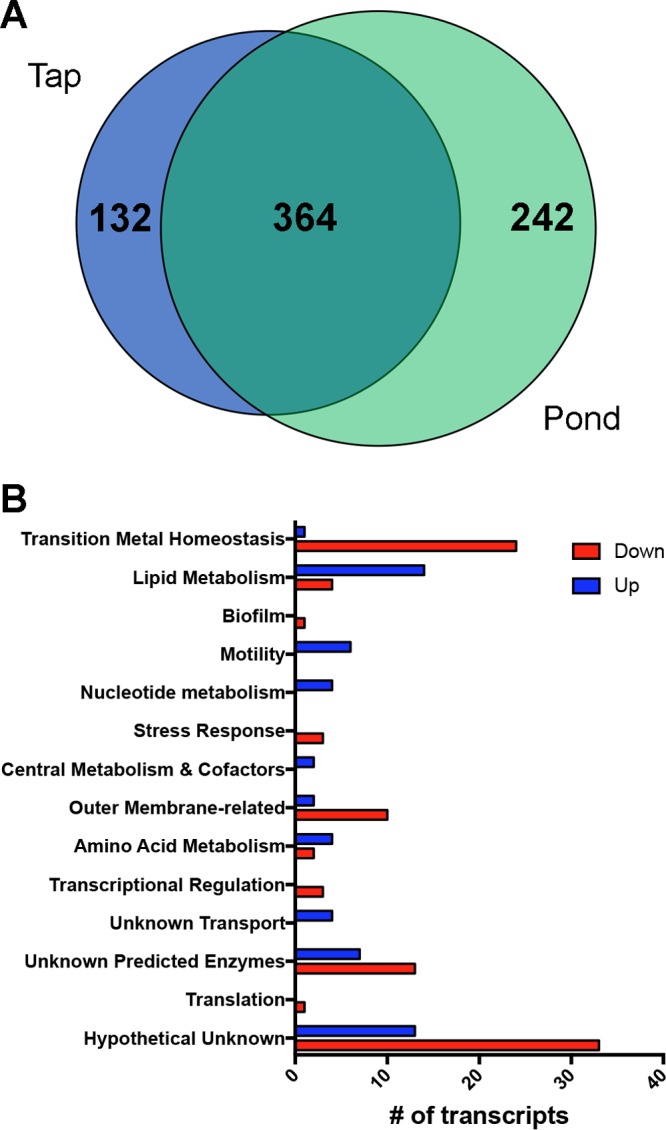
Pseudomonas aeruginosa transcriptome changes comparing a freshwater pond to tap water. (A) Venn diagram showing the similarity of transcriptome changes in tap water or pond water each compared to 1/2R2B media. Numbers represent number of transcripts. (B) Transcripts that were changed comparing freshwater to tap water clustered by function showing number of transcripts that were higher in pond water (blue bars) or lower in pond water (red bars).
There were 151 transcripts differentially expressed between PW and pond water. In pond water, there was increased expression of transcripts involved in lipid and head group metabolism, branched-chain amino acid uptake, aerotaxis, chemotaxis, nucleotide utilization, and a number of uncharacterized genes (Fig. 2B and Data Set S4). The expression of transcripts involved in metal stress and detoxification, drug efflux and outer membrane permeability, and a number of uncharacterized genes were decreased in pond water compared to PW. In particular, the induction of metal stress genes only in the PW treatment is not surprising given the high concentration of metals in that system and their comparative rarity in most surface waters. With similar reasoning, the expression of genes for SDS metabolism and drug resistance were decreased in pond water compared to PW. Given the frequent carriage of these compounds in PW, this is not surprising. Important to note here is that Shelburne Pond does not drain a large watershed and has very little residential or industrial inputs; thus, while it has a high nutrient load, there is relatively little input of consumer products.
Conservation of select metabolic gene responses in diverse water sources and in environmental strains.
Our microarrays were based on a single source of PW and a single freshwater pond. To test the broader applicability of our findings, we quantified the induction of three transcripts in two additional PW sources: a municipal and a well water source, and three additional freshwater sources: a mesotrophic impoundment (Indian Brook Reservoir), a eutrophic lake (Colchester Pond), and a river (Winooski River). The betB, dht, and eutB transcripts were strongly induced under all of these conditions compared to the 1/2R2B control and normalized to the ppiD transcript (46, 47) (Fig. 3A). There was some variability between sites, but they were small compared to the scale of the changes. The difference in normalization method for microarrays and these qRT-PCR experiments likely explains their difference in the scale of transcriptional changes, as the microarray is based on a whole-chip signal rather than a single control transcript. Given the large drop in the expression of many abundant translation-related transcripts in the PW and freshwater samples (Fig. 1 and 2 and Data Sets S2 and S3), this would lead to decreasing apparent differences in the microarray.
FIG 3.
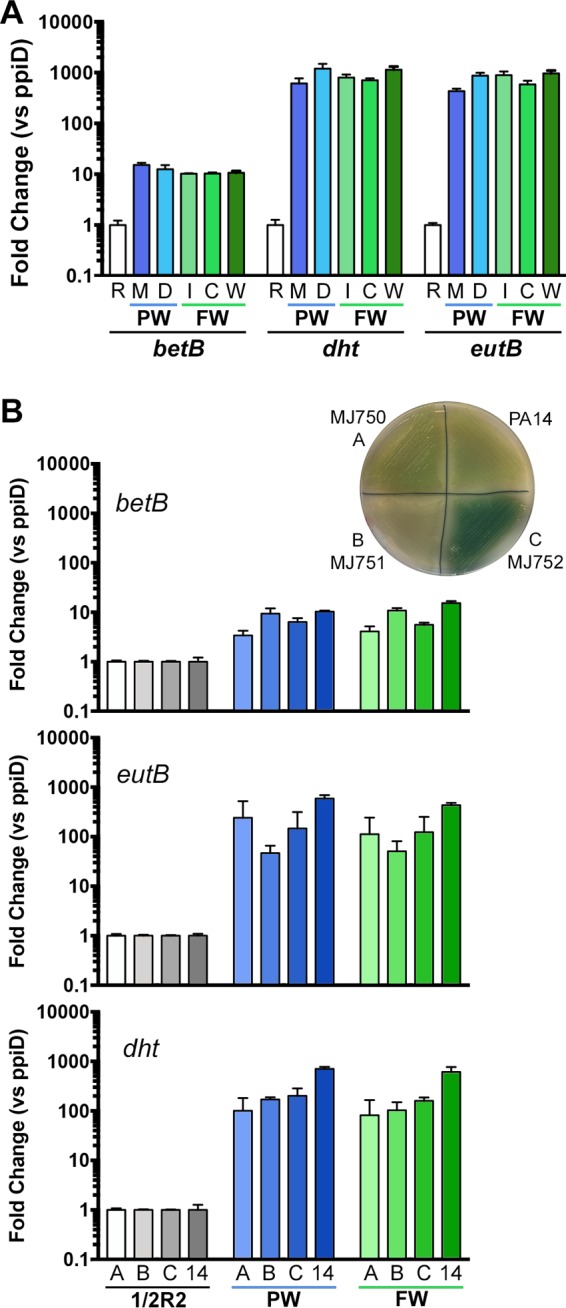
Measuring P. aeruginosa transcript changes in other sources of potable water and freshwater and in environmental isolates. (A) Induction of the bet, eutB, and dht transcripts occurs in a variety of potable and surface water sources. (B) The betB, eutB, and dht transcripts are induced in potable and pond water in three environmental isolates. Inset shows the environmental isolates grown on PIA. For both panels, the betB, dht, and eutB transcripts were measured by qRT-PCR normalizing to the ppiD transcripts and setting the 1/2R2B ratio to one for each transcript. R, 1/2R2B; M, municipal tap water; 1/2R, 1/2R2B; D, well water; I, Indian Brook Reservoir; C, Colchester Pond; W, Winooski River; PW, potable water; FW, freshwater. Blue bars denote potable water samples, and green bars denote freshwater samples. The PW for panel B is identical to the M sample in panel A, and the FW in panel B is identical to the C samples of panel A. The PA14 data shown in panel B are directly pulled from panel A to enable easier comparison.
We also tested the conservation of these metabolism-related transcriptional responses in three wild isolates of P. aeruginosa isolated from water fixtures on our campus that were phenotypically distinct from PA14 (see inset, Fig. 3B). In all three environmental isolates, the betB, eutB, and dht transcripts were induced with some variation in levels between strains (Fig. 3B). These data suggest that the responses of the wound isolate, PA14, are representative of plumbing-derived strains, at least for these transcripts.
Roles of ethanolamine, choline, and pyrimidine catabolic genes in potable water and freshwater.
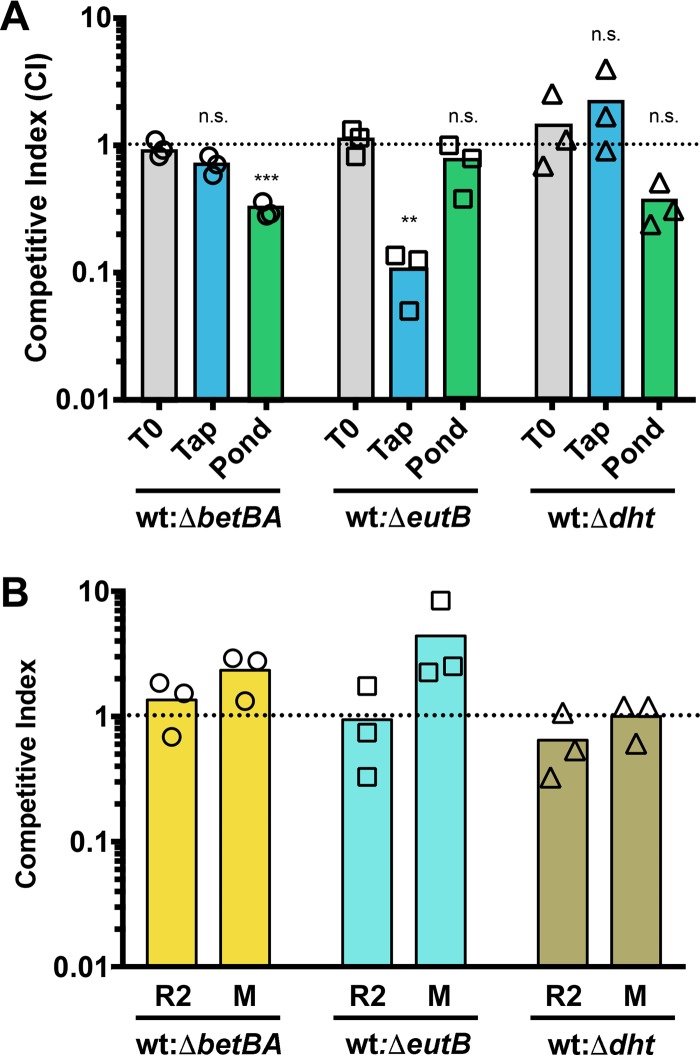
One of our goals was to identify specific AOC components via the P. aeruginosa transcriptional response and test their relative importance using competition assays. As described in Materials and Methods, standard chromosomal lacZ or antibiotic resistance markers showed strong defects when wild-type (WT)-versus-WT competitions were conducted in PW. Therefore, we used serial dilution plating onto two different plate types, one permissive to WT and mutants, and the other restrictive to mutant growth. Using this system, we tested the importance of choline (betBA), ethanolamine (eutB), and pyrimidine (dht) metabolism in PW and surface water from a pond. As shown in Fig. 4A, a loss of choline oxidation to glycine betaine was important for competition versus wild type in surface water from a pond but not in PW. Conversely, ethanolamine deamination was important in PW but not in freshwater from a pond. Pyrimidine metabolism showed a trend toward importance in pond water, but this change was not significant, though the mean effect size was similar to that seen for loss of choline metabolism. None of these deletions impacted competition in 1/2R2B or morpholinepropanesulfonic acid (MOPS) medium with pyruvate and glucose as carbon sources (Fig. 4B).
FIG 4.
Competition experiments show the relative importance of P. aeruginosa metabolic genes in tap water and freshwater. (A) Competition in tap and pond water. (B) Competition in 1/2R2B (R2) and MOPS pyruvate glucose media (M). Competition experiments were conducted comparing wild-type PA14 to isogenic deletion mutants with deletions of betBA, eutB, or dht. The ratios of WT to mutants at time zero (T0) are shown in the gray bars, while tap water (blue bars) and freshwater (green bars) are shown for 24 h. The dotted line marks a WT-to-mutant ratio of 1:1. Data were analyzed using a one-way ANOVA with a Dunnett's posttest, as described in Materials and Methods comparing each competition to the T0 ratio. n.s., not significant; **, P < 0.01; ***, P < 0.001. All ratios in panel B are not significantly different from the input ratio.
DISCUSSION
The importance of the preinfection environment on acquisition rate and pathogenicity, including molecular details of bacterial response to the preinfection niche, has been well studied for the opportunistic pathogens Legionella pneumophila, Vibrio cholerae, and Listeria monocytogenes, as well as for the foodborne enterics E. coli and Salmonella enterica. A similar examination is lacking for P. aeruginosa. Little is known about the physiologic or transcriptional programs used by P. aeruginosa to survive in its preinfection water niches, despite ample evidence for PW systems, recreational waters, and freshwater systems as important infection sources.
Mendis and colleagues have done the most comprehensive assessment to date of P. aeruginosa interaction with a model drinking water medium (48). They showed that P. aeruginosa exposed to the tap water model Fraquil was still able to invade epithelial cells and was more cytotoxic than P. aeruginosa exposed to rich media, but Fraquil did not support strong biofilm formation. As these authors point out, real tap water has many nutrients, including organic nitrogen and carbon sources, which are not present in Fraquil (48). The reason Mendis et al. chose Fraquil, and the drawback of using real tap water, is that tap water is both geographically and temporally variable. Here, we have used snapshot samples of tap water, well water, and surface water from a few freshwater ponds and a river, representing different sources and nutrient loads, to make an argument about the broad conclusions that can still be reached regardless of the likely numerous differences that will exist between disparate sources of water. We also utilized environmental strains to ensure that responses of PA14 could be extrapolated to wild P. aeruginosa isolates (Fig. 3B), and while there was variation in the absolute induction levels between these strains, the three metabolic transcripts of interest were all strongly induced in both tap water and pond water in these strains.
All natural and municipal water sources contain dissolved nutrients, and part of the aim of municipal water treatment is to limit the dissolved organic carbon and nitrogen sources to improve water taste and safety, in part by keeping growth of heterotrophic bacterial counts low. Increases in dissolved carbon, particularly AOC, have been associated with increased heterotrophic growth and some instances of waterborne Enterobacteriaceae outbreaks. While groundwater and surface water can have relatively high levels of AOC (0.1 to 9 mg · liter−1, and often much higher), most treated water has AOC levels between 7 and 200 μg · liter−1, and guidelines are set to keep AOC at <50 μg · liter−1 in some jurisdictions for public health (36). With nitrate in the 0.5 to 5 mg · liter−1 range and phosphate often present at between 0.05 and 0.5 mg · liter−1, AOC appears to be the growth-limiting resource for heterotrophs, like P. aeruginosa, in the potable water system (35, 38). Excellent studies were conducted defining the range of AOC and the growth of bacteria under these conditions (see, e.g., references 38, 43, and 37), but the identities of the AOC compounds remained largely unknown. Here, we show that choline, and perhaps pyrimidines, are important components of AOC in surface freshwater, while ethanolamine appears to be an important component in tap water (Fig. 4). These competition studies were guided by our transcriptional profiles, and we propose that these transcriptomes contain information regarding the identities of other important components of the AOC pool.
The field of Gram-negative bacterial pathogenesis is revisiting the literature of slow growth that has been the cornerstone of environmental microbiology and mycobacteriology, as it has been appreciated that bacteria in many host-associated niches, particularly chronic infections, are growing very slowly. The ability of P. aeruginosa to grow exceptionally slowly in tap water may be one of the traits or preconditions that allow this bacterium to establish chronic infections in the lung, which also often show slow growth (49), though under dramatically different nutrient regimes. One of the signals of slow growth and carbon hoarding we see in tap water and pond water is the induction of transcripts encoding enzymes of the glyoxylate shunt, which has been shown to be important in slow growth, particularly under host-like conditions, such as cystic fibrosis sputum (50) and growth on mucin (51). Further examination of the molecular details of P. aeruginosa interaction with freshwater and tap water will likely yield more clues about its basic biology and the metabolic flexibility that enables its pathogenesis.
MATERIALS AND METHODS
Bacterial strains and maintenance conditions.
P. aeruginosa PA14 wild-type and deletion strains described below were maintained on Lennox broth (LB) or Pseudomonas isolation agar (PIA). E. coli DH5α and S17λpir, used in cloning and conjugation, respectively, were maintained on LB with appropriate antibiotics when needed. Environmental strains of P. aeruginosa (MJ750, MJ751, and MJ752) were isolated from plumbing fixtures within buildings on the University of Vermont campus using PIA with pyocyanin added and incubated at 42°C to enrich for P. aeruginosa (52). All strains collected in this way were verified as P. aeruginosa using the genus- and species-specific PCR primers described by LiPuma and colleagues (53).
Generation of deletion strains.
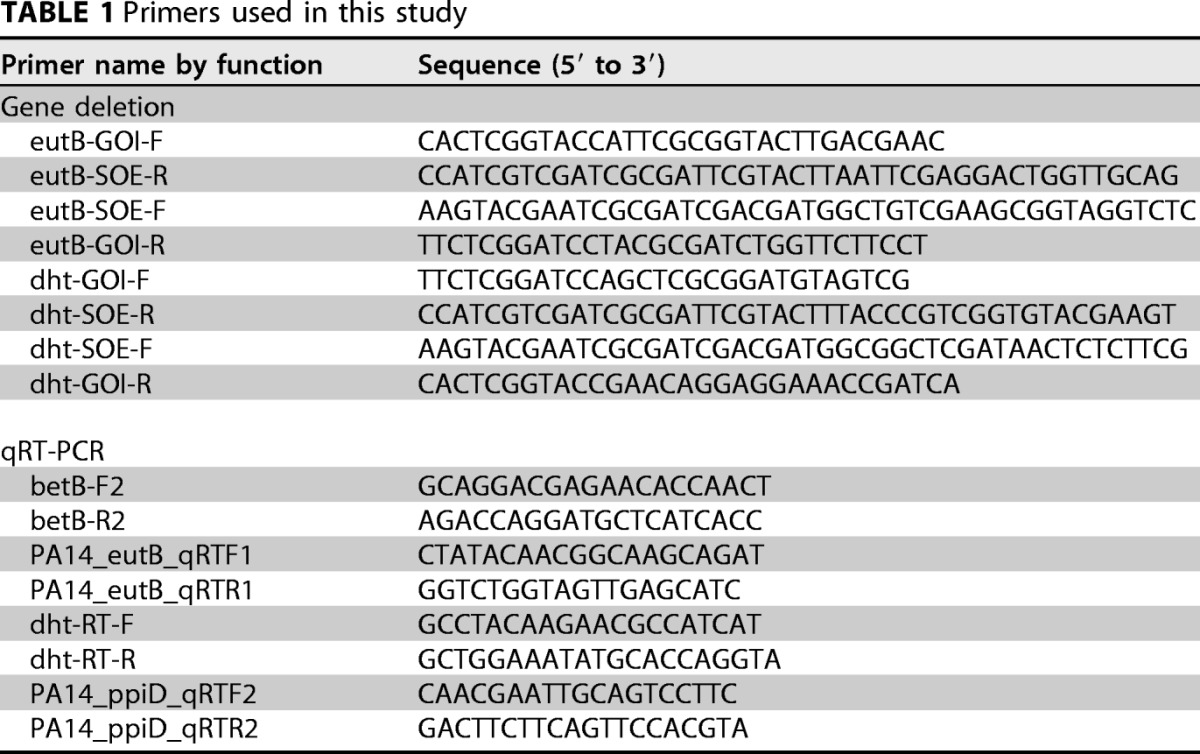
Our betBA deletion mutant of PA14 was previously reported (54). Mutants with deletions of dht and eutB were generated by amplifying the flanking regions of each gene with the primers listed in Table 1. The amplified fragments were digested with BamH1 and KpnI and ligated into similarly cut pMQ30 (55). After validation, the resulting plasmids were transformed into S17λpir, which was conjugated with P. aeruginosa PA14 wild-type cells and single-crossover integrants selected on PIA with 50 μg/ml gentamicin. After restreaking of gentamicin-resistant colonies on plates with gentamicin, cells were grown in LB without antibiotic for 3 h, and double-crossover recombinants were selected on LB with no NaCl and 5% sucrose at 30°C. Deletions were verified by PCR and phenotypic analysis of growth on uracil and ethanolamine as sole nitrogen sources for dht and eutB mutants, respectively (note that dht mutants are not completely deficient for growth on uracil as a sole nitrogen source but are severely compromised).
TABLE 1.
Primers used in this study
Growth conditions and RNA preparation for microarrays.
Cells were exposed to four environments: diluted R2B, diluted LB, potable water from a lab faucet, and water from a eutrophic pond. Half-strength R2B (1/2R2B) and half-strength LB (1/2LB) media were made and sterilized by autoclaving. Potable water was sampled from a faucet in our laboratory by collecting the first 30 ml of the day and then vortexing the faucet's aeration disk in the water sample for 20 s. About half the sample was decanted for use in the experiment. Pond water was collected from the shore near the boat launch on Shelburne Pond in Shelburne, VT, on 28 May 2014 (the day of exposure for transcriptomics) and consisted of 30 ml of water with a small amount of sediment. This sample was taken during a light bloom of the cyanobacterium Microcystis aeruginosa.
Before the addition of cells, 12-well Transwell tissue culture plates (Costar; 0.4 μm pore size) were prepared by presoaking wells and filters with 1.5 ml of a nonsterile aliquot of their respective treatment condition in the lower chamber and 450 μl of heat-treated and 0.22-μm-pore-size sterile-filtered inducing condition in the upper chamber for 1 h at 22°C. The filtration was done to remove native bacteria and eukaryotes from the upper chamber in the potable water and pond samples, but sterile medium was also filtered to control for any leachate from the filters. After filtration, all samples were heated at 70°C for 30 min to inactivate any bacteria that passed through the filter. Such heat treatments are standard in municipal water studies, as many waterborne bacteria pass through a 0.22-μm-pore-size filter.
PA14 was inoculated from a fresh LB plate into 3 ml of 1/2R2B medium and grown overnight at 37°C on a rotating wheel. Cells were collected by centrifugation and resuspended with room temperature 1/2R2B to an optical density at 600 nm (OD600) of 4.0. Fifty microliters of each strain was then added to the top chamber of a Transwell tissue culture plate, prepared as described above, to achieve a final OD600 of 0.4. Inductions were carried out for 3 h at 22°C. Following induction, cells were gathered by scraping the filter and walls of the upper chamber and removing all of the upper chamber media and added to 1,000 μl of RNAprotect bacterial reagent (Qiagen). After 5 min, the protected cells were collected by centrifugation, and the supernatant was decanted before the pellets were frozen at −80°C.
RNA was prepared using a Qiagen RNeasy kit, according to the manufacturer's protocol, with the following changes. Prior to extraction, cell pellets were resuspended in 200 μl of Tris-EDTA (TE) supplemented with 3 mg/ml lysozyme and incubated at room temperature for 20 min. An on-column DNase I treatment was performed before the RNA was eluted in RNase-free water. Samples were then treated a second time with RNase-free DNase I (NEB) and incubated for 1 h at 37°C before a second round of RNeasy column purification was performed.
Microarray methodology.
Microarray analysis was performed by the Vermont Genetics Network Microarray Facility using Affymetrix Pseudomonas aeruginosa PAO1 gene chips and DNA probes generated by the NuGEN Pico system. Tap water (PW) was assessed in biological triplicate and the other conditions in biological duplicate, and signals from all probes for a given gene were averaged into one probe intensity using the Expression Console and Transcriptome Analysis Console software package version 2.0 (Affymetrix). Transcript changes were identified as those exhibiting at least a 3-fold change in signal between compared conditions using reliability, maintainability, and availability (RMA) analysis and a P value of <0.05.
Growth conditions and RNA preparation for quantitative reverse transcription-PCR.
All of these samples were frozen within an hour of collection and stored at −20°C before being thawed, filtered through 0.22-μm-pore-size filters, and heat treated at 70°C as described for the microarray experiments. Cells were pregrown in 1/2R2B overnight at 30°C with shaking, collected by centrifugation, washed under the conditions to which they would be exposed, resuspended at an OD600 of 0.4 in 600 μl in a 24-well dish, and incubated at 22°C for 3 h. Water samples were taken from a municipal water-supplied tap in the corresponding author's residence (Essex, VT), a well water-supplied tap (Jericho, VT), the mesotrophic Indian Brook Reservoir (Essex, VT), the eutrophic Colchester Pond (Colchester, VT), and the Winooski River (Williston, VT).
Quantitative RT-PCR.
Growth conditions and RNA preparations were carried out as described above (in biological triplicate). cDNA was generated using Superscript II with random hexamers and 100 ng of total RNA isolated from each strain under each condition. Quantitative RT-PCR was performed with technical duplicates using NEB's Q5 2× minimal medium (MM) supplemented with SYBR green I nucleic acid gel stain (Thermo Fisher Scientific) at a final concentration of 0.2×, as previously described (56), and the primers are listed in Table 1. For each gene, transcript abundance was determined using a genomic DNA standard curve dilution series. Each sample for each transcript was normalized to its cognate ppiD abundance before conversion to relative expression based on the average expression level in the noninduced control sample (1/2R2B).
Competition experiments.
Cells were grown in MOPS medium with 5 mM pyruvate and 0.5 mM glucose overnight with shaking at 30°C. Cells were collected by centrifugation, resuspended at an OD600 of 0.1 in modified MOPS medium lacking both carbon and nitrogen sources, and incubated for 3 h with shaking at 30°C. After this starvation period, cells from the WT and the respective mutants were mixed and diluted 100-fold, and this cell mixture served as a 100× stock added to municipal tap water (Essex, VT), surface water from the mesotrophic Indian Brook Reservoir (Essex, VT), 1/2R2B, or MOPS medium with 20 mM pyruvate and 5 mM glucose. The tap and pond water samples had been previously filter sterilized and heat treated for 30 min at 70°C, as described in “Microarray methodology” above, the MOPS medium was filter sterilized, and the 1/2R2B was sterilized by autoclaving. An aliquot was taken from the inoculum stock to calculate input ratios, and samples were incubated in their respective environments with shaking at 30°C for 24 h. After this incubation, samples were taken from each replicate. To differentially count WT and mutant CFU, we utilized the known auxotrophies of each mutant strain. All three mutant strains and WT could grow equally well on MOPS medium with 20 mM glucose as the main carbon source and 2 mM glycine betaine as the sole nitrogen source. To differentiate the two strains in competition, the mixtures were also plated on MOPS with 20 mM glucose as the main carbon source and 2 mM choline, ethanolamine, or uracil as the sole nitrogen source, which did not allow growth of the ΔbetBA, ΔeutB, or Δdht mutant, respectively. Using this system, WT CFU were calculated from the restrictive plates, and mutant cell numbers were calculated by subtracting the WT CFU from the total CFU on the permissive MOPS-glycine betaine plates. Mutant CFU was then divided by the WT CFU to calculate the competitive index normalized to that ratio from the time zero (T0) point.
This method of differentiation was required because we determined from pilot experiments that the nutrient/growth stress in these environments compared to standard lab conditions led to any cells expressing a chromosomal lacZ or an antibiotic resistance marker to have a decreased competitive index in WT-versus-WT competition experiments.
Statistical analysis and data visualization.
Microarray statistics were calculated as described in “Microarray methodology” above using RMA. There were three biological replicate tap (PW) arrays and two each for the other conditions. BioVenn (57) was used to generate the Venn diagram precursor. All other statistical analyses and figure generation were conducted in GraphPad Prism. Gene functional classification was done by manually combining related GO, COG, and KEGG predictions into more general functions.
Statistics for Fig. 4 were calculated using one-way ANOVA with a Dunnett's corrected posttest. This statistical framework was used because our hypothesis was that the competitive index was altered compared to the input ratio within each mutant/WT mixture.
Accession number(s).
Array data are available in the GEO database under accession number GSE104819.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dimitri Krementsov for providing a water sample from the well water supplying his house, and we thank the anonymous reviewers for excellent experimental suggestions.
This study was supported by grants from the National Institute of Allergy and Infectious Disease (grant AI103003), the National Institute of General Medicine (grants GM103496 and GM103449), and NASA cooperative agreement NNX16ZHA001C, as well as by T32 HL076122 fellowship support of G.G.W.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02350-17.
REFERENCES
- 1.Trautmann M, Lepper PM, Haller M. 2005. Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am J Infect Control 33:S41–S49. doi: 10.1016/j.ajic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Pirnay JP, Matthijs S, Colak H, Chablain P, Bilocq F, Van Eldere J, De Vos D, Zizi M, Triest L, Cornelis P. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ Microbiol 7:969–980. doi: 10.1111/j.1462-2920.2005.00776.x. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Wingender J, Muller H, Tummler B. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol 60:1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remold SK, Brown CK, Farris JE, Hundley TC, Perpich JA, Purdy ME. 2011. Differential habitat use and niche partitioning by Pseudomonas species in human homes. Microb Ecol 62:505–517. doi: 10.1007/s00248-011-9844-5. [DOI] [PubMed] [Google Scholar]
- 5.Whitby JL, Rampling A. 1972. Pseudomonas aeruginosa contamination in domestic and hospital environments. Lancet i:15–17. doi: 10.1016/S0140-6736(72)90006-2. [DOI] [PubMed] [Google Scholar]
- 6.Bédard E, Prévost M, Déziel E. 2016. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiologyopen 5:937–956. doi: 10.1002/mbo3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas P, Moore M, Bell E, Friedman S, Decker J, Shayegani M, Martin K. 1985. Pseudomonas dermatitis associated with a swimming pool. JAMA 253:1156–1159. doi: 10.1001/jama.1985.03350320080022. [DOI] [PubMed] [Google Scholar]
- 8.Zichichi L, Asta G, Noto G. 2000. Pseudomonas aeruginosa folliculitis after shower/bath exposure. Int J Dermatol 39:270–273. doi: 10.1046/j.1365-4362.2000.00931.x. [DOI] [PubMed] [Google Scholar]
- 9.Salmen P, Dwyer DM, Vorse H, Kruse W. 1983. Whirlpool-associated Pseudomonas aeruginosa urinary tract infections. JAMA 250:2025–2026. doi: 10.1001/jama.1983.03340150067029. [DOI] [PubMed] [Google Scholar]
- 10.Dziuban EJ, Liang JL, Craun GF, Hill V, Yu PA, Painter J, Moore MR, Calderon RL, Roy SL, Beach MJ, Centers for Disease Control and Prevention. 2006. Surveillance for waterborne disease and outbreaks associated with recreational water–United States, 2003–2004. MMWR Surveill Summ 55:1–30. [PubMed] [Google Scholar]
- 11.Schelstraete P, Van Daele S, De Boeck K, Proesmans M, Lebecque P, Leclercq-Foucart J, Malfroot A, Vaneechoutte M, De Baets F. 2008. Pseudomonas aeruginosa in the home environment of newly infected cystic fibrosis patients. Eur Respir J 31:822–829. doi: 10.1183/09031936.00088907. [DOI] [PubMed] [Google Scholar]
- 12.Heirali A, McKeon S, Purighalla S, Storey DG, Rossi L, Costilhes G, Drews SJ, Rabin HR, Surette MG, Parkins MD. 2016. Assessment of the microbial constituents of the home environment of individuals with cystic fibrosis (CF) and their association with lower airways infections. PLoS One 11:e0148534. doi: 10.1371/journal.pone.0148534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barben J, Hafen G, Schmid J, Swiss Paediatric Respiratory Research Group. 2005. Pseudomonas aeruginosa in public swimming pools and bathroom water of patients with cystic fibrosis. J Cyst Fibros 4:227–231. doi: 10.1016/j.jcf.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Speert DP, Campbell ME, Henry DA, Milner R, Taha F, Gravelle A, Davidson AG, Wong LT, Mahenthiralingam E. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am J Respir Crit Care Med 166:988–993. doi: 10.1164/rccm.2203011. [DOI] [PubMed] [Google Scholar]
- 15.Hammond JH, Hebert WP, Naimie A, Ray K, Van Gelder RD, DiGiandomenico A, Lalitha P, Srinivasan M, Acharya NR, Lietman T, Hogan DA, Zegans ME. 2016. Environmentally endemic Pseudomonas aeruginosa strains with mutations in lasR are associated with increased disease severity in corneal ulcers. mSphere 1:e00140-. doi: 10.1128/mSphere.00140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petignat C, Francioli P, Nahimana I, Wenger A, Bille J, Schaller MD, Revelly JP, Zanetti G, Blanc DS. 2006. Exogenous sources of Pseudomonas aeruginosa in intensive care unit patients: implementation of infection control measures and follow-up with molecular typing. Infect Control Hosp Epidemiol 27:953–957. doi: 10.1086/506409. [DOI] [PubMed] [Google Scholar]
- 17.Trautmann M, Halder S, Hoegel J, Royer H, Haller M. 2008. Point-of-use water filtration reduces endemic Pseudomonas aeruginosa infections on a surgical intensive care unit. Am J Infect Control 36:421–429. doi: 10.1016/j.ajic.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Cuttelod M, Senn L, Terletskiy V, Nahimana I, Petignat C, Eggimann P, Bille J, Prod'hom G, Zanetti G, Blanc DS. 2011. Molecular epidemiology of Pseudomonas aeruginosa in intensive care units over a 10-year period (1998–2007). Clin Microbiol Infect 17:57–62. doi: 10.1111/j.1469-0691.2010.03164.x. [DOI] [PubMed] [Google Scholar]
- 19.Cholley P, Thouverez M, Floret N, Bertrand X, Talon D. 2008. The role of water fittings in intensive care rooms as reservoirs for the colonization of patients with Pseudomonas aeruginosa. Intensive Care Med 34:1428–1433. doi: 10.1007/s00134-008-1110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertrand X, Thouverez M, Talon D, Boillot A, Capellier G, Floriot C, Helias JP. 2001. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med 27:1263–1268. doi: 10.1007/s001340100979. [DOI] [PubMed] [Google Scholar]
- 21.Berthelot P, Grattard F, Mahul P, Pain P, Jospe R, Venet C, Carricajo A, Aubert G, Ros A, Dumont A, Lucht F, Zeni F, Auboyer C, Bertrand JC, Pozzetto B. 2001. Prospective study of nosocomial colonization and infection due to Pseudomonas aeruginosa in mechanically ventilated patients. Intensive Care Med 27:503–512. doi: 10.1007/s001340100870. [DOI] [PubMed] [Google Scholar]
- 22.Bonten MJ, Bergmans DC, Speijer H, Stobberingh EE. 1999. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. Am J Respir Crit Care Med 160:1212–1219. doi: 10.1164/ajrccm.160.4.9809031. [DOI] [PubMed] [Google Scholar]
- 23.Kropec A, Huebner J, Riffel M, Bayer U, Benzing A, Geiger K, Daschner FD. 1993. Exogenous or endogenous reservoirs of nosocomial Pseudomonas aeruginosa and Staphylococcus aureus infections in a surgical intensive care unit. Intensive Care Med 19:161–165. doi: 10.1007/BF01720533. [DOI] [PubMed] [Google Scholar]
- 24.Blanc DS, Nahimana I, Petignat C, Wenger A, Bille J, Francioli P. 2004. Faucets as a reservoir of endemic Pseudomonas aeruginosa colonization/infections in intensive care units. Intensive Care Med 30:1964–1968. doi: 10.1007/s00134-004-2389-z. [DOI] [PubMed] [Google Scholar]
- 25.Grundmann H, Kropec A, Hartung D, Berner R, Daschner F. 1993. Pseudomonas aeruginosa in a neonatal intensive care unit: reservoirs and ecology of the nosocomial pathogen. J Infect Dis 168:943–947. doi: 10.1093/infdis/168.4.943. [DOI] [PubMed] [Google Scholar]
- 26.Rogues AM, Boulestreau H, Lasheras A, Boyer A, Gruson D, Merle C, Castaing Y, Bebear CM, Gachie JP. 2007. Contribution of tap water to patient colonisation with Pseudomonas aeruginosa in a medical intensive care unit. J Hosp Infect 67:72–78. doi: 10.1016/j.jhin.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Garvey MI, Bradley CW, Tracey J, Oppenheim B. 2016. Continued transmission of Pseudomonas aeruginosa from a wash hand basin tap in a critical care unit. J Hosp Infect 94:8–12. doi: 10.1016/j.jhin.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Blanc DS, Francioli P, Zanetti G. 2007. Molecular epidemiology of Pseudomonas aeruginosa in the intensive care units–a review. Open Microbiol J 1:8–11. doi: 10.2174/1874285800701010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkinham JO III, Hilborn ED, Arduino MJ, Pruden A, Edwards MA. 2015. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ Health Perspect 123:749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mena KD, Gerba CP. 2009. Risk assessment of Pseudomonas aeruginosa in water. Rev Environ Contam Toxicol 201:71–115. [DOI] [PubMed] [Google Scholar]
- 31.Kerr KG, Snelling AM. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect 73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Ferroni A, Nguyen L, Pron B, Quesne G, Brusset MC, Berche P. 1998. Outbreak of nosocomial urinary tract infections due to Pseudomonas aeruginosa in a paediatric surgical unit associated with tap-water contamination. J Hosp Infect 39:301–307. doi: 10.1016/S0195-6701(98)90295-X. [DOI] [PubMed] [Google Scholar]
- 33.Kossow A, Kampmeier S, Willems S, Berdel WE, Groll AH, Burckhardt B, Rossig C, Groth C, Idelevich EA, Kipp F, Mellmann A, Stelljes M. 2017. Control of multidrug-resistant Pseudomonas aeruginosa in allogeneic hematopoietic stem cell transplant recipients by a novel bundle including remodeling of sanitary and water supply systems. Clin Infect Dis 65:935–942. doi: 10.1093/cid/cix465. [DOI] [PubMed] [Google Scholar]
- 34.Bicking KC, Koirala S, Solomon B, Rosenberg J, Robinson BF, Neri A, Laufer Halpin A, Arduino MJ, Moulton-Meissner H, Noble-Wang J, Chea N, Gould CV. 2017. Pseudomonas aeruginosa outbreak in a neonatal intensive care unit attributed to hospital tap water. Infect Control Hosp Epidemiol 38:801–808. doi: 10.1017/ice.2017.87. [DOI] [PubMed] [Google Scholar]
- 35.van der Kooij D, Oranje JP, Hijnen WA. 1982. Growth of Pseudomonas aeruginosa in tap water in relation to utilization of substrates at concentrations of a few micrograms per liter. Appl Environ Microbiol 44:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Kooij D, Visser A, Hijnen WAM. 1982. Determining the concentration of easily assimilable organic-carbon in drinking-water. J Am Water Works Assoc 74:540–545. [Google Scholar]
- 37.Nissinen TK, Miettinen IT, Martikainen PJ, Vartiainen T. 2001. Molecular size distribution of natural organic matter in raw and drinking waters. Chemosphere 45:865–873. doi: 10.1016/S0045-6535(01)00103-5. [DOI] [PubMed] [Google Scholar]
- 38.LeChevallier MW, Schulz W, Lee RG. 1991. Bacterial nutrients in drinking water. Appl Environ Microbiol 57:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legnani P, Leoni E, Rapuano S, Turin D, Valenti C. 1999. Survival and growth of Pseudomonas aeruginosa in natural mineral water: a 5-year study. Int J Food Microbiol 53:153–158. doi: 10.1016/S0168-1605(99)00151-8. [DOI] [PubMed] [Google Scholar]
- 40.Lehtola MJ, Miettinen IT, Vartiainen T, Myllykangas T, Martikainen PJ. 2001. Microbially available organic carbon, phosphorus, and microbial growth in ozonated drinking water. Water Res 35:1635–1640. doi: 10.1016/S0043-1354(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 41.Liu W, Wu H, Wang Z, Ong SL, Hu JY, Ng WJ. 2002. Investigation of assimilable organic carbon (AOC) and bacterial regrowth in drinking water distribution system. Water Res 36:891–898. doi: 10.1016/S0043-1354(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 42.Polanska M, Huysman K, van Keer C. 2005. Investigation of assimilable organic carbon (AOC) in Flemish drinking water. Water Res 39:2259–2266. doi: 10.1016/j.watres.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Vital M, Hammes F, Egli T. 2008. Escherichia coli O157 can grow in natural freshwater at low carbon concentrations. Environ Microbiol 10:2387–2396. doi: 10.1111/j.1462-2920.2008.01664.x. [DOI] [PubMed] [Google Scholar]
- 44.Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol 2:103. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comolli JC, Donohue TJ. 2004. Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol Microbiol 51:1193–1203. doi: 10.1046/j.1365-2958.2003.03904.x. [DOI] [PubMed] [Google Scholar]
- 46.Wargo MJ. 2013. Choline catabolism to glycine betaine contributes to Pseudomonas aeruginosa survival during murine lung infection. PLoS One 8:e56850. doi: 10.1371/journal.pone.0056850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wargo MJ, Ho TC, Gross MJ, Whittaker LA, Hogan DA. 2009. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect Immun 77:1103–1111. doi: 10.1128/IAI.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendis N, Lin YR, Faucher SP. 2014. Comparison of virulence properties of Pseudomonas aeruginosa exposed to water and grown in rich broth. Can J Microbiol 60:777–781. doi: 10.1139/cjm-2014-0519. [DOI] [PubMed] [Google Scholar]
- 49.Kopf SH, Sessions AL, Cowley ES, Reyes C, Van Sambeek L, Hu Y, Orphan VJ, Kato R, Newman DK. 2016. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flynn JM, Phan C, Hunter RC. 2017. Genome-wide survey of Pseudomonas aeruginosa PA14 reveals a role for the glyoxylate pathway and extracellular proteases in the utilization of mucin. Infect Immun. doi: 10.1128/IAI.00182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ringen LM, Drake CH. 1952. A study of the incidence of Pseudomonas aeruginosa from various natural sources. J Bacteriol 64:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spilker T, Coenye T, Vandamme P, LiPuma JJ. 2004. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fitzsimmons LF, Flemer S Jr, Wurthmann AS, Deker PB, Sarkar IN, Wargo MJ. 2011. Small-molecule inhibition of choline catabolism in Pseudomonas aeruginosa and other aerobic choline-catabolizing bacteria. Appl Environ Microbiol 77:4383–4389. doi: 10.1128/AEM.00504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willsey GG, Wargo MJ. 2016. Sarcosine catabolism in Pseudomonas aeruginosa is transcriptionally regulated by SouR. J Bacteriol 198:301–310. doi: 10.1128/JB.00739-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.