ABSTRACT
Besides metabolic pathways and regulatory networks, transport systems are also pivotal for cellular metabolism and hyperproduction of biochemicals using microbial cell factories. The identification and characterization of transporters are therefore of great significance for the understanding and engineering of transport reactions. Herein, a novel l-glutamate exporter, MscCG2, which exists extensively in Corynebacterium glutamicum strains but is distinct from the only known l-glutamate exporter, MscCG, was discovered in an industrial l-glutamate-producing C. glutamicum strain. MscCG2 was predicted to possess three transmembrane helices in the N-terminal region and located in the cytoplasmic membrane, which are typical structural characteristics of the mechanosensitive channel of small conductance. MscCG2 has a low amino acid sequence identity (23%) to MscCG and evolved separately from MscCG with four transmembrane helices. Despite the considerable differences between MscCG2 and MscCG in sequence and structure, gene deletion and complementation confirmed that MscCG2 also functioned as an l-glutamate exporter and an osmotic safety valve in C. glutamicum. Besides, transcriptional analysis showed that MscCG2 and MscCG genes were transcribed in similar patterns and not induced by l-glutamate-producing conditions. It was also demonstrated that MscCG2-mediated l-glutamate excretion was activated by biotin limitation or penicillin treatment and that constitutive l-glutamate excretion was triggered by a gain-of-function mutation of MscCG2 (A151V). Discovery of MscCG2 will enrich the understanding of bacterial amino acid transport and provide additional targets for exporter engineering.
IMPORTANCE The exchange of matter, energy, and information with surroundings is fundamental for cellular metabolism. Therefore, studying transport systems that are essential for these processes is of great significance. Besides, transport systems of bacterial cells are usually related to product excretion as well as product reuptake, making transporter engineering a useful strategy for strain improvement. The significance of our research is in identifying and characterizing a novel l-glutamate exporter from the industrial workhorse Corynebacterium glutamicum, which will enrich the understanding of l-glutamate excretion and provide a new target for studying bacterial amino acid transport and engineering transport reactions.
KEYWORDS: l-glutamate exporter, mechanosensitive channel, MscCG2, Corynebacterium glutamicum, inducing treatment
INTRODUCTION
Corynebacterium glutamicum, a nonpathogenic Gram-positive bacterium, was originally isolated as an l-glutamate-producing strain and has been engineered to serve as an industrial workhorse for production of various amino acids (1). Within the amino acid market, l-glutamate, which is widely used as a flavor enhancer, represents the largest product segment, with a giant market size of approximately 2.5 million tons per year (2). Amino acid transport systems are increasingly attracting attention for improving the industrial amino acid-producing strains (3–5). In the past few years, a number of amino acid exporters were identified in C. glutamicum, such as LysE, ThrE, and BrnFE, which participate in transporting l-lysine, l-threonine, l-serine, l-isoleucine, l-leucine, l-valine, and l-methionine (6–9). The nature of the l-glutamate exporter has long been unclear; recently, however, a mechanosensitive channel of small conductance (MscS)-like protein (MscCG, also known as NCgl1221) was proven to contribute to l-glutamate excretion in C. glutamicum (10).
The MscS-like channel superfamily is present in diverse cell-walled organisms, and its members display physiological functions in bacterial osmoregulation, Ca2+ regulation, cell division, and amino acid export (11). The mscCG gene of C. glutamicum encodes an MscS homolog, and its disruption causes an approximately 80% decrease in l-glutamate excretion compared to that of the wild-type strain (10, 12). It has been proposed that inducing treatments on C. glutamicum, such as biotin limitation, Tween 40 addition, and penicillin addition, cause an alteration in membrane tension, and MscCG is then activated, leading to l-glutamate excretion (10, 13). MscCG has an N-terminal region homologous to that of well-characterized Escherichia coli MscS and an additional C-terminal region unlike any known MscS-like proteins (10, 14). Previous studies suggest that the N-terminal domain containing three transmembrane (TM) helices necessary and sufficient for l-glutamate excretion and the C-terminal domain containing a fourth TM helix have a regulatory role in the release of intracellular l-glutamate (12, 15–17). Although the physiological importance of MscCG in l-glutamate excretion is acknowledged, MscCG gene-disrupted C. glutamicum strains still produced significant amounts of l-glutamate, implying the existence of other long-elusive l-glutamate exporters (10, 12).
In this study, genomic analysis of C. glutamicum Z188, a strain for industrial l-glutamate production in China, revealed an mscCG2 gene that existed in many but not all C. glutamicum strains. The localization and physiological functions of MscCG2 were then investigated, demonstrating that MscCG2 was confined to the cytoplasmic membrane and involved in l-glutamate excretion and osmoregulation. MscCG2 represents a novel l-glutamate exporter that is different from MscCG.
RESULTS
Demonstration of mscCG2 in C. glutamicum by genomic and transcriptional analyses.
C. glutamicum Z188 is an industrial strain that has been widely used in l-glutamate production in China. Biotin limitation or an excess of biotin with penicillin addition is required to induce l-glutamate excretion. C. glutamicum Z188 has been sequenced, and the draft genome sequence has been deposited at DDBJ/EMBL/GenBank under accession number NZ_AKXP00000000. Comparative analysis of its genome sequence across the available C. glutamicum genome sequences revealed an MscS-like channel (named MscCG2) that existed in many but not all C. glutamicum strains (Fig. 1). According to the prediction of InterProScan software (18), MscCG2 possesses the typical structural characteristics of the MscS family.
FIG 1.
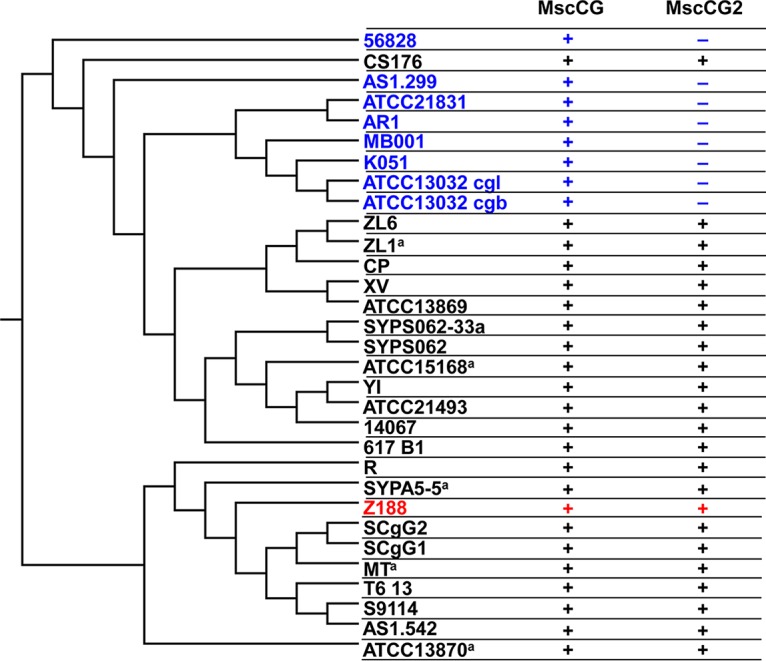
Phylogenetic analysis of C. glutamicum strains and l-glutamate exporters. The dendrogram was calculated by the CVTree Web interface using a composition vector approach (28). The tree was constructed using the MEGA 7 program (34) and displayed only topology. Plus and minus represent the existence and absence of a gene in a C. glutamicum genome sequence, respectively. Strains highlighted in blue are those that possess only MscCG. C. glutamicum Z188 is highlighted in red. Strains followed by a superscript “a” were previously classified as Brevibacterium flavum (ZL1 and ATCC 15168), Corynebacterium crenatum (SYPA5-5 and MT), and Corynebacterium acetoacidophilum (ATCC 13870). However, a recent average nucleotide identity analysis of their genome sequences suggests that they should be reclassified as members of the species C. glutamicum (37). Therefore, these strains were also analyzed here.
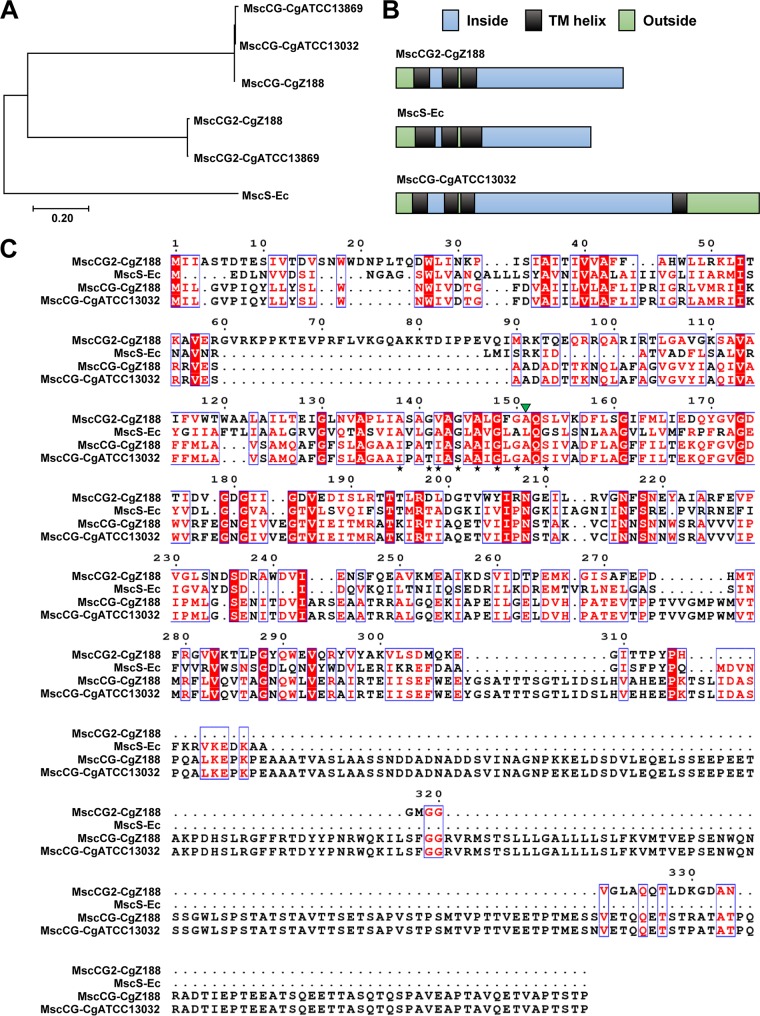
C. glutamicum Z188 and ATCC 13869 are two of those strains that possess both mscCG2 and mscCG genes, while C. glutamicum ATCC 13032 possesses only mscCG. The mscCG2 gene of C. glutamicum Z188 is 1,005 bp long with a deduced amino acid sequence that shares 20% identity with MscS of E. coli, 23% identity with MscCG of C. glutamicum strains Z188, ATCC 13032, and ATCC 13869, and 99% identity with MscCG2 of C. glutamicum ATCC 13869. Phylogenetic analysis clearly shows that MscCG2 and MscCG belong to different branches of the MscS family, suggesting that MscCG2 has evolved separately from MscCG (Fig. 2A). Although MscCG2 does not have significant sequence similarities with MscCG and E. coli MscS, all of these proteins share similar secondary structures in the N-terminal region (Fig. 2B) (10, 12). Their N-terminal regions all consist of three TM helices with the N terminus outside the cytoplasm (Fig. 2B).
FIG 2.
(A) Phylogenetic analysis of amino acid sequences of MscS-like proteins. The tree was constructed with the MEGA 7 program using the neighbor-joining method (29). CgATCC13869, CgATCC13032, and CgZ188 represent C. glutamicum strains ATCC 13869, ATCC 13032, and Z188, respectively. Ec, E. coli. (B) Membrane topology of MscS-like proteins. Membrane topology of MscCG2 from C. glutamicum Z188 was predicted by TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM) (38). Membrane topologies of MscS from E. coli and MscCG from C. glutamicum ATCC 13032 were proposed based on previous studies (10, 12, 14). (C) Multiple amino acid sequence alignment of MscS-like proteins (numbering relative to MscCG2 from C. glutamicum Z188). The alignment was conducted with the ESPript 3 program (http://espript.ibcp.fr) (39). Black stars represent the conserved l-alanine and glycine present in the third TM helix of E. coli MscS. The green triangle represents the residue involved in a gain-of-function mutation in gating of MscCG (20).
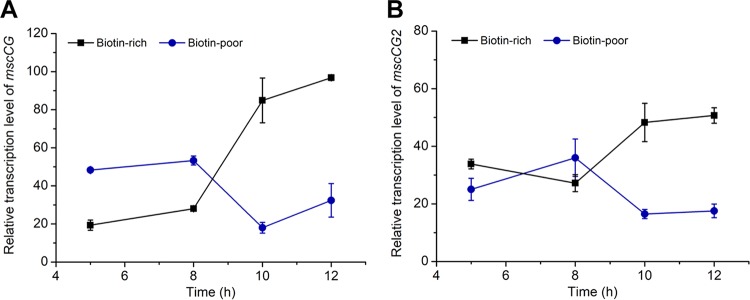
To further verify whether mscCG2 was functionally expressed in C. glutamicum Z188, we analyzed the transcription levels of mscCG2 as well as mscCG under l-glutamate-nonproducing and -producing conditions by using quantitative PCR (qPCR). mscCG and mscCG2 were transcribed under both biotin-rich and biotin-poor conditions. Notably, the transcription levels of mscCG and mscCG2 displayed similar trends (Fig. 3). All the results demonstrate that mscCG2 is transcribed in C. glutamicum Z188.
FIG 3.
Transcription levels of mscCG (A) and mscCG2 (B) of C. glutamicum Z188 under l-glutamate-nonproducing and -producing conditions. C. glutamicum Z188 was cultivated in biotin-rich or biotin-poor fermentation media, and cells were collected during the first 12 h. Three parallel experiments were performed. Total RNA was extracted, treated with DNase I, and used for cDNA synthesis. qPCR was conducted using cDNA as a template and the primers listed in Table 4. Error bars indicate standard deviations of results from three parallel experiments.
Localization of MscCG2 in C. glutamicum cells.
It has been reported that both MscCG and E. coli MscS locate in the cytoplasmic membrane (10, 12, 19). To investigate the localization of MscCG2, MscCG2 fused with enhanced green fluorescent protein (EGFP) in its C terminus was constructed and expressed in C. glutamicum Z188, resulting in strain Z188(mscCG2-egfp). Strain Z188(pTRCmob) harboring an empty vector and strain Z188(egfp) expressing EGFP were used as controls. Under fluorescence, EGFP signals were hardly detectable for strain Z188(pTRCmob). In strain Z188(egfp), EGFP proteins that were distributed in the cytoplasm were observed. Regarding strain Z188(mscCG2-egfp), the MscCG2-EGFP fusion protein located in the periphery of the cells, suggesting that MscCG2 is associated with the cytoplasmic membrane (Fig. 4).
FIG 4.
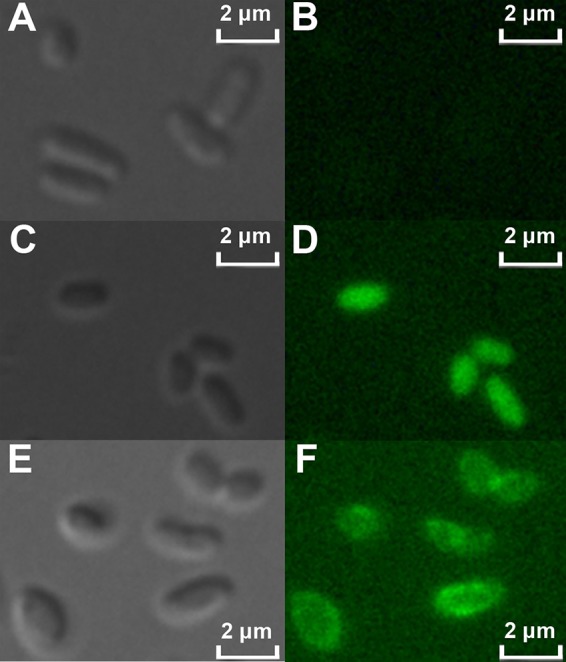
Localization of MscCG2-EGFP fusion protein in C. glutamicum Z188 under a confocal laser scanning microscope. (A) Strain Z188(pTRCmob) under visible light; (B) strain Z188(pTRCmob) under fluorescence (488-nm laser illumination); (C) strain Z188(egfp) under visible light; (D) strain Z188(egfp) under fluorescence; (E) strain Z188(mscCG2-egfp) under visible light; (F) strain Z188(mscCG2-egfp) under fluorescence. All of the images were generated under the same intensities of excitation.
Effects of MscCG2 on l-glutamate excretion.
Since MscCG2 is functionally expressed in l-glutamate-producing strain Z188 and shares a similar secondary structure with the l-glutamate exporter MscCG, it may play a part in l-glutamate excretion. mscCG2 gene-disrupted mutant Z188ΔmscCG2 was then constructed. Single deletion of mscCG2 did not lead to a significant decrease in l-glutamate excretion in biotin-poor medium (Fig. 5A; see Table S1 in the supplemental material), which was possibly due to the existence of mscCG. To eliminate the influence of mscCG on l-glutamate excretion, mscCG-deleted mutants Z188ΔmscCG and Z188ΔmscCG2ΔmscCG were constructed. Z188ΔmscCG still produced significant amounts of l-glutamate. However, l-glutamate excretion by the double mutant Z188ΔmscCG2ΔmscCG was almost eliminated, demonstrating the noticeable role of mscCG2 in l-glutamate excretion (Fig. 5A). To confirm its function, mscCG2 was expressed in Z188ΔmscCG2ΔmscCG using the pTRCmob vector, generating Z188ΔmscCG2ΔmscCG(mscCG2). mscCG was also complemented in Z188ΔmscCG2ΔmscCG as a control, generating Z188ΔmscCG2ΔmscCG(mscCG). As expected, complementation of mscCG2 or mscCG caused l-glutamate excretion to be resumed (Fig. 5A).
FIG 5.
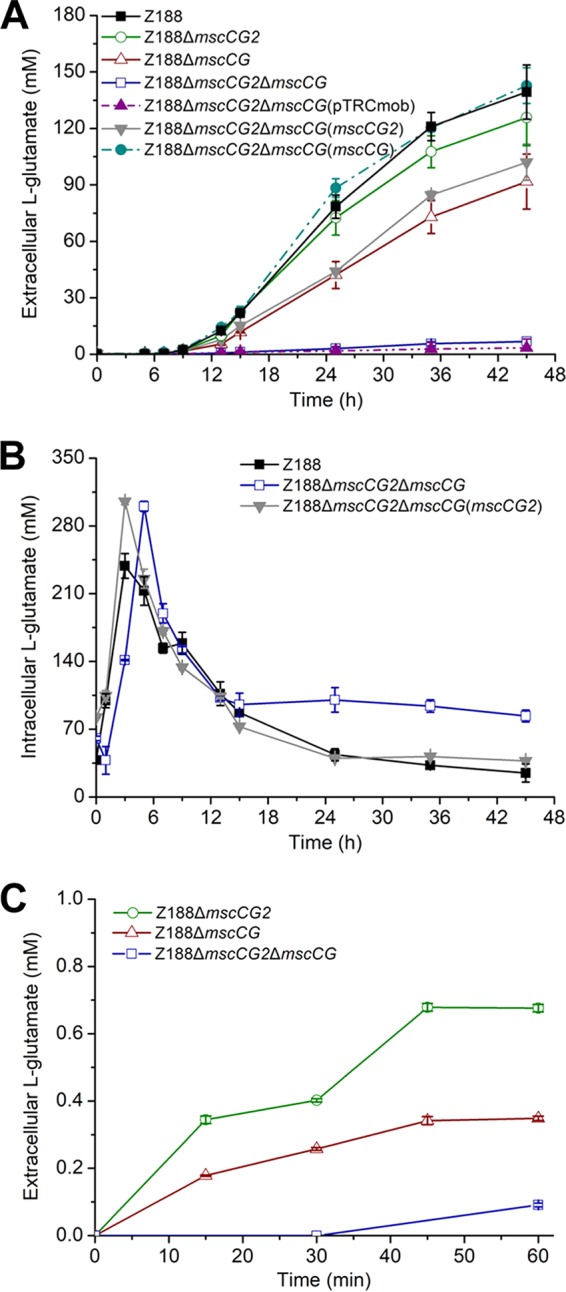
Export of l-glutamate by C. glutamicum Z188 and its derivatives. (A) Extracellular l-glutamate concentrations of C. glutamicum Z188 and its derivatives. Strains were cultivated in biotin-poor fermentation medium at 30°C and with shaking at 220 rpm. (B) Intracellular l-glutamate concentrations of C. glutamicum Z188 and its derivatives. Strains were cultivated in biotin-poor fermentation medium at 30°C and with shaking at 220 rpm. (C) l-Glutamate export rates of MscCG and MscCG2. After being washed twice with ice-cold CGXII medium, the pregrown cells in fermentation medium were resuspended in prewarmed CGXII medium (30°C) containing 2 mM Glu-Glu to an OD600 of 10.0. The cells were incubated at 30°C and with shaking at 220 rpm. Error bars indicate standard deviations of results from three parallel experiments.
Detection of an intracellular l-glutamate pool also provided evidence that MscCG2 was involved in l-glutamate excretion (Fig. 5B). The intracellular l-glutamate levels of the wild-type strain Z188 and its mutants all were high during the nonproducing period. It was noticed that the maximum intracellular l-glutamate levels of Z188ΔmscCG2ΔmscCG and Z188ΔmscCG2ΔmscCG(mscCG2) were higher than that of Z188. We speculated that the lack of mscCG or mscCG2 might be the cause for this phenomenon. During the producing period, the intracellular l-glutamate levels of strain Z188 and its mutants all decreased gradually. However, the intracellular l-glutamate level of Z188ΔmscCG2ΔmscCG was 2- to 4-fold higher than the levels of Z188 and Z188ΔmscCG2ΔmscCG(mscCG2) (Fig. 5B), suggesting that in the absence of MscCG, the cells still can export l-glutamate efficiently because MscCG2 functioned as an l-glutamate exporter.
To compare the export rates of l-glutamate by MscCG and MscCG2, a short-term peptide uptake and amino acid export experiment was conducted. Addition of 2 mM Glu-Glu in the cultures of Z188ΔmscCG and Z188ΔmscCG2 resulted in export of l-glutamate at different rates. Strain Z188ΔmscCG2 (still harboring MscCG) excreted l-glutamate at a higher rate than strain Z188ΔmscCG (still harboring MscCG2) (Fig. 5C), suggesting that MscCG possesses a higher l-glutamate export activity than MscCG2. The results were consistent with the fact that strain Z188ΔmscCG2ΔmscCG(mscCG) excreted l-glutamate into the culture at a higher rate than Z188ΔmscCG2ΔmscCG(mscCG2) during the fermentation (Fig. 5A).
Activation of MscCG2 by biotin limitation and penicillin addition.
In a model of induction of l-glutamate excretion in C. glutamicum, treatments such as biotin limitation and penicillin addition alter membrane tension and lead to l-glutamate excretion by activating MscCG (10). Since C. glutamicum Z188 produces a considerable amount of l-glutamate under biotin-limited conditions, it is possible that l-glutamate excretion through MscCG2 also requires activation. To confirm our hypothesis, C. glutamicum Z188 and its derivatives were tested for l-glutamate excretion under different conditions. As expected, under the condition of biotin excess, wild-type strain Z188, Z188ΔmscCG2, and Z188ΔmscCG2ΔmscCG(mscCG2) produced little l-glutamate (Fig. 6A). When treated with biotin limitation or penicillin addition, these strains produced large amounts of l-glutamate, suggesting that MscCG2 needs to be activated by the alteration of membrane tension for l-glutamate excretion, as is the case for MscCG. Regarding strain Z188ΔmscCG2ΔmscCG, little but detectable extracellular l-glutamate was produced under either biotin excess or biotin-limited conditions. Addition of penicillin did not induce Z188ΔmscCG2ΔmscCG to produce l-glutamate markedly (Fig. 6A). On one hand, the results further proved that both MscCG2 and MscCG played crucial roles in l-glutamate excretion. On the other hand, some other exporters may nonspecifically mediate l-glutamate excretion in the absence of MscCG2 and MscCG.
FIG 6.
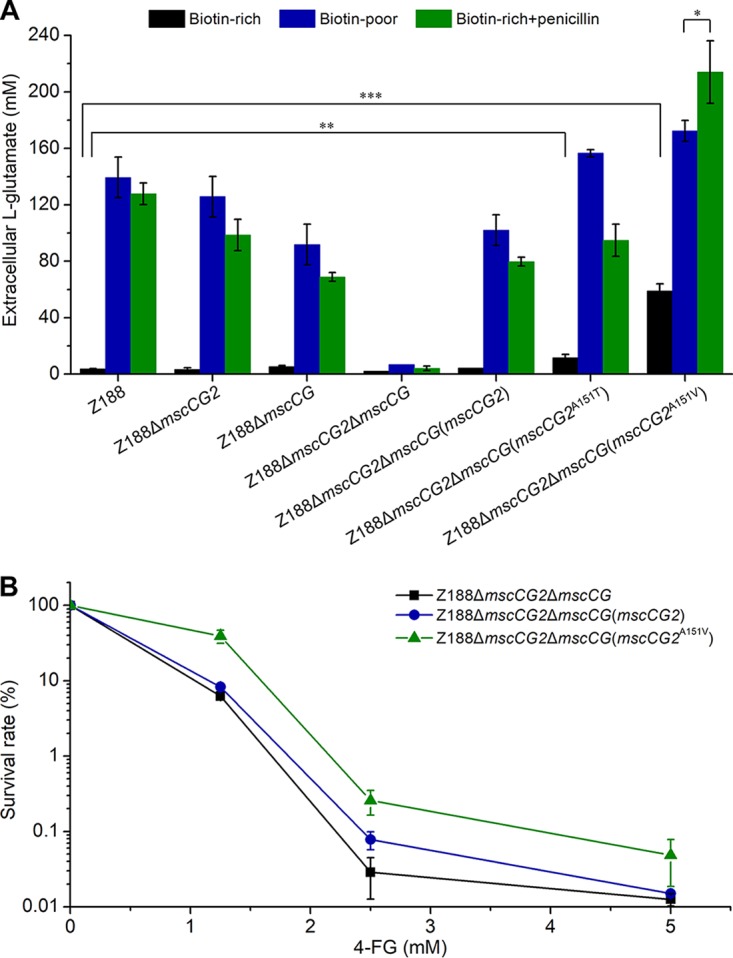
Effects of inducing treatments and gain-of-function mutations of MscCG2 on excretion of l-glutamate and its analog. (A) Effects of inducing treatments on l-glutamate excretion of C. glutamicum Z188 and its derivatives. When biotin-rich fermentation medium was used, 60 μg/liter biotin was added. For penicillin treatments, cells were cultured in biotin-rich fermentation medium and 60 U/ml penicillin was added when the OD600 reached about 12. Extracellular l-glutamate concentrations were analyzed after 45 h of cultivation. Asterisks indicate significant changes in extracellular l-glutamate concentration based on a comparison between different strains. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Student's two-tailed t test). (B) 4-FG resistance of MscCG2 mutants. C. glutamicum Z188 derivatives were inoculated into CGXII medium with different concentrations of 4-FG and cultivated at 30°C and with shaking at 220 rpm. When the OD600 of each strain without 4-FG reached 1.0, the cultures were diluted and plated on solid LBG medium. The viability of the diluted cells was determined by counting the number of colonies on the plates. Error bars indicate standard deviations of results from three parallel experiments.
Constitutive l-glutamate excretion by gain-of-function mutation of MscCG2.
Both MscCG- and MscCG2-mediated l-glutamate excretion needs activation by the alteration of membrane tension. However, previous studies demonstrated that A111V and A111T mutations in the third TM helix of MscCG may cause a structural transformation and confer constitutive l-glutamate leakage phenotypes even under biotin-rich conditions, so-called gain-of-function mutations (10, 20). Multiple amino acid sequence alignment shows that the l-alanine residue at position 111 of MscCG (A111), purported to be involved in a gain-of-function mutation, is conserved in MscCG2 (A151) (Fig. 2C). Therefore, two MscCG2 mutants (MscCG2A151T and MscCG2A151V) were constructed and tested for their roles in l-glutamate excretion in strain Z188ΔmscCG2ΔmscCG. In the presence of excess biotin, different degrees of l-glutamate leakage were observed for MscCG2A151T and MscCG2A151V. Interestingly, MscCG2A151V not only exported l-glutamate constitutively but also resulted in a higher extracellular l-glutamate level under penicillin addition conditions (Fig. 6A). It has been reported that gain-of-function mutations of MscCG render cells resistant to the l-glutamate analog, 4-fluoroglutamate (4-FG) (10). We therefore examined the effect of the MscCG2(A151V) mutation on resistance to 4-FG. In the presence of 1.25 mM 4-FG, the cell survival rate of strain Z188ΔmscCG2ΔmscCG that lacks the two MscS-like channels was only 6.3% ± 0.5%. Expression of the wild-type MscCG2 did not largely improve the resistance to 4-FG (cell survival rate of 8.3% ± 0.9%). However, the cell survival rate of strain Z188ΔmscCG2ΔmscCG(mscCG2A151V) reached 39.0% ± 7.5% (Fig. 6B). The results verify the capability of MscCG2 to excrete l-glutamate and its analog, 4-FG.
Application of MscCG2 in l-glutamate excretion of C. glutamicum ATCC 13032.
Since MscCG2 participates in l-glutamate excretion of C. glutamicum Z188, it is speculated that MscCG2 can also serve as an l-glutamate exporter in C. glutamicum strains that do not possess MscCG2, such as strain ATCC 13032. To test our hypothesis, mscCG2 and mscCG from C. glutamicum Z188 were expressed in an mscCG-deleted mutant of C. glutamicum ATCC 13032 (ATCC 13032ΔmscCG) separately. The resultant strains, ATCC 13032ΔmscCG(mscCG2) and ATCC 13032ΔmscCG(mscCG), produced levels of l-glutamate similar to those of wild-type C. glutamicum ATCC 13032 in biotin limitation (Table 1). The results suggest that both MscCG2 and MscCG from strain Z188 are usable for l-glutamate excretion in C. glutamicum ATCC 13032. To test whether overexpression of l-glutamate exporters can improve l-glutamate production, mscCG2 and mscCG from strain Z188 were overexpressed in wild-type strain ATCC 13032, resulting in strains ATCC 13032(mscCG2) and ATCC 13032(mscCG), respectively. However, no significant increase was observed when mscCG or mscCG2 was overexpressed (Table 1).
TABLE 1.
Effects of mscCG and mscCG2 on l-glutamate excretion in C. glutamicum ATCC 13032a
| Strain | l-Glutamate concn (mM) | Dry cell wt (g/liter) |
|---|---|---|
| ATCC 13032 | 44.2 ± 4.8 | 8.3 ± 0.6 |
| ATCC 13032(pTRCmob) | 39.2 ± 6.9 | 7.1 ± 0.3 |
| ATCC 13032(mscCG) | 42.4 ± 1.4 | 7.1 ± 0.1 |
| ATCC 13032(mscCG2) | 39.2 ± 1.0 | 6.8 ± 0.2 |
| ATCC 13032ΔmscCG(pTRCmob) | 6.8 ± 0.0*** | 7.9 ± 0.2 |
| ATCC 13032ΔmscCG(mscCG) | 42.8 ± 3.7 | 8.1 ± 0.5 |
| ATCC 13032ΔmscCG(mscCG2) | 37.4 ± 4.8 | 8.3 ± 0.0 |
Samples were picked and analyzed after 45 h of cultivation in shake flasks. Data represent averages and standard deviations of results of three independent cultures. Asterisks indicate significant changes in extracellular l-glutamate concentration based on a comparison between the engineered strain and wild-type strain ATCC 13032. ***, P ≤ 0.001 (Student's two-tailed t test).
Roles of MscCG2 in osmoregulation.
MscCG has been proven to be a mechanosensitive channel involved in the fine-tuning of osmotic adaptation (21, 22). To investigate the role of MscCG2 in osmoregulation, we determined the cell survival rates of C. glutamicum Z188 and its mutants upon osmotic downshock. When mscCG or mscCG2 was deleted individually in strain Z188, only the mscCG-deleted strain Z188ΔmscCG showed a statistically significant decrease in cell survival rate upon osmotic downshock, verifying the important role of MscCG in osmoregulation (Fig. 7). Deletion of mscCG2 in strain Z188ΔmscCG further decreased survival significantly, suggesting that MscCG2 also functions as a mechanosensitive channel but plays a minor physiological role in osmoregulation.
FIG 7.
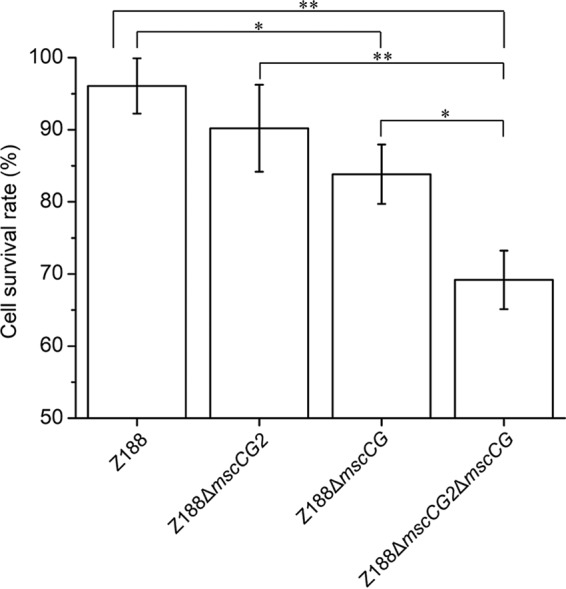
Cell survival rates of C. glutamicum Z188 derivatives upon osmotic downshock. Cells cultivated in a high-osmolarity LBG medium containing 1.17 M NaCl (2,350 mosmol) were transferred into normal LBG medium containing 0.17 M NaCl (410 mosmol) or high-osmolarity LBG medium containing 1.17 M NaCl (2,350 mosmol). The cultures were incubated at 30°C and with shaking at 220 rpm for another 40 min, diluted using isosmotic medium, and plated on solid LBG medium. The viability of the diluted cells was determined by counting the number of colonies on the plates. Stars indicate significant changes in cell survival rates based on a comparison between different strains. *, P ≤ 0.05; **, P ≤ 0.01 (Student's two-tailed t test). Error bars indicate standard deviations of results from three parallel experiments.
DISCUSSION
To transform microorganisms into efficient cell factories, manipulation of metabolic pathways and regulatory networks is a commonly used strategy (23, 24). Recently, the significance of a bacterial transport system for the bioproduction of various chemicals, especially amino acids, has become increasingly evident. Engineering of transport reactions is also becoming an attractive strategy for strain improvement (13, 25). Therefore, identification and characterization of transporters are important not only for understanding the mechanism of hyperproduction of specific chemicals but also for engineering the producers. Previous studies suggest the existence of more than one l-glutamate exporter in C. glutamicum, whereas MscCG is the only one verified to date (10, 12). In this study, MscCG2 of C. glutamicum Z188 was identified as a cytoplasmic membrane protein and another functional l-glutamate exporter. The bioinformatic analysis reveals the extensive coexistence of MscCG and MscCG2 in C. glutamicum strains (Fig. 1), which is consistent with the discovery that bacteria usually possess multiple channel homologs with overlapping functions (26). Although both MscCG and MscCG2 belong to the MscS-like channel superfamily and have similar biological functions in l-glutamate excretion, MscCG2 has a low amino acid sequence identity (23%) to MscCG and seems to evolve separately from MscCG (Fig. 2A). MscCG was proven to have three TM helices in the N-terminal region and an additional fourth TM helix in the C-terminal region with both N and C termini outside the cytoplasm (10, 12). However, MscCG2 does not have the fourth TM helix in the C-terminal region, and its C terminus locates inside the cytoplasm (Fig. 2B). All the evidence suggests that MscCG2 is a novel l-glutamate exporter that is distinct from MscCG. Interestingly, C. glutamicum ATCC 13032 does not harbor mscCG2, but its mscCG-disrupted mutant still shows significant l-glutamate excretion (Table 1) (12), implying the possible existence of other l-glutamate exporters.
Despite the differences in sequence and structure, some interesting similarities between MscCG and MscCG2 were also observed. First, both MscCG and MscCG2 need to be activated for l-glutamate excretion by the alteration of membrane tension caused by biotin limitation or penicillin addition. Moreover, gain-of-function mutations on the third TM helix of both MscCG and MscCG2 could confer constitutive export of l-glutamate and its analog, 4-FG (Fig. 6) (10, 20). Second, mscCG and mscCG2 genes were transcribed in similar patterns under l-glutamate-nonproducing and -producing conditions. No induction phenomenon caused by l-glutamate-producing conditions was observed (Fig. 3). Variation in the mscCG and mscCG2 transcription levels during the first 12 h might be due to different cell growth and l-glutamate accumulation statuses. Both MscCG and MscCG2 are MscS-like channels, and osmoregulation is supposed to be their primary function. The present and previous studies also confirmed their roles in osmoregulation (Fig. 7) (21, 22). However, our results suggest that MscCG plays a more important part than MscCG2. This may provide an explanation for why bacteria of the same species differ in equipment with such a fundamental protein as an MscS-like channel; that is, all the C. glutamicum strains harbor MscCG, while only some harbor both MscCG and MscCG2.
MscS-like channels usually have multiple biological functions (11). MscCG has been suggested to excrete not only l-glutamate but also l-aspartic acid and possibly l-phenylalanine (10, 27). E. coli MscS not only serves as an osmotic safety valve in E. coli but also mediates l-glutamate excretion when expressed in C. glutamicum (12). Therefore, it will not be surprising if MscCG2 has other undiscovered functions. Further study of the functional diversity of MscCG2 will help to elucidate its physiological roles and unleash its potential of exporter engineering.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 2. E. coli DH5α was used for general cloning and cultivated aerobically at 37°C in Luria-Bertani (LB) broth. Kanamycin (Km; 50 μg/ml) was added to LB broth as required. C. glutamicum strains were cultivated aerobically at 30°C in LBG medium, seed medium, or fermentation medium. The LBG medium is LB medium supplemented with 5 g/liter glucose. The seed medium contains the following, per liter: 25 g glucose, 15 g corn steep liquor, 0.6 g MgSO4, 2.0 g K2HPO4, 0.02 g MnSO4, 0.01 g FeSO4, 10 g (NH4)2SO4, and 0.2 mg thiamine hydrochloride. The fermentation medium contains the following, per liter: 120 g glucose, 4 g corn steep liquor, 0.6 g MgSO4, 2.0 g K2HPO4, 0.02 g MnSO4, 0.01 g FeSO4, 10 g (NH4)2SO4, and 0.2 mg thiamine hydrochloride. When biotin-rich fermentation medium was used, 60 μg/liter biotin was added. Km (25 μg/ml) was added to the medium as required.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | General cloning host | TaKaRa |
| C. glutamicum | ||
| Z188 | l-Glutamate-producing strain | Lab collection |
| ATCC 13032 | Wild-type strain | ATCC |
| Z188(pTRCmob) | Z188 derivative harboring pTRCmob | This study |
| Z188(egfp) | Z188 derivative harboring pTRCmob-egfp | This study |
| Z188(mscCG2-egfp) | Z188 derivative harboring pTRCmob-mscCG2-egfp | This study |
| Z188ΔmscCG2 | Z188 derivative with its mscCG2 deleted | This study |
| Z188ΔmscCG | Z188 derivative with its mscCG deleted | This study |
| Z188ΔmscCG2ΔmscCG | Z188 derivative with its mscCG2 and mscCG deleted | This study |
| Z188ΔmscCG2ΔmscCG(pTRCmob) | Z188ΔmscCG2ΔmscCG derivative harboring pTRCmob | This study |
| Z188ΔmscCG2ΔmscCG(mscCG) | Z188ΔmscCG2ΔmscCG derivative harboring pTRCmob-mscCG | This study |
| Z188ΔmscCG2ΔmscCG(mscCG2) | Z188ΔmscCG2ΔmscCG derivative harboring pTRCmob-mscCG2 | This study |
| ATCC 13032(pTRCmob) | ATCC 13032 derivative harboring pTRCmob | This study |
| ATCC 13032(mscCG) | ATCC 13032 derivative harboring pTRCmob-mscCG | This study |
| ATCC 13032(mscCG2) | ATCC 13032 derivative harboring pTRCmob-mscCG2 | This study |
| ATCC 13032ΔmscCG | ATCC 13032 derivative with its mscCG deleted | This study |
| ATCC 13032ΔmscCG(pTRCmob) | ATCC 13032ΔmscCG derivative harboring pTRCmob | This study |
| ATCC 13032ΔmscCG(mscCG2) | ATCC 13032ΔmscCG derivative harboring pTRCmob-mscCG2 | This study |
| ATCC 13032ΔmscCG(mscCG) | ATCC 13032ΔmscCG derivative harboring pTRCmob-mscCG | This study |
| Plasmids | ||
| pTRCmob | E. coli-C. glutamicum shuttle expression vector with a derepressed Ptrc promoter, Kmr | 40 |
| pK18mobsacB | Gene deletion/insertion vector derived from pK18; mob sacB Kmr | 30 |
| pET21a-egfp | pET21a derivative, carries an enhanced green fluorescent protein-encoding gene, Ampr | 32 |
| pTRCmob-egfp | pTRCmob derivative, carries egfp | This study |
| pTRCmob-mscCG2-egfp | pTRCmob derivative, carries mscCG2-egfp | This study |
| pK18mobsacB-ΔmscCG2 | pK18mobsacB derivative, carries a mutant allele of mscCG2 from Z188 | This study |
| pK18mobsacB-ΔmscCG | pK18mobsacB derivative, carries a mutant allele of mscCG from Z188 | This study |
| pTRCmob-mscCG2 | pTRCmob derivative, carries mscCG2 from Z188 | This study |
| pTRCmob-mscCG | pTRCmob derivative, carries mscCG from Z188 | This study |
| pTRCmob-mscCG2A151V | pTRCmob derivative, carries a mutant of mscCG2 from Z188 | This study |
| pTRCmob-mscCG2A151T | pTRCmob derivative, carries a mutant of mscCG2 from Z188 (mscCG2A151T) | This study |
Kmr and Ampr, resistance to kanamycin and ampicillin, respectively.
Phylogenetic analysis.
The dendrogram of C. glutamicum strains was calculated by the whole-genome-based Web server CVTree using a composition vector approach (28). Genome sequences of 31 strains shown in Table 3 were uploaded to http://tlife.fudan.edu.cn/cvtree3/ and used for analysis. The phylogenetic tree was constructed using the MEGA 7 program (29).
TABLE 3.
Descriptions of strains analyzed in Fig. 1
| Strain | Synonyma | Description | Ancestorb | ENA database accession no. |
|---|---|---|---|---|
| ATCC 13032 cgl | C. glutamicum type strain (Kyowa Hakko) | GCA_000011325.1 | ||
| ATCC 13032 cgb | C. glutamicum type strain (Bielefeld) | GCA_000196335.1 | ||
| K051 | Substrain of ATCC 13032 | ATCC 13032 | GCA_000382905.1 | |
| MB001 | Prophage-free variant of ATCC 13032 with a 6% reduced genome | ATCC 13032 | GCA_000445015.1 | |
| ATCC 13869 | B. lactofermentum | Wild-type B. lactofermentum | GCA_001687645.1 | |
| ATCC 13870 | C. acetoacidophilum | Wild-type C. acetoacidophilum | GCA_001912755.2 | |
| ATCC 14067 | B. flavum | Wild-type B. flavum | GCA_002243555.1 | |
| ATCC 21493 | B. flavum | Derivative of ATCC 14067 (SIIM B234), producing arginine | ATCC 14067 | GCA_001912805.1 |
| SYPS-062 | l-Serine-overproducing strain | GCA_000819915.1 | ||
| SYPS-062-33a | l-Serine-overproducing strain, derived from SYPS-062 | GCA_000934815.1 | ||
| ATCC 15168 | B. flavum | l-Isoleucine-producing strain | GCA_000987865.1 | |
| R | C. glutamicum isolated in Japan from a meadow soil sample | GCA_000010225.1 | ||
| AS1.299 | C. pekinense | Wild-type C. pekinense, producing l-glutamate (CICC 10119, SIIM B3) | GCA_001912725.1 | |
| 617 B1 | A l-glutamate-producing strain previously used in China (CICC 10117, SIIM B1) | GCA_001912785.1 | ||
| T6-13 | B. tianjinese | Wild-type B. tianjinese (CICC 20182, SIIM B226) | GCA_001912155.1 | |
| S9114 | A strain for industrial production of l-glutamate | T6-13 | GCA_000224315.2 | |
| AS1.542 | C. crenatum | Wild-type C. crenatum (CICC10124, SIIM B6) | GCA_001912735.1 | |
| MT | C. crenatum | A mutant of AS1.542, producing l-arginine | AS1.542 | GCA_000380545.1 |
| SYPA5-5 | C. crenatum | A mutant of AS1.542, producing l-arginine | AS1.542 | GCA_000732145.1 |
| ATCC 21831(AR0) | l-Arginine-producing strain | GCA_000742715.1 | ||
| AR1 | l-Arginine-producing strain, derived from ATCC 21831 | GCA_000742735.1 | ||
| CP | l-Leucine-producing strain | GCA_001447865.2 | ||
| 56828 | GCA_001518935.2 | |||
| YI | l-Isoleucine-producing strain | GCA_001643035.1 | ||
| CS176 | GCA_002003445.1 | |||
| XV | l-Valine-producing strain | GCA_001936195.1 | ||
| ZL1 | l-Lysine-producing stain | GCA_001683055.1 | ||
| SCgG1 | l-Glutamate-hyperproducing strain | GCA_000404145.1 | ||
| SCgG2 | l-Glutamate-hyperproducing strain | GCA_000404185.1 | ||
| Z188 | l-Glutamate-hyperproducing strain | GCA_000417765.1 | ||
| ZL6 | l-Lysine-producing strain | GCA_001683115.1 |
Brevibacterium and Corynebacterium species are included.
According to references, ATCC/CGMCC record, or DDBJ/EMBL/GenBank record.
Determination of relative transcriptional levels.
C. glutamicum Z188 was cultivated in biotin-rich or biotin-poor fermentation medium, and cells were collected during the first 12 h. RNA was isolated from the cell pellet using an RNAprep Pure Cell/Bacteria kit (Tiangen Biotech, Beijing, China). After treatment with DNase I (Tiangen Biotech, Beijing, China), cDNA was synthesized using random primers and a FastQuant RT kit (Tiangen Biotech, Beijing, China). The resultant cDNA was used as a template for qPCR analysis. The RNA samples were used as the templates for PCR to confirm that genomic DNA contamination during RNA extraction was minimal. Specific primers for qPCR were designed with Beacon Designer software (Table 4). qPCR was performed by using a SuperReal Premix SYBR green kit (Tiangen Biotech, Beijing, China) and the Applied Biosystems 7500 real-time PCR system (Thermo Fisher Scientific, USA) according to the manufacturer's instructions.
TABLE 4.
Primers used in this study
| Relevance | Primer | Sequence (5′-3′) |
|---|---|---|
| pK18mobsacB-ΔmscCG2 construction | ΔmscCG2-P1 | TCGGGATCCACCACACCAATGTGCTTGC |
| ΔmscCG2-P2 | CAGAGATGCCGCGGGACTTCAGTCTTTGGC | |
| ΔmscCG2-P3 | GAAGTCCCGCGGCATCTCTGCATTCGAACC | |
| ΔmscCG2-P4 | AAACTGCAGGTGACGCCGATGGGGTAGTTGG | |
| pK18mobsacB-ΔmscCG construction | ΔmscCG-P1 | TCCCCCGGGGGACCCGTCCAAGCCAAG |
| ΔmscCG-P2 | TGTCGGCTCGCGTGGAATCAAAAACGCCAA | |
| ΔmscCG-P3 | TGATTCCACGCGAGCCGACACCATCGAAC | |
| ΔmscCG-P4 | TGCTCTAGAAAGGGAGTTGAAGGTGACGCC | |
| pTRCmob-mscCG2 construction | mscCG2-F | TACGGGATCCTTAAGCATTGTCGTTTTAC |
| mscCG2-R | GCAACTGCAGTTAGTTGGCGTCGCCTTTATCC | |
| pTRCmob-mscCG construction | mscCG-F | TAGCGAGCTCCACGACTTTCTGGCTCCTTT |
| mscCG-R | CGGGATCCGAAATGGGACACGTCTGTAATC | |
| pTRCmob-mscCG2-egfp construction | pT-mscCG2-F | GCATGGACGAGCTGTACAAGTAACTGCAGGCATGCAAGCTTGGC |
| pT-mscCG2-R | AGCTCCTCGCCCTTGCTCACGTTGGCGTCGCCTTTATCCAGC | |
| egfp-F | CGCTGGATAAAGGCGACGCCAACGTGAGCAAGGGCGAGGAG | |
| egfp-R | CAGCCAAGCTTGCATGCCTGCAGTTACTTGTACAGCTCGTCCAT | |
| pTRCmob-egfp construction | pT-egfp-F | CGTGGGCACCGATGGTGAGCAAGGGCGAGGAGCTG |
| pT-egfp-R | CTTGCTCACCATCGGTGCCCACGATAGTGATAGTG | |
| pTRCmob-mscCG2A151V construction | mscCG2A151V-F | GCTTGGTTTTGGCGTGCAGTCGCTGGTGAAGGACTTTCTCAG |
| mscCG2A151V-R | CACCAGCGACTGCACGCCAAAACCAAGCGCAACACCAG | |
| pTRCmob-mscCG2A151T construction | mscCG2A151T-F | GCTTGGTTTTGGCACCCAGTCGCTGGTGAAGGACTTTCTCAG |
| mscCG2A151T-R | CACCAGCGACTGGGTGCCAAAACCAAGCGCAACACCAG | |
| qPCR | q16S-F | ATAACTTGAGTGCTGTAGG |
| q16S-R | TTGGTGTTCCTCCTGATA | |
| qmscCG-F | TTGAAGGCACAGTCATTG | |
| qmscCG-R | GGATAATCACGGTCTCTTG | |
| qmscCG2-F | TCGCTAGTACAGATACAG | |
| qmscCG2-R | GCTGATTGGCTTATTGAT |
Gene deletion in C. glutamicum.
Gene deletion mutants of C. glutamicum were constructed via allele exchange using the suicide plasmid pK18mobsacB (30). pK18mobsacB-ΔmscCG2 containing a mutant allele of mscCG2 was constructed to disrupt the mscCG2 gene of C. glutamicum Z188. The mutant allele of mscCG2 was generated by connecting a left and a right homologous flank of mscCG2. Initially, the left and right flanks were amplified from the genomic DNA of C. glutamicum Z188 by using primer pairs ΔmscCG2-P1/ΔmscCG2-P2 and ΔmscCG2-P3/ΔmscCG2-P4, respectively (Table 4). The two fragments were joined into a single fragment by overlap extension PCR. The BamHI- and PstI-digested fragment was ligated to pK18mobsacB digested with the same endonucleases to construct pK18mobsacB-ΔmscCG2. pK18mobsacB-ΔmscCG was constructed using the same procedure. Primer pairs ΔmscCG-P1/ΔmscCG-P2 and ΔmscCG-P3/ΔmscCG-P4 were used to amplify the homologous flanks (Table 4). SmaI and XbaI were used to digest the DNA fragment and pK18mobsacB. Mutants Z188ΔmscCG2, Z188ΔmscCG, Z188ΔmscCGΔmscCG2, and ATCC 13032ΔmscCG were then constructed by transforming the generated plasmid into C. glutamicum Z188 or C. glutamicum ATCC 13032 by electroporation transformation and procedures described previously (30, 31).
Gene overexpression in C. glutamicum.
pTRCmob was used for protein overexpression in C. glutamicum. The coding regions and native promoters of mscCG2 and mscCG were amplified by PCR from the genomic DNA of C. glutamicum Z188 by using primer pairs mscCG2-F/mscCG2-R and mscCG-F/mscCG-R, respectively (Table 4). The mscCG2 and mscCG fragments were digested with BamHI/PstI and SacI/BamHI, respectively. The fragments were ligated to pTRCmob digested with the same endonucleases, resulting in pTRCmob-mscCG2 and pTRCmob-mscCG, respectively. To construct pTRCmob-mscCG2-egfp that harbors a mscCG2-egfp fusion gene, inverse PCR was carried out to linearize the plasmid pTRCmob-mscCG2 and remove the termination codon of mscCG2 using the primer pair pT-mscCG2-F/pT-mscCG2-R. The egfp gene was amplified from pET21a-egfp using the primer pair egfp-F/egfp-R (32) and was ligated to linearized pTRCmob-mscCG2 using a CloneExpress II one-step cloning kit (Vazyme, China). pTRCmob-egfp was constructed by using inverse PCR to remove mscCG2 from pTRCmob-mscCG2-egfp using the primer pair pT-egfp-F/pT-egfp-R. pTRCmob-mscCG2A151V and pTRCmob-mscCG2A151T were constructed by inverse PCR using pTRCmob-mscCG2 as a template and primer pairs mscCG2A151V-F/mscCG2A151V-R and mscCG2A151T-F/mscCG2A151T-R, respectively. The plasmids were transformed into C. glutamicum by electroporation transformation, producing the derivatives listed in Table 2.
Localization of MscCG2-EGFP fusion protein under confocal laser scanning microscope.
Strains Z188(pTRCmob), Z188(egfp), and Z188(mscCG2-egfp) were cultivated, and cells were harvested in the stationary phase. Images were acquired on a confocal laser scanning microscope based on an Olympus IX71 inverted microscope frame fitted with a 100×, 1.49-numerical-aperture oil immersion objective. Excitation light at 488 nm from a solid-state laser (Coherent Inc.) was used to excite EGFP and MscCG2-EGFP proteins.
Shake flask fermentation.
Seed cultures of C. glutamicum strains were prepared by transferring the overnight cultures prepared in LBG medium into 500-ml baffled flasks containing 50 ml seed medium at an initial optical density at 600 nm (OD600) of 0.1. Cells at the exponential growth phase were transferred into 500-ml baffled shake flasks with 30 ml fermentation medium with an inoculum size of 10% (vol/vol). The cells were cultivated at 30°C and with shaking at 220 rpm. The pH of the medium was adjusted by supplementation with ammonia. For penicillin treatments, cells were cultured in biotin-rich fermentation medium and 60 U/ml penicillin was added when the OD600 reached about 12.
Peptide uptake and amino acid export assay.
Pregrown cells in fermentation medium were harvested by centrifugation at 5,000 × g for 10 min and washed twice with ice-cold CGXII medium (33). The peptide uptake and amino acid excretion were then initiated by resuspending the cells in prewarmed CGXII medium (30°C) containing 2 mM Glu-Glu (Sigma-Aldrich Co.). The resulting cell density (OD600) was 10.0. The cells were incubated at 30°C and with shaking at 220 rpm. Samples were taken every 15 min, and extracellular amino acids were quantified as described below.
Analysis of 4-FG resistance of MscCG2 mutants.
The resistance of MscCG2 mutants to l-glutamate analog 4-FG was examined by using a previously described method with modifications (10). C. glutamicum Z188 derivatives were inoculated into CGXII medium with different concentrations of 4-FG (0, 1.25, 2.5, and 5.0 mM) and an initial OD600 of 0.05. The cells were cultivated at 30°C and with shaking at 220 rpm. When the OD600 of each strain in the control medium without 4-FG reached 1.0, all cultures, with and without 4-FG, were plated on solid LBG medium. The viability of the diluted cells was determined by counting the number of colonies on the plates.
Osmotic downshock and analysis of cell viability.
The cell survival rates of C. glutamicum strains upon osmotic downshock were examined by using previously described methods with modifications (22, 34). The overnight cultures of C. glutamicum prepared in LBG medium were transferred into a high-osmolarity LBG medium containing 1.17 M NaCl (2,350 mosmol) with an initial OD600 of 0.05. The cells were cultivated at 30°C and with shaking at 220 rpm. When the OD600 reached 0.65, the cultivation was stopped and 20 μl culture was transferred into 980 μl normal LBG medium containing 0.17 M NaCl (410 mosmol) or high-osmolarity LBG medium containing 1.17 M NaCl (2,350 mosmol). The cultures were incubated at 30°C and with shaking at 220 rpm for another 40 min. Then the cultures were plated on solid LBG medium. The viability of the cells was determined by counting the number of colonies on the plates.
Analytical methods.
For measurement of the extracellular l-glutamate concentration of shake flask fermentation, 500-μl cultures were centrifuged at 13,000 × g for 5 min, and the supernatant was used for detection after appropriate dilution. Regarding analysis of intracellular l-glutamate concentration, 500-μl cultures were centrifuged at 14,000 × g and 4°C for 10 min with 500 μl silicon oil (specific gravity, 1.07) (Shinetsu Kagaku Kogyo, Tokyo, Japan) as the separation layer and 300 μl 21% HClO4 as an acid fixation layer (35). The sediment cells in the acid layer were further disrupted using a freeze-thaw procedure. The resulting extracts were removed by suction with an autopipette, and the samples were neutralized with an equal amount of 2 M Na2CO3. The neutralized samples were then subjected to appropriate analysis with an SBA-40D biosensor analyzer (Institute of Biology of Shandong Province Academy of Sciences, Shandong, China). The cytoplasmic volume was assumed to be 1.6 μl/mg dry cell weight (36). Regarding peptide uptake and amino acid export assay, the concentration of l-glutamate was measured by using a Prominence UFLC (Shimadzu, Japan) equipped with a Zorbax Eclipse AAA column (4.6 by 150 mm, 5 μm) and a UV detector. A gradient of 50 mM sodium acetate buffer at pH 6.4 with a gradient solution containing acetonitrile-water (50:50, vol/vol) was used as the eluent. Amino acids were detected as their 2,4-dinitrofluorobenzene derivatives at 360 nm by following the precolumn derivation method.
Accession number(s).
The C. glutamicum Z188 whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession number NZ_AKXP00000000. The mscCG2 gene locates at bp 212785 to bp 213789 of NZ_AKXP01000027. The protein ID of C. glutamicum Z188 MscCG2 in NCBI is WP_004567541.1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31700044), the Key Research Program of the Chinese Academy of Sciences (ZDRW-ZS-2016-2), and the first Special Support Plan for Talents Development and High-level Innovation and Entrepreneurship Team of the Tianjin Municipal City.
Y.W., G.C., P.Z., and J.S. conceived and designed the experiments. Y.W., D.X., L.F., and X.W. performed the experiments. Y.W., G.C., L.F., and X.N. analyzed the data. P.Z., J.S., S.Z., and Y.M. contributed reagents and analytic tools. All authors contributed to the writing of the article.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02691-17.
REFERENCES
- 1.Becker J, Wittmann C. 2012. Systems and synthetic metabolic engineering for amino acid production—the heartbeat of industrial strain development. Curr Opin Biotechnol 23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Becker J, Wittmann C. 2016. Industrial microorganisms: Corynebacterium glutamicum, p 183–220. In Wittmann C, Liao JC (ed), Industrial biotechnology: microorganisms. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 3.Eggeling L. 2017. Exporters for production of amino acids and other small molecules. Adv Biochem Eng Biotechnol 159:199–225. doi: 10.1007/10_2016_32. [DOI] [PubMed] [Google Scholar]
- 4.Marin K, Krämer R. 2007. Amino acid transport systems in biotechnologically relevant bacteria, p 289–325. In Wendisch VF. (ed), Amino acid biosynthesis—pathways, regulation and metabolic engineering. Springer, Berlin, Germany. [Google Scholar]
- 5.Park JH, Lee SY. 2008. Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol 19:454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Vrljic M, Sahm H, Eggeling L. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 7.Simic P, Sahm H, Eggeling L. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J Bacteriol 183:5317–5324. doi: 10.1128/JB.183.18.5317-5324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennerknecht N, Sahm H, Yen MR, Patek M, Saier MH Jr, Eggeling L. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trotschel C, Deutenberg D, Bathe B, Burkovski A, Kramer R. 2005. Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187:3786–3794. doi: 10.1128/JB.187.11.3786-3794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura J, Hirano S, Ito H, Wachi M. 2007. Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce l-glutamic acid production. Appl Environ Microbiol 73:4491–4498. doi: 10.1128/AEM.02446-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox CD, Nakayama Y, Nomura T, Martinac B. 2015. The evolutionary ‘tinkering’ of MscS-like channels: generation of structural and functional diversity. Pflugers Arch 467:3–13. doi: 10.1007/s00424-014-1522-2. [DOI] [PubMed] [Google Scholar]
- 12.Becker M, Borngen K, Nomura T, Battle AR, Marin K, Martinac B, Kramer R. 2013. Glutamate efflux mediated by Corynebacterium glutamicum MscCG, Escherichia coli MscS, and their derivatives. Biochim Biophys Acta 1828:1230–1240. doi: 10.1016/j.bbamem.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Wachi M. 2013. Amino acid exporters in Corynebacterium glutamicum, p 335–349. In Yukawa H, Inui M (ed), Corynebacterium glutamicum. Springer, Berlin, Germany. [Google Scholar]
- 14.Bass RB, Strop P, Barclay M, Rees DC. 2002. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita C, Hashimoto K, Kumagai K, Maeda T, Takada A, Yabe I, Kawasaki H, Wachi M. 2013. l-Glutamate secretion by the N-terminal domain of the Corynebacterium glutamicum NCgl1221 mechanosensitive channel. Biosci Biotechnol Biochem 77:1008–1013. doi: 10.1271/bbb.120988. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama Y, Becker M, Ebrahimian H, Konishi T, Kawasaki H, Kramer R, Martinac B. 2016. The impact of the C-terminal domain on the gating properties of MscCG from Corynebacterium glutamicum. Biochim Biophys Acta 1858:130–138. doi: 10.1016/j.bbamem.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Becker M, Kramer R. 2015. MscCG from Corynebacterium glutamicum: functional significance of the C-terminal domain. Eur Biophys J 44:577–588. doi: 10.1007/s00249-015-1041-x. [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller S, Bartlett W, Chandrasekaran S, Simpson S, Edwards M, Booth IR. 2003. Domain organization of the MscS mechanosensitive channel of Escherichia coli. EMBO J 22:36–46. doi: 10.1093/emboj/cdg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama Y, Yoshimura K, Iida H. 2012. A gain-of-function mutation in gating of Corynebacterium glutamicum NCgl1221 causes constitutive glutamate secretion. Appl Environ Microbiol 78:5432–5434. doi: 10.1128/AEM.01310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borngen K, Battle AR, Moker N, Morbach S, Marin K, Martinac B, Kramer R. 2010. The properties and contribution of the Corynebacterium glutamicum MscS variant to fine-tuning of osmotic adaptation. Biochim Biophys Acta 1798:2141–2149. doi: 10.1016/j.bbamem.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto K, Nakamura K, Kuroda T, Yabe I, Nakamatsu T, Kawasaki H. 2010. The protein encoded by NCgl1221 in Corynebacterium glutamicum functions as a mechanosensitive channel. Biosci Biotechnol Biochem 74:2546–2549. doi: 10.1271/bbb.100636. [DOI] [PubMed] [Google Scholar]
- 23.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. 2012. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol 8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Tao F, Xin B, Liu H, Gao Y, Zhou NY, Xu P. 2017. Switch of metabolic status: redirecting metabolic flux for acetoin production from glycerol by activating a silent glycerol catabolism pathway. Metab Eng 39:90–101. doi: 10.1016/j.ymben.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Burkovski A, Kramer R. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol 58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 26.Naismith JH, Booth IR. 2012. Bacterial mechanosensitive channels—MscS: evolution's solution to creating sensitivity in function. Annu Rev Biophys 41:157–177. doi: 10.1146/annurev-biophys-101211-113227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto K, Murata J, Konishi T, Yabe I, Nakamatsu T, Kawasaki H. 2012. Glutamate is excreted across the cytoplasmic membrane through the NCgl1221 channel of Corynebacterium glutamicum by passive diffusion. Biosci Biotechnol Biochem 76:1422–1424. doi: 10.1271/bbb.120366. [DOI] [PubMed] [Google Scholar]
- 28.Zuo G, Hao B. 2015. CVTree3 web server for whole-genome-based and alignment-free prokaryotic phylogeny and taxonomy. Genomics Proteomics Bioinformatics 13:321–331. doi: 10.1016/j.gpb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 31.Ruan Y, Zhu L, Li Q. 2015. Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity. Biotechnol Lett 37:2445–2452. doi: 10.1007/s10529-015-1934-x. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Li Q, Zheng P, Zhang Z, Liu Y, Sun C, Cao G, Zhou W, Wang X, Zhang D, Zhang T, Sun J, Ma Y. 2015. Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb Cell Fact 14:121. doi: 10.1186/s12934-015-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keilhauer C, Eggeling L, Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahome PG, Setlow P. 2008. Growth, osmotic downshock resistance and differentiation of Bacillus subtilis strains lacking mechanosensitive channels. Arch Microbiol 189:49–58. doi: 10.1007/s00203-007-0292-z. [DOI] [PubMed] [Google Scholar]
- 35.Ishizaki A, Yamamoto K, Furuta Y. 1995. A new method for the accurate and rapid determination of the concentrations of intracellular metabolites in cells during fermentation. Biotechnol Tech 9:409–412. doi: 10.1007/BF00160827. [DOI] [Google Scholar]
- 36.Gutmann M, Hoischen C, Kramer R. 1992. Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim Biophys Acta 1112:115–123. doi: 10.1016/0005-2736(92)90261-J. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Yang S. 2017. Comparative analysis of Corynebacterium glutamicum genomes: a new perspective for the industrial production of amino acids. BMC Genomics 18:940. doi: 10.1186/s12864-016-3255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 39.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Ouyang SP, Kim J, Chen GQ. 2007. The impact of PHB accumulation on l-glutamate production by recombinant Corynebacterium glutamicum. J Biotechnol 132:273–279. doi: 10.1016/j.jbiotec.2007.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.