ABSTRACT
To overcome the adverse impacts of environmental stresses during growth, different adaptive regulation mechanisms can be activated in Lactococcus lactis. In this study, the transcription levels of eight transcriptional regulators of L. lactis subsp. lactis F44 under acid stress were analyzed using quantitative reverse transcription-PCR. Eight gene-overexpressing strains were then constructed to examine their influences on acid-resistant capability. Overexpressing ythA, a PspC family transcriptional regulator, increased the survival rate by 3.2-fold compared to the control at the lethal pH 3.0 acid shock. Moreover, the nisin yield was increased by 45.50%. The ythA-overexpressing strain FythA appeared to have higher intracellular pH stability and nisin-resistant ability. Subsequently, transcriptome analysis revealed that the vast majority of genes associated with amino acid biosynthesis, including arginine, serine, phenylalanine, and tyrosine, were predominantly upregulated in FythA. Arginine biosynthesis (argG and argH), arginine deiminase pathway, and polar amino acid transport (ysfE and ysfF) were proposed to be the main regulation mechanisms of YthA. Furthermore, the transcription of genes associated with pyrimidine and exopolysaccharide biosynthesis were upregulated. The transcriptional levels of nisIPRKFEG genes were substantially higher in FythA, which directly contributed to the yield and resistance of nisin. Three potential DNA-binding sequences were predicted by computer analysis using the upstream regions of genes with prominent changes. This study showed that YthA could increase acid resistance and nisin yield and revealed a putative regulation mechanism of YthA.
IMPORTANCE Nisin, produced by Lactococcus lactis subsp. lactis, is widely used as a safe food preservative. Acid stress becomes the primary restrictive factor of cell growth and nisin yield. In this research, we found that the transcriptional regulator YthA was conducive to enhancing the acid resistance of L. lactis F44. Overexpressing ythA could significantly improve the survival rate and nisin yield. The stability of intracellular pH and nisin resistance were also increased. Transcriptome analysis showed that nisin immunity and the biosynthesis of some amino acids, pyrimidine, and exopolysaccharides were enhanced in the engineered strain. This study elucidates the regulation mechanism of YthA and provides a novel strategy for constructing robust industrial L. lactis strains.
KEYWORDS: Lactococcus lactis, transcriptional regulator, YthA, acid stress, nisin yield
INTRODUCTION
Lactococcus lactis, a member of lactic acid bacteria (LAB), is extensively employed in the food industry and usually used as a primary building block for applications in biomedicine and other fields (1–3). Nisin production has been optimized, distributed, and used widely in food and biomedical industries for the last 50 years (4, 5). During fermentation, L. lactis is confronted with many types of stress, especially acid stress because of the conversion of pyruvate to lactate (6), which probably leads to the suboptimal growth and the decrease in nisin yield. Otherwise, pH values decrease, and the corresponding increase in hydrogen ion concentrations in the cytoplasm might contribute to the stability of nisin (7). Thus, studying acid resistance mechanism and constructing acid-tolerant industrial strains are essential. L. lactis has developed a multifactorial response to cope with acid stress, including the ATP-dependent expulsion of protons, the production of basic compounds, the repair of impaired macromolecules, and protection of the cell envelope from various types of damage (8). Moreover, bacteria have established a complicated regulatory network via integrating and coordinating the expression levels of different genes.
A series of transcriptional regulators has been found to contribute to acid resistance. Transcriptional activator GadE controls the most important acid resistance system, the glutamate-dependent (Gad) system, by directly activating the expression of gadA and gadBC in Escherichia coli (9). As a major partner of GadE, regulator RcsB can also directly activate gadA transcription via the glutamate decarboxylase (GAD) box (10). In Streptococcus mutans, two Spx global regulators, spxA and spxB, are associated with stress tolerance, and an ΔspxA strain shows decreased survival rate under acid stress (11). The transcriptional regulator Ldb0677 has been identified as an acid stress-related regulator by proteomics approach, and its targets have been revealed via bacterial one-hybrid technology in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 (12). In Lactobacillus acidophilus, inactivation of the transcriptional regulator La867 has a remarkable influence on acid tolerance (13).
In L. lactis, a range of transcriptional regulation mechanisms have been preliminarily studied. CodY and CcpA are two widely known global regulators in L. lactis. CodY controls various cellular functions, including nutrient transport and nitrogen metabolism, especially branched-chain amino acid metabolism (14, 15). CodY also fulfills a prominent role in carbon starvation and near-zero growth (16, 17). Moreover, the catabolite control protein CcpA can repress putrescine synthesis and the utilization of galactose and fructose (18, 19). CcpA also activates the central metabolism and the expression of prolidase PepQ, constituting a link between carbon and nitrogen metabolism regulations. In addition, a significant number of operon-specific transcriptional regulators have been observed. For instance, ClaR can regulate cellobiose and lactose metabolism, and MalR is an activator of the maltose transport system (20, 21). The transcriptional regulator FabT affects fab gene expression in fatty acid biosynthesis (22). The transcriptional regulator GadR can control the GAD pathway by activating the transcription of gadB and gadC (23). CtsR shows a negative regulation of the clp genes clpC, clpP, clpE, and clpB (24).
The survival of bacteria depends on their ability to sense and respond to cell envelope disturbances. The phage shock protein (Psp) system plays a pivotal role in envelope stress response (25). In E. coli, PspC family members participate in the stress response system to maintain the integrity and function of bacterial cell envelope (26). When the cell envelope is damaged, the Psp response cannot be established, and the proton motive force is dissipated (27). A physical and/or biochemical change gives rise to the cell stress response and results in increased expression levels of psp genes. PspA, PspB, PspC, and PspF form a signal transduction system in the regulation of psp gene expression. PspB and PspC, two cytoplasmic membrane proteins, can form an integral complex to act as sensory activators of the Psp response in both Yersinia enterocolitica and E. coli (28). The three-gene ythC-ythB-ythA operon exists in L. lactis, and YthA and YthB contain the conserved region of PspC family transcriptional regulators. YthC is a hypothetical protein and has not been explored. Nevertheless, no other gene related to Psp response system in L. lactis has been observed, and the regulation mechanism of YthA remains unclear.
In this study, which is based on a transcription level analysis of L. lactis F44 under acid stress using quantitative reverse transcription-PCR (qRT-PCR), eight transcriptional regulators with pronounced expression changes were overexpressed to determine which can increase the acid resistance. Overexpressing ythA could increase the survival rate and nisin yield. The stability of intracellular pH and nisin resistance were also confirmed to be improved in FythA. To further investigate the regulation mechanism of YthA, transcriptome sequencing (RNA-seq) was performed. The vast majority of genes in the metabolisms of some amino acids, pyrimidine, exopolysaccharides (EPS), nisin biosynthesis, and nisin immunity were remarkably upregulated. By combining transcriptome data with in silico motif predictions, three potential DNA binding motifs were identified in the promoter regions of genes with prominent changes. Thus, we show that YthA contributes to acid resistance and nisin yield and provides a putative acid tolerance mechanism for YthA.
RESULTS
Determination of transcriptional regulators contributing to acid resistance in L. lactis F44.
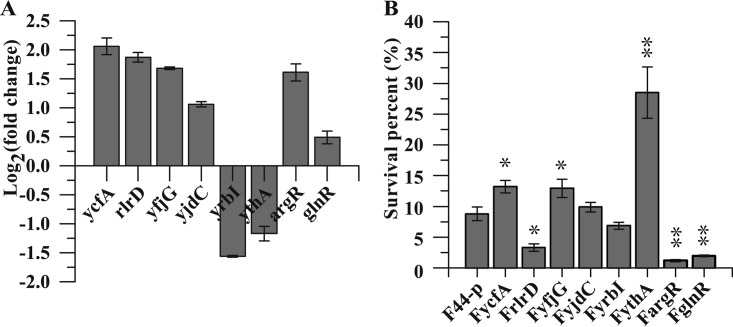
Based on transcriptional level analysis of eight transcriptional regulators in L. lactis F44 after a pH 4.0 acid shock for 1 h using qRT-PCR (Fig. 1A), these genes were overexpressed in L. lactis F44 (Tables 1 and 2). Subsequently, to clarify the effects on the acid resistance of these engineered strains, an acid resistance assay was performed, and strain F44-p harboring an empty vector pLEB124 was used as a control. These analyses showed different survival rates after lethal pH 3.0 acid shock for 2.5 h (Fig. 1B). Notably, ythA overexpression enhanced the acid-resistant ability, and the engineered strain FythA showed a 3.2-fold increase of survival rate compared to the control under acid shock. Moreover, the survival rates of FyjdC and FyrbI were slightly higher than that of the control, and FyjdC and FyrbI showed no significant changes. However, the survival rates of FrlrD, FargR, and FglnR were all remarkably lower than that of F44-p. Due to the pronounced improvement in acid resistance, we focused on further investigating the physiological characteristics of FythA during fermentation.
FIG 1.
(A) Relative transcription levels of eight regulators under acid shock analyzed by qRT-PCR. The fold changes of qRT-PCR were normalized using 16S rRNA as an internal control gene. The error bars, indicating standard deviations (SD), are from three replicate flasks. (B) Acid tolerance of the strains overexpressing transcriptional regulators. The survival percentage was the ratio of the CFU per milliliter counted after the acid challenge to the CFU/ml at T0 (start of the challenge). Cells were cultured in fermentation medium (pH 7.2) and challenged in fermentation medium (pH 3.0). Error bars indicate the SD of three independent experiments. *, P < 0.01; **, P < 0.001 (t test).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| L. lactis F44 | Parental strain, derived from L. lactis YF11 (China General Microbiological Culture Collection Center accession no. CGMCC7.52) | Laboratory stock (64) |
| L. lactis F44-p | L. lactis F44 carrying empty pLEB124; Emr | This study |
| L. lactis FycfA | ycfA-overexpressing strain; Emr | This study |
| L. lactis FrlrD | rlrD-overexpressing strain; Emr | This study |
| L. lactis FyfjG | yfjG-overexpressing strain; Emr | This study |
| L. lactis FyjdC | yjdC-overexpressing strain; Emr | This study |
| L. lactis FyrbI | yrbI-overexpressing strain; Emr | This study |
| L. lactis FythA | ythA-overexpressing strain; Emr | This study |
| L. lactis FargR | argR-overexpressing strain; Emr | This study |
| L. lactis FglnR | glnR-overexpressing strain; Emr | This study |
| E. coli TG1 | Plasmid preparation | Laboratory stock |
| M. flavus ATCC 10240 | Indicator (the Gram-positive pathogen sensitive to nisin) | Laboratory stock |
| Plasmids | ||
| pLEB124 | Expression vector; Emr | Laboratory stock |
| pLEBycfA | Derivative of vector pLEB124 carrying ycfA from L. lactis F44; Emr | This study |
| pLEBrlrD | Derivative of vector pLEB124 carrying rlrD from L. lactis F44; Emr | This study |
| pLEByfjG | Derivative of vector pLEB124 carrying yfjG from L. lactis F44; Emr | This study |
| pLEByjdC | Derivative of vector pLEB124 carrying yjdC from L. lactis F44; Emr | This study |
| pLEByrbI | Derivative of vector pLEB124 carrying yrbI from L. lactis F44; Emr | This study |
| pLEBythA | Derivative of vector pLEB124 carrying ythA from L. lactis F44; Emr | This study |
| pLEBargR | Derivative of vector pLEB124 carrying argR from L. lactis F44; Emr | This study |
| pLEBglnR | Derivative of vector pLEB124 carrying glnR from L. lactis F44; Emr | This study |
Emr, erythromycin resistance.
TABLE 2.
Candidates of transcriptional regulators
| Gene | Identification no. | Gene length (bp) | Accession no. | Product |
|---|---|---|---|---|
| ycfA | F44_g0218 | 648 | ATY86861.1 | TetR family transcriptional regulator |
| rlrD | F44_g0345 | 822 | ATY86977.1 | LysR family transcriptional regulator |
| yfjG | F44_g0506 | 627 | ATY87125.1 | TetR family transcriptional regulator |
| yjdC | F44_g0905 | 465 | ATY87505.1 | MarR family transcriptional regulator |
| yrbI | F44_g1581 | 834 | ATY88165.1 | XRE family transcriptional regulator |
| ythA | F44_g1877 | 465 | ATY88441.1 | Stress-responsive transcriptional regulator, PspC family |
| argR | F44_g2007 | 459 | ATY88561.1 | Arginine operon repressor |
| glnR | F44_g2177 | 369 | ATY88875.1 | MerR family transcriptional regulator |
Effects of YthA on physiological characters and nisin yield of L. lactis F44.
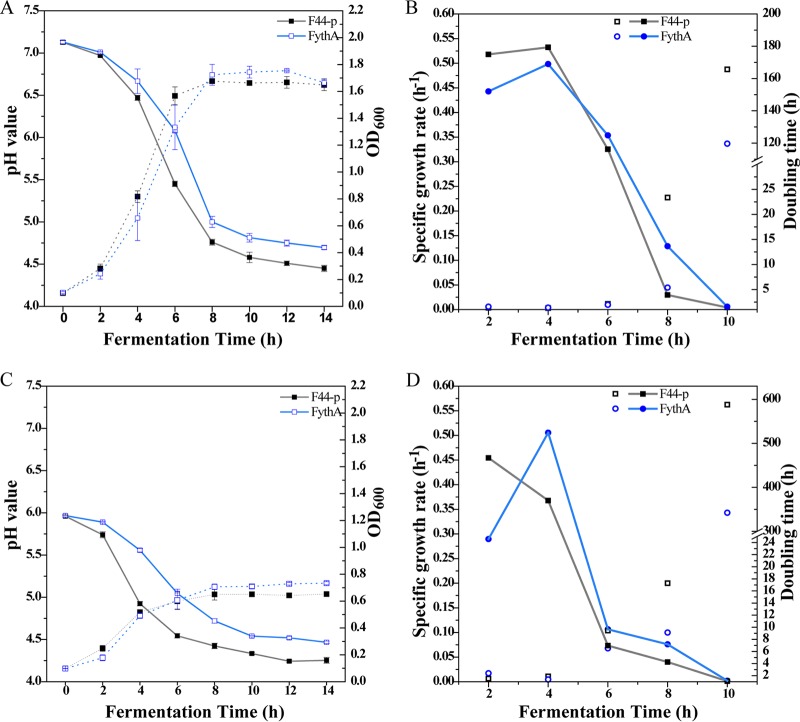
Cell density, pH value, and nisin yield were monitored every 2 h to evaluate the effects of YthA during fermentation. In the fermentation with an initial pH of 7.2, FythA presented a lower growth rate compared to the control after 6 h of incubation (Fig. 2A). Subsequently, the biomass accumulation sharply increased from 6 to 8 h, and the specific growth rate exceeded that of F44-p after 6 h (Fig. 2B). The biomass of FythA peaked (optical density [OD] = 1.76 ± 0.01) at 10 h, and it was 4.65% higher than that of the control (OD = 1.68 ± 0.01) at 8 h. The pH value of culture declined due to the production and accumulation of lactic acid. Intriguingly, the pH values of FythA fermentation broth were higher than that of the control during the entire fermentation. The final pH reached 4.70 ± 0.23 at 14 h, whereas the control was pH 4.49 ± 0.04 (Fig. 2A).
FIG 2.
Physiological characteristics of a ythA-overexpressing strain. (A) Cell density (dashed lines) and pH value (solid lines) during fermentation at the initial pH 7.2. OD600 indicates the optical density measured at 600 nm. (B) Specific growth rate and doubling time during fermentation at the initial pH 7.2. (C) Cell density (dashed lines) and pH value (solid lines) during fermentation at the initial pH 6.0. OD600 indicates the optical density measured at 600 nm. (D) Specific growth rate and doubling time during fermentation at the initial pH 6.0.
To further elucidate the effects of YthA on acid resistance, the cell density and the pH value were measured in an acidic medium with an initial pH 6.0. As expected, the engineered strain exhibited a 12.6% higher total biomass than the control, and pH values decreased slower than that in the control throughout the fermentation (Fig. 2C). In contrast to the fermentation with an initial pH of 7.2, the specific growth rate of FythA increased from 0.29 to 0.51 h−1 (from 2 to 4 h) and was higher than that of the control (0.37 h−1) after 4 h (Fig. 2D). The doubling time of FythA (9.13 h) was lower than that of the control (17.28 h) after 8 h. The difference in physiological characteristics between FythA and F44-p was more significant than that for the initial pH 7.2 (P < 0.01, after 10 h). These results further confirmed that YthA was conducive to resisting acid stress. We presumed that YthA might promote the production of certain alkaline substances, such as alkaline amino acids and NH3.
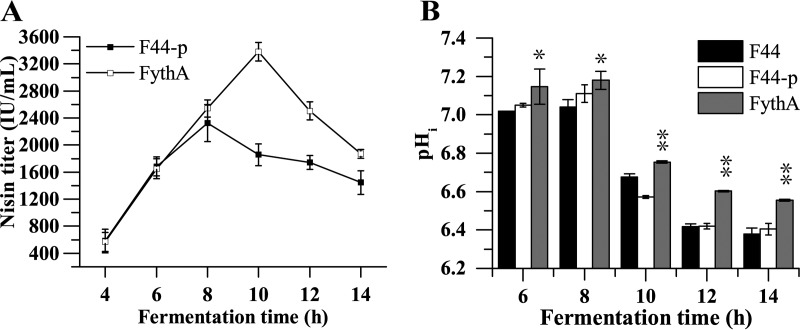
The nisin yield was determined every 2 h during fermentation with an initial pH of 7.2. Notably, the nisin titer of FythA peaked at 3,380.20 ± 135.40 IU/ml, which was 45.50% higher than that of the control at 8 h, and decreased thereafter (Fig. 3A). This demonstrated that the nisin yield could be improved by increasing the acid resistance, a finding consistent with our previous study (7).
FIG 3.
Nisin yield and pHi of ythA-overexpressing strain. (A) Effects of transcriptional regulator YthA on the nisin titer during fermentation at an initial pH of 7.2. Samples were taken every 2 h, ranging from 4 to 14 h. (B) pHi of FythA, F44, and F44-p during fermentation. Samples were taken every 2 h for 6 to 14 h. Error bars indicate the SD from three independent experiments. *, P < 0.05; **, P < 0.01 (t test).
YthA enhances the intracellular pH stability.
Intracellular pH (pHi) homeostasis plays a major role in maintaining physiological stability under acid stress in LAB. To further investigate the influence of regulator YthA on the acid resistance, the pHi values of the FythA strain and wild-type strain F44 were measured during fermentation. As shown in Fig. 3B, the pHi values of both strains decreased, along with the fermentation, and obvious differences of pHi were detected between FythA and F44. The engineered strain FythA exhibited a higher pHi than that in F44 during fermentation, especially at the stationary phase. In other words, FythA maintained a relatively stable pHi during the process. These results indicated that the transcriptional regulator YthA could contribute to maintaining the stability of pHi in L. lactis and guaranteed the growth of strains, which might be beneficial for acid tolerance and nisin yield.
Transcriptome analysis of the YthA-overexpressing strain.
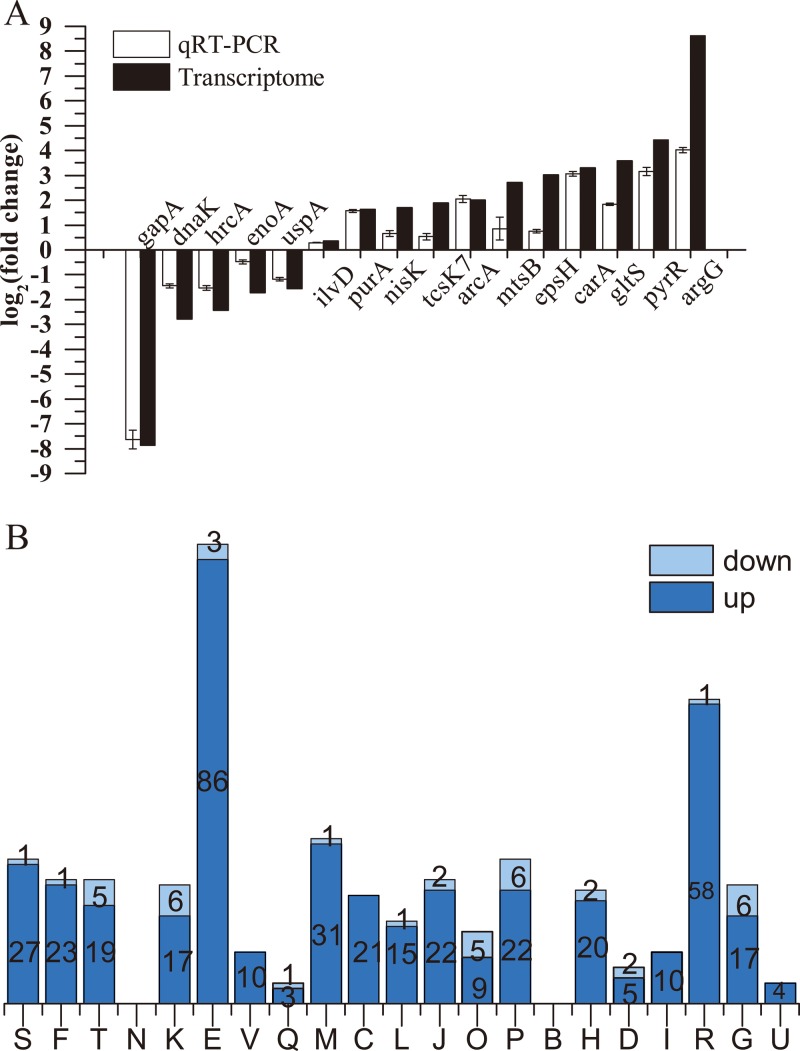
To investigate the possible regulation mechanism mediated by YthA, a transcriptomic approach was used with an Illumina HiSeq 4000. FythA and F44 grown in fermentation medium were collected at the exponential phase (OD600 ≈ 0.8). For transcriptomic analysis, at a cutoff of 1.5-fold change (log2-fold change) and a P value of <0.05, 304 and 45 genes, respectively, were upregulated and downregulated compared to the L. lactis F44 with empty plasmid. A total of 16 genes were selected to be validated by qRT-PCR. Among these genes, 10 were significantly upregulated, 5 were downregulated, and 1 showed no obvious change. Apparent positive correlations were verified between the transcriptomic results and qRT-PCR for these genes (Fig. 4A). The functional distribution of significantly changed proteins, according to the Cluster of Orthologous Groups (COG) classification for proteins, is shown in Fig. 4B. A number of genes in class E (amino acid transport and metabolism) were upregulated, which suggested that YthA mainly regulated transport and metabolism of amino acids to improve acid resistance.
FIG 4.
(A) The relative mRNA level changes of 16 genes in FythA were compared between transcriptome data and qRT-PCR results. The fold changes of qRT-PCR were normalized using 16S rRNA as an internal control gene. Error bars indicating the SD were from three replicate flasks. (B) Distribution of significantly changed genes according to the COG classification. The y axis indicates the number of genes in various COG categories. Columns: S, function unknown; F, nucleotide transport and metabolism; T, signal transduction mechanisms; N, cell motility; K, transcription; E, amino acid transport and metabolism; V, defense mechanisms; Q, secondary metabolites biosynthesis, transport, and catabolism; M, cell wall/membrane/envelope biogenesis; C, energy production and conversion; L, replication, recombination, and repair; J, translation, ribosomal structure, and biogenesis; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; B, chromatin structure and dynamics; H, coenzyme transport and metabolism; D, cell cycle control, mitosis, and meiosis; I, lipid transport and metabolism; R, general function prediction only; G, carbohydrate transport and metabolism; U, intracellular trafficking and secretion.
YthA improves the metabolism and transport of amino acids.
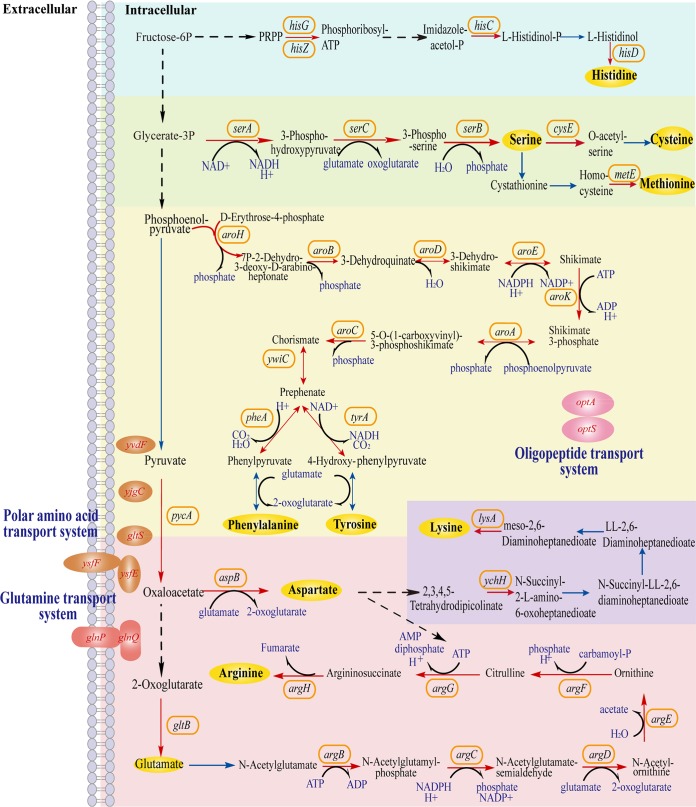
We found here that the metabolism of some amino acids, especially arginine, serine, phenylalanine, and tyrosine, was notably enhanced in FythA (Fig. 5 and Table 3). Most genes in the glutamate and arginine biosynthesis pathway, including gltB, argB, argC, argD, argE, argF, argG, and argH, were consistently upregulated. Among them, the argininosuccinate synthase gene argG and the argininosuccinate lyase gene argH, respectively, showed dramatic 8.63- and 7.67-fold upregulations. The arginine deiminase (ADI) pathway is a known pivotal mechanism that can withstand acid stress in LAB. Genes associated with the ADI pathway—arcA (ADI), arcB (ornithine carbamoyltransferase), arcC2 (carbamate kinase), arcC3 (carbamate kinase), and arcD1 (arginine/ornithine antiporter)—were all abundant. Three genes, namely, proA, proB, and proC, that are involved in proline biosynthesis were expressed at elevated levels. The expression of aspB gene, which encodes aspartate aminotransferase to generate aspartate from oxaloacetate in the aspartate biosynthesis pathway, was significantly increased by ∼1.87-fold.
FIG 5.
Effects of transcriptional regulator YthA on the metabolism of some amino acids. The upregulated genes are represented by red fonts or solid arrows. Genes with no significant change are represented by blue solid arrows.
TABLE 3.
Significantly changed genes involved in the transport and metabolism of amino acids
| Pathway | Gene | Log2 ratioa | Description |
|---|---|---|---|
| Arginine biosynthesis | argG | 8.62 | Argininosuccinate synthase |
| argH | 7.67 | Argininosuccinate lyase | |
| argF | 2.56 | Ornithine carbamoyltransferase | |
| argE | 3.45 | Acetylornithine deacetylase | |
| argD | 1.65 | Acetylornithine aminotransferase | |
| argC | 1.69 | N-Acetyl-gammaglutamyl-phosphate reductase | |
| argB | 2.65 | Acetylglutamate kinase | |
| Arginine deiminase (ADI) pathway | arcD2 | 1.86 | Arginine/ornithine antiporter |
| arcD1 | 2.29 | Arginine/ornithine antiporter | |
| arcC3 | 2.70 | Carbamate kinase | |
| arcC2 | 2.27 | Carbamate kinase | |
| arcB | 2.24 | Ornithine carbamoyltransferase | |
| arcA | 2.00 | Arginine deiminase | |
| Aromatic amino acids biosynthesis | aroK | 2.52 | Shikimate kinase |
| aroH | 3.16 | Phospho-2-dehydro-3-deoxyheptonate aldolase | |
| aroF | 2.21 | 3-Deoxy-7-phosphoheptulonate synthase | |
| aroE | 2.07 | Shikimate 5-dehydrogenase | |
| aroD | 2.37 | 3-Dehydroquinate dehydratase | |
| aroC | 1.97 | Chorismate synthase | |
| aroB | 2.39 | 3-Dehydroquinate synthase | |
| aroA | 1.99 | 3-Phosphoshikimate 1-carboxyvinyltransferase | |
| Proline biosynthesis | proA | 2.24 | Gamma-glutamyl phosphate reductase |
| proB | 1.51 | Glutamate 5-kinase | |
| proC | 1.91 | Pyrroline-5-carboxylate reductase | |
| Serine biosynthesis | serC | 2.74 | MFS transporter |
| serB | 2.21 | Phosphoserine phosphatase | |
| serA | 2.14 | 3-Phosphoglycerate dehydrogenase | |
| Lysine biosynthesis | ychH | 1.86 | 2,3,4,5-Tetrahydropyridine-2,6-carboxylate N-succinyltransferase |
| yciA | 2.01 | N-Acetyldiaminopimelate deacetylase | |
| lysA | 2.31 | Diaminopimelate decarboxylase | |
| Histidine biosynthesis | hisC | 1.67 | Histidinol-phosphate aminotransferase |
| hisD | 1.58 | Histidinol dehydrogenase | |
| hisG | 1.86 | ATP phosphoribosyltransferase | |
| hisZ | 1.97 | ATP phosphoribosyltransferase, regulatory subunit | |
| Glutamate transport system | glnQ | 1.61 | Glutamine ABC transporter ATP-binding protein |
| glnP | 1.70 | Amino acid ABC transporter permease | |
| Methionine biosynthesis | metE | 2.19 | Methyltetrahydropteroyltriglutamate/homocysteine S-methyltransferase |
| metA | 1.87 | Homoserine O-succinyltransferase | |
| Polar amino acid transport system | ysfE | 6.32 | Glutamate ABC transporter ATP-binding protein |
| ysfF | 4.45 | Glutamate ABC transporter permeas | |
| gltS | 3.58 | Glutamate or arginine ABC transporter substrate binding protein | |
| yjgC | 1.53 | Amino acid ABC transporter substrate binding protein | |
| yvdF | 1.60 | ABC transporter substrate-binding protein | |
| Oligopeptide transport system | optS | 1.97 | ABC transporter substrate-binding protein |
| optA | 1.55 | ABC transporter substrate-binding protein | |
| Others | tyrA | 1.94 | Prephenate dehydrogenase |
| asnB | 2.15 | Asparagine synthase | |
| pheA | 2.34 | Prephenate dehydratase | |
| thrB | 2.09 | Homoserine kinase | |
| aspB | 1.87 | Aspartate aminotransferase | |
| yddD | 1.57 | Glyoxalase | |
| araT | 1.51 | Aminotransferase A | |
| murA | 1.51 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | |
| pepN | 1.50 | Aminopeptidase N | |
| axe | −1.80 | Acetyl esterase | |
| lysP1 | 1.99 | Lysine specific permease | |
| cysE | 1.99 | Serine acetyltransferase | |
| gltB | 1.79 | Glutamate synthase |
Values are expressed as the fold change in a log2 scale.
In addition to the biosynthesis of arginine, the pathways of the other two alkaline amino acids, histidine and lysine, were also improved. Four genes that refer to histidine biosynthesis (hisZ, hisG, hisC, and hisD) were also highly upregulated. The genes hisZ and hisG, which encode ATP-phosphoribosyltransferase, increased 1.86- and 1.97-fold, respectively. These two enzymes catalyze the condensation of ATP with phosphoribosyl pyrophosphate, the first step of histidine biosynthesis (29). Moreover, several lysine biosynthesis genes, including ychH, yciA, and lysA, were upregulated. The expression of the diaminopimelate decarboxylase gene lysA, which catalyzes the final step in the lysine biosynthesis pathway of bacteria, was increased by 2.31-fold.
The genes encoding the enzymes responsible for serine biosynthesis (serA, serC, and serB) were upregulated, suggesting that YthA positively regulated the serine biosynthesis. The homoserine kinase gene thrB that converts homoserine to O-phosphohomoserine in threonine biosynthesis yielded a 2.09-fold increase in abundance. The serine acetyltransferase gene cysE, which catalyzes the conversion of serine to O-acetyl-l-serine, was upregulated 1.99-fold. Notably, genes associated with methionine biosynthesis (metA and metE) were also upregulated (1.87- and 2.19-fold, respectively).
Moreover, the shikimate pathway (including aroA, aroB, aroC, aroD, aroE, aroF, aroK, and aroH), which is the early step in the biosynthesis of aromatic amino acids, including phenylalanine, tyrosine, and tryptophan, was integrally upregulated. Among these genes, aroA, aroK, and aroC, which encode enzymes that catalyze shikimate to chorismite, were upregulated. The production of pivotal enzyme 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHPS) is not only the first step in the synthesis of aromatic compounds, but DAHPS is also the rate-limiting enzyme. The introduction of E. coli aroH encoding DAHPS resulted in a significant increase in phenylalanine in Corynebacterium glutamicum (30). In the present study, the transcription of aroH was dramatically upregulated by 3.16-fold.
Several genes implicated in the polar amino acid transport system (ysfEF, gltS, yvdF, and yjgC), the glutamate transport system (glnPQ), and the oligopeptide transport system (optS and optA) were also upregulated. ysfE, ysfF, and gltS expression levels were increased. Overall, the results demonstrate that transcriptional regulator YthA positively mediates the biosynthesis and transport systems of multiple amino acids.
Effects of YthA on EPS synthesis and cell division.
Exopolysaccharides (EPS) can protect bacterial cells against extreme stresses, including pH, temperature, and osmotic stress. In this study, transcriptional analysis revealed that most EPS synthesis genes (epsABCDEFGHIJK) were induced in FythA (Table 4). The epsA, epsB, epsC, epsD, and epsK genes were all upregulated (>1.5-fold and <2.0-fold). The transcriptional levels of epsEFGHIJ increased by more than 2.0-fold. The polysaccharide chain-length-determining genes epsF and epsG showed 2.51-fold and 2.86-fold abundance. Intriguingly, epsH encoding glycosyltransferase showed a 3.01-fold increase in abundance.
TABLE 4.
Significantly changed genes involved in exopolysaccharide synthesis, cell division, nisin biosynthesis and resistance, and nucleotide metabolism
| Pathway | Gene | Log2 ratioa | Description |
|---|---|---|---|
| Exopolysaccharide synthesis | epsH | 3.01 | Group 1 glycosyltransferase |
| epsI | 2.81 | Hypothetical protein | |
| epsJ | 2.30 | Polysaccharide biosynthesis protein | |
| epsG | 2.86 | Family 2 glycosyltransferase | |
| epsF | 2.51 | Hypothetical protein | |
| epsE | 2.29 | Group 1 glycosyltransferase | |
| epsK | 1.57 | Polysaccharide biosynthesis protein | |
| epsD | 1.87 | Sugar transferase | |
| epsC | 1.93 | Tyrosine protein phosphatase | |
| epsB | 1.74 | Tyrosine protein kinase | |
| epsA | 1.87 | Polysaccharide biosynthesis protein | |
| Cell division | ftsX | 1.77 | Cell division permease FtsX |
| ftsQ | 1.55 | Cell division initiation protein DivIB | |
| ftsE | 1.56 | Cell division ATP-binding protein | |
| smc | 2.16 | Chromosome segregation protein SMC | |
| Nisin synthesis and immunity | nisI | 1.46 | Nisin immunity protein |
| nisP | 1.46 | Peptidase | |
| nisK | 1.69 | Nisin biosynthesis sensor protein NisK | |
| nisR | 1.50 | Nisin biosynthesis two-component system, response regulator NisR | |
| nisF | 1.29 | Lantibiotic ABC transporter ATP-binding protein | |
| nisE | 1.76 | Lantibiotic ABC transporter permease | |
| nisG | 2.10 | Transporter | |
| Pyrimidine metabolism | carA | 3.29 | Carbamoyl-phosphate synthase small subunit |
| pyrB | 1.50 | Uridine phosphorylase | |
| pyrC | 2.99 | Uracil permease | |
| pyrZ | 2.89 | Dihydroorotate dehydrogenase electron transfer subunit | |
| pyrD | 3.29 | Aspartate carbamoyltransferase catalytic subunit | |
| pyre | 3.22 | Dihydroorotate dehydrogenase | |
| pyrF | 2.69 | ADP-ribose pyrophosphatase | |
| pyrR | 4.41 | Phosphoribosyl transferase | |
| pyrP | 3.19 | 5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase | |
| upp | 2.51 | Orotidine 5′-phosphate decarboxylase | |
| udp | 1.62 | Adenylosuccinate synthetase |
Values are expressed as the fold change in a log2 scale.
A series of genes related to cell division were considerably induced. Gene smc, which encodes a chromosome segregation protein, increased by 2.16-fold. The cell division protein gene ftsQ displayed a 1.56-fold upregulation. The gene transcription levels of ftsE and ftsX, which encode ATP-binding protein and permease protein of the cell division transport system, increased 1.56-fold and 1.78-fold, respectively, in abundance.
YthA increases the biosynthesis and immunity of nisin.
The nisin gene cluster nisABTCIPRKFEG was observed in our strain. The majority of these genes were upregulated in the engineered strain (Table 4). The nisin immunity gene nisI and the nisin leader peptide-processing gene nisP both increased in expression by 1.46-fold. The nisR-nisK two-component system (TCS) genes showed 1.50- and 1.69-fold abundance. Nisin transport system genes nisFEG, which are also important for nisin immunity, were induced 1.29-, 1.76-, and 2.10-fold, respectively. To further investigate immunity to nisin, we performed nisin resistance assays for FythA, F44-p, and F44. FythA was more resistant to nisin than either F44-p or F44 (Fig. 6). The maximum valid dilution of FythA on the plate with 18,000 IU/ml nisin was 1:106, whereas those of F44 and F44-p were only 1:103 (Fig. 6A). Moreover, FythA showed better viability than the control on the plate containing 20,000 IU/ml nisin (Fig. 6B). These results showed that YthA had a detectable effect on the ability of biosynthesis and immunity of nisin.
FIG 6.
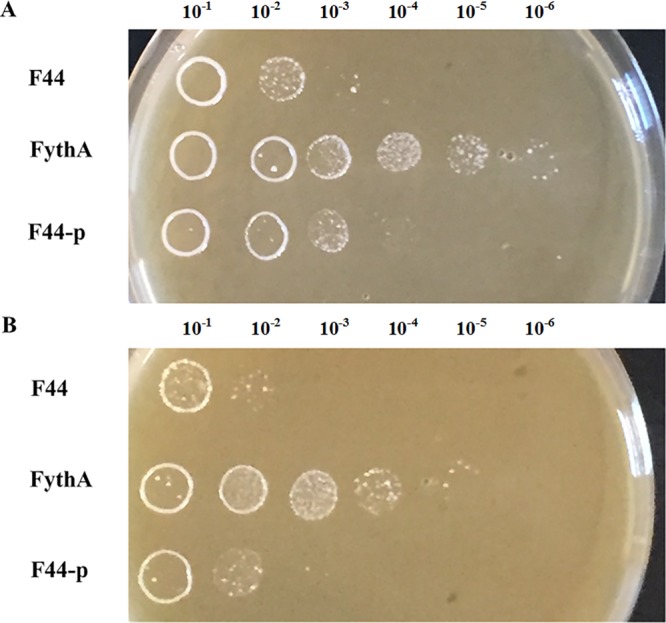
Nisin resistance of FythA, F44-p, and F44. The nisin-resistant capacity was determined by the serial dilutions plated on the fermentation medium agar plates containing 18,000 IU/ml nisin (A) and 20,000 IU/ml nisin (B).
Pyrimidine metabolism and other effects of YthA.
A total of 11 genes implicated in pyrimidine metabolism (carA, pyrBCDEFPRZ, upp, and udp) were prominently upregulated. The pyrB, pyrD, pyrE, pyrF, pyrP, and pyrR genes increased by >3-fold (Table 4). The aminoacyl-tRNA synthetase genes (cysS, asnC, argS, trpS, and lysS1) were enhanced (see Table S1 in the supplemental material). Moreover, ylgC (RNA methyltransferase) was upregulated 3.49-fold. Two ribosomal protein genes, rpmG3 and rpsN2, showed 2.85- and 2.55-fold increases in abundance, respectively, indicating high potential for protein translation.
In addition to NisR/NisK, the KdpD/KdpE and TcsR7/TcsK7 TCSs also yielded increases. Several transcriptional regulators, including yebF, yfdD, scrR, ybjK, rcfB, yfdC, rex, rlrG, argR, and zitR, were also upregulated (see Table S1 in the supplemental material). Among them, the first five genes increased by >2-fold. YfdD and YfdC were both annotated as regulatory protein Spx. On the contrary, four regulators—yohC, hrcA, tcsR8, and ysgA—were decreased by >2-fold. In particular, yohC was downregulated by 3.02-fold. Interestingly, three genes (uspA2, yahB, and yjaB) that encode universal stress proteins were repressed by ∼1.5-fold. Moreover, the carbon starvation cstA protein decreased 2.05-fold.
Carbohydrate utilization was also significantly affected by YthA (see Table S1 in the supplemental material). Two sucrose utilization genes, transcriptional regulator scrR and β-fructofuranosidase scrB, were abundant. Two genes related to the glycolysis pathway pgi (glucose-6-phosphate isomerase) and yrjI (probable phosphoglycerate mutase) were upregulated. Nevertheless, the gapA gene that encodes the NADP+-dependent glyceraldehyde 3-phosphate dehydrogenase decreased dramatically by 7.86-fold. In addition, glk and enoA were both downregulated.
Metal irons implicated in many vital biological processes are essential for the survival of bacteria. As shown in Table S1 in the supplemental material, the expression of the manganese/zinc transport system genes mtsA, mstB, and mtsC dramatically increased (3.71- and 3.29-fold). The expression of era (GTPase), which is essential for the cell viability of bacteria, increased by 1.8-fold. Four genes—secY, secA1, secA2, and secE—that encode preprotein translocase were all enhanced. In particular, secE demonstrated a 3.0-fold increase in abundance. Astonishingly, four chaperone genes (grpE, dnaK, groES, and groEL) were downregulated.
Proposed binding sites of YthA.
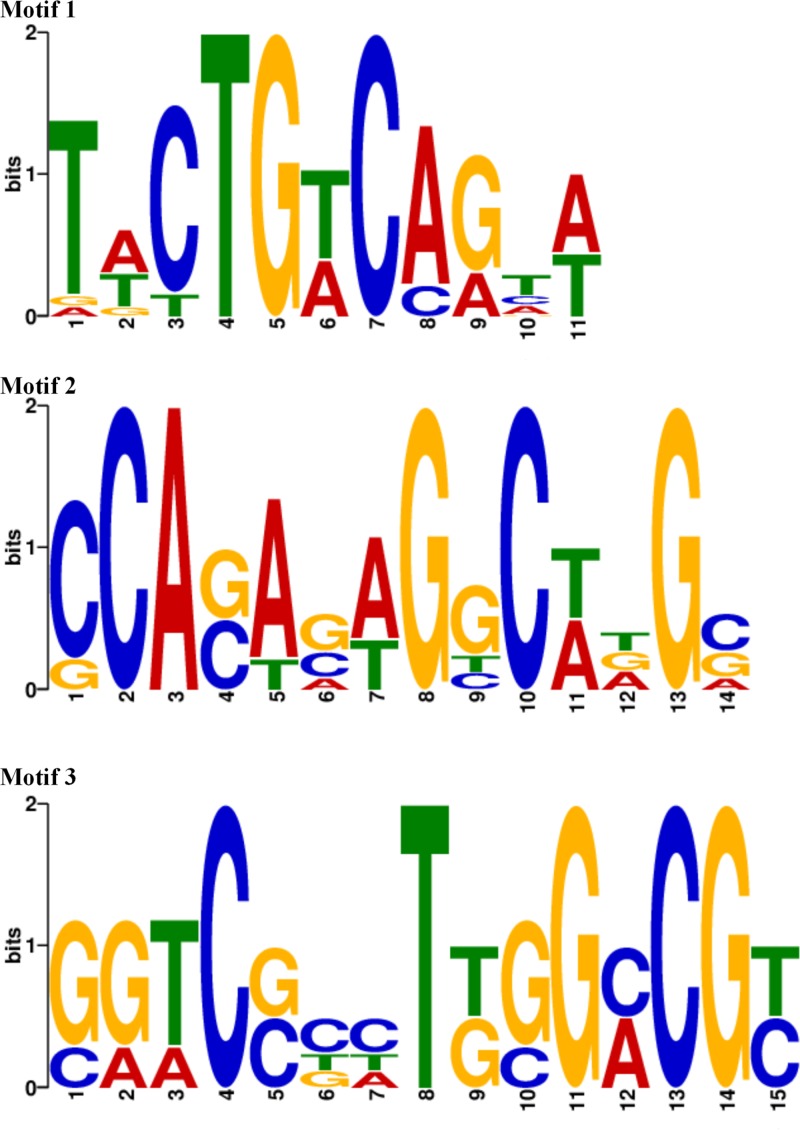
To gain further insight into the possible DNA-binding motifs of YthA regulation, 15 promoter regions of the most highly changed genes (most of them up- or downregulated by >3-fold) were analyzed to find putative binding sites of YthA using the motif finder MEME (see Table S2 in the supplemental material). Three putative motifs containing 11 to 15 bp were detected, as shown in Fig. 7. Motif 1, TWCTGWCAGTW, was identified at 18 sites upstream of five candidates: pyrR, pyrE, pyrB-carA, pyrZD, and ylgC. Motif 2, CCASASWGGCWDGS, was found upstream of argGH, pyrR, pyrE, pyrB-carA and pyrZD. Motif 3 (GTCSBHTKGGMCGY) was only found upstream of genes, such as argG, argH, gltS, argE, aroH, and gapA, that are related to amino acid biosynthesis. These motifs may be associated with YthA regulation in L. lactis F44, and further verification is needed.
FIG 7.
Visualization of putative DNA binding motifs of YthA predicted by MEME. Motif 1 (TWCTGWCAGTW), motif 2 (CCASASWGGCWDGS), and motif 3 (GTCSBHTKGGMCGY) are depicted by sequence logos generated by the MEME suite tools. The relative heights of the letters represent the frequencies of nucleotides at each position.
DISCUSSION
The protein YthA is a stress-responsive transcriptional regulator of the PspC family, with 154 amino acids and a calculated molecular mass of 18.3 kDa. The overall approach used in the present study research is depicted in Fig. 8. FythA displayed a slow pH decrease, whereas it exhibited a higher total biomass and growth rate compared to the control under acidic fermentation. Moreover, FythA showed an increased acid resistance and nisin yield. A transcriptomic approach was used to identify the regulation mechanism of YthA in L. lactis (Fig. 9). We observed that YthA was strongly targeted in amino acid biosynthesis, pyrimidine metabolism, exopolysaccharide biosynthesis, and nisin immunity. The stress-responsive transcriptional regulator YthA was found to contribute to acid resistance, and its regulation mechanism is preliminarily revealed in L. lactis.
FIG 8.
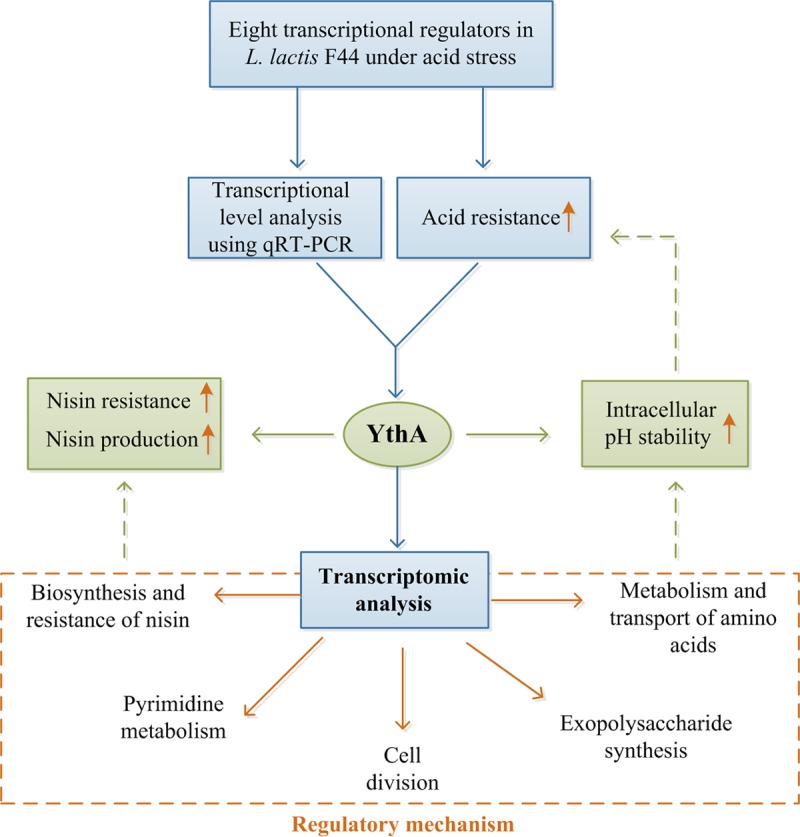
Overall approach used in this research. A summary of the various steps involved in our research workflow is depicted.
FIG 9.
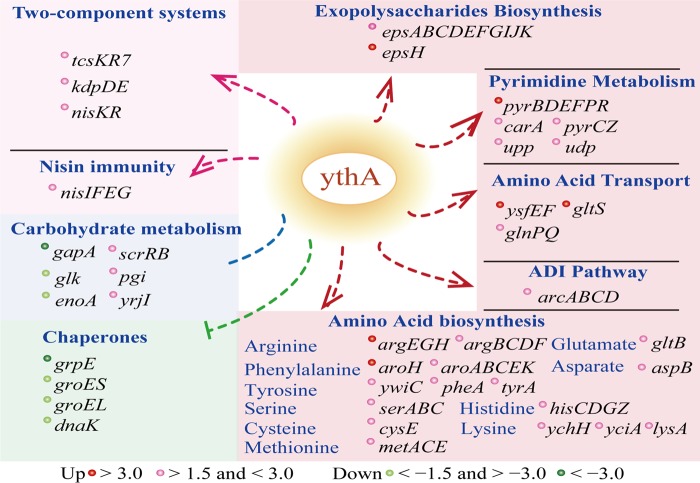
Proposed transcription mechanism of transcriptional regulator YthA on L. lactis F44. According to transcriptome analysis, YthA was presumed to regulate the amino acid transport and biosynthesis, ADI pathway, pyrimidine metabolism, exopolysaccharide synthesis, two-component systems, nisin immunity, and chaperones. Red spots represent more abundant genes >3.0-fold. Pink spots represent upregulated genes (between 1.5- and 3.0-fold). Green spots represent downregulated genes (>3.0-fold). Light green spots represent downregulated genes (between 1.5- and 3.0-fold).
L. lactis encounters acid stress because of the accumulation of lactic acid, which may inhibit or even cease growth. A previous study found that a range of transcriptional regulators could assist bacteria in increasing acid resistance (12). In our study, overexpressing ythA improved the acid resistance of L. lactis. As shown in Fig. 2C, the decreased pH values in FythA were lower throughout the fermentation, whereas the total biomass was higher than that of the control. Moreover, the engineered strain FythA contributed to the stability of the pHi value (Fig. 3B). Maintaining the relative pHi stability is vital to bacteria and guarantees normal physiology and cell metabolism (31, 32). To investigate the regulation mechanism, a transcriptomic approach was used. The expression levels of a large number of genes involved in amino acid biosynthesis increased in FythA. Amino acids participate in a series of LAB physiological process, including protein synthesis, intracellular pH regulation, metabolic energy generation, and stress resistance (33). Six genes in arginine synthesis were significantly upregulated. The levels of argG and argH, in particular, dramatically increased 8.62- and 7.67-fold, respectively. Previous studies found that argG and argH were acid inducible, and their upregulation might enhance the metabolic flux from aspartate to arginine, which could increase the production of ATP and ammonia and the consumption of acidic amino acids (aspartate and glutamate) (34, 35). Moreover, the biosynthesis of the basic amino acids histidine (hisZ, hisG, hisC, and hisD) and lysine (ychH, yciA, and lysA) was strongly induced. Cytoplasmic buffering, which can sequester or release protons, is important in pH homeostasis (36). In our study, YthA could activate the biosynthesis of basic amino acids (arginine, histidine, and lysine) to sequester protons. In addition, YthA enhanced the entire arginine deiminase (ADI) pathway consisting of arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC2 and ArcC3). The ADI pathway can convert arginine into ornithine via citrulline. This pathway is one of the most representative mechanisms for cells to cope with acid stress and energy deficiency by producing ammonia and ATP (37). Overall, these results might provide clues to explain the physiological characteristics of the ythA-overexpressing strain (Fig. 2). Furthermore, YthA could induce the biosynthesis of arginine, histidine, lysine, and ADI pathway to maintain intracellular pH homeostasis in response to acid stress.
Also, EPS, which are important components influencing cell surface characteristics, are vital in pathogenesis and symbiosis and protect the cell from various environmental stresses (38). A epsXABCDEFGHIJKL cluster in L. lactis F44 has been observed. EpsA is essential for the EPS biosynthesis and is a positive regulator for the EPS production (39). EspH is a key enzyme that catalyzes the biosynthesis of oligosaccharide-repeating units (40). The protein EpsF, a membrane protein, participates in exporting saccharide, and the protein EpsG exhibits autophosphorylation (41). In Methylovorus sp. strain MP688, it was demonstrated that EPS are indispensable to bacterial survival in adverse environments (42). Thus, we presumed that YthA can help bacteria survive acid stress by promoting the EPS biosynthesis.
In addition, YthA might activate several other mechanisms and possibly contribute to withstand acid stress. The two-component system (TCS) for signal transduction acts as an essential mechanism for environmental sensing and signal transduction in most bacteria. In the present study, the transcription levels of TCS KdpD/KdpE were notably upregulated. The system is highly conserved across more than 1,000 bacterial species and is connected to K+ homeostasis, the enhancement of virulence, and survival against acid, salt, and oxidative stresses (43, 44). In addition, the levels of manganese/zinc transport system genes, namely, mtsA, mstB, and mtsC, were dramatically increased. These genes are responsible for transporting zinc, manganese, and iron in Streptococcus pyogenes (45). However, MtsA fulfills a role in the homeostasis of iron and manganese, without controlling zinc homeostasis in vivo (46). Manganese is indispensable for enzymatic catalysis, and a connection between manganese homeostasis and oxidative stress has been found (46).
In the present study, fermentation results suggest that overexpressing ythA was beneficial to the cell growth of L. lactis under acid stress (Fig. 2). According to the transcriptome results, YthA could promote the synthesis and transport of some amino acids (Fig. 5). Except for arginine, almost all of the genes involved in the synthesis of serine, phenylalanine, and tyrosine were markedly upregulated in our data sets. Besides its role in protein synthesis, l-serine also works as an important precursor of several essential compounds, such as phosphatidylserine and d-serine. A previous study reported that a deficiency in serine biosynthesis led to a low growth rate of Mycobacterium tuberculosis (47). Moreover, several genes in the biosynthesis of threonine, cysteine, histidine, methionine, and lysine were abundant. Notably, genes involved in the transport system of polar amino acids (ysfEF, gltS, yvdF, yjgC, and glnPQ) and oligopeptide transport system (opts and optA) were upregulated. Among these examples, GltS is a transporter for glutamate uptake in Helicobacter pylori, E. coli, and C. glutamicum (48, 49). YthA may facilitate glutamate uptake, which could also influence arginine synthesis indirectly.
In addition, the six genes (carA and pyrBCDEF) responsible for transferring l-glutamine to UMP, the de novo synthesis of pyrimidines, were upregulated. UMP, which can be further converted into UTP, CTP, dCTP, and dTTP, plays a vital role in pyrimidine synthesis (50). The pyrimidine biosynthetic pathway is also associated with arginine metabolism through carbamoyl phosphate (51). Furthermore, GTPase Era functions as an RNA chaperone by controlling the processing and maturation of 16S rRNA and the 30S (small) ribosomal subunit (52). Aside from participating in ribosome assembly, GTPases are involved in other cellular processes, including DNA replication, cell metabolism, cell division, and stress response (53). Moreover, the abundant transcripts of ftsQ, ftsE, and ftsX suggested faster cell division compared to the control. The gene ftsQ, which encodes a cell division protein, is essential in the assembly of all of the components of the septal region (54). The FtsEX complex is one of the PG hydrolase regulatory systems that recruit and control PG hydrolases during growth (55).
In L. lactis, the nisABTCIPRKFEG cluster is responsible for the synthesis, modification, regulation, immunity, and secretion of nisin. NisR/NisK is supposed to be a transcription activator of nisin biosynthesis and immunity in L. lactis (56, 57). NisR can directly activate nisA, nisR, and nisF and implies a vital role in the virulence of Streptococcus suis serotype 2 (58). In our study, FythA exhibited higher nisin resistance and yield than the control (Fig. 6 and 3A). Furthermore, the transcription enhancements of nisin cluster genes were verified. Previous studies have demonstrated that the Psp system is pivotal in bacteriocin resistance, especially in nisin resistance (59–61). Enhancing nisin resistance is a strategy for improving nisin yield (62). The nisin producer L. lactis employs two immunity systems, NisI and NisFEG, to protect against nisin (63, 64). In our study, the increase in the level of NisIFEG activated by YthA might be another reason for the high nisin yield. Moreover, the TcsK7/TcsR7 system is conserved with BceSR and BraSR and serves as the key regulator for the resistance of bacitracin and nisin in Bacillus subtilis and Staphylococcus aureus, respectively (65, 66). The upregulation of the TCS TcsK7/TcsR7 system induced by YthA would be important in nisin resistance. Thus, similar to our previous study, improving the nisin-resistant ability of L. lactis F44 could increase the nisin yield significantly (67).
To improve acid tolerance, recent studies have focused on manipulating regulatory genes other than metabolic genes in various industrial microbes (68). However, the regulatory systems related to the acid tolerance of L. lactis are rarely known. To further elucidate the issue, eight transcriptional regulators were overexpressed, and the transcriptional regulator YthA was verified to improve acid tolerance, maintain intracellular pH homeostasis, and increase the nisin resistance and nisin yield of L. lactis F44. Using transcriptome analysis coupled with qRT-PCR verification, we further investigated the possible regulation mechanism of YthA and assumed three potential DNA-binding motifs. The direct target genes and the DNA-binding motifs need to be further determined.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
All of the strains and plasmids used in this study are listed in Table 1. L. lactis subsp. lactis F44 was used for gene engineering. E. coli TG1 was used as the host for genetic manipulation. E. coli was incubated in Luria-Bertani (LB) medium (tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter) at 37°C, with shaking at 180 rpm. L. lactis was cultivated in a fermentation medium containing peptone at 1.5 g/liter, yeast extract at 1.5 g/liter, sucrose at 1.5 g/liter, KH2PO4 at 2.0 g/liter, NaCl at 0.15 g/liter, corn steep liquor at 0.3 g/liter, cysteine at 0.26 g/liter, and MgSO4·7H2O at 0.015 g/liter at an initial pH 7.2, followed by incubation at 30°C without shaking. Micrococcus flavus ATCC 10240, used as an indicator strain for nisin assay, was grown in LB medium at 37°C with 180-rpm agitation. Media were supplemented with the antibiotic erythromycin for the selection at concentrations of 100 μg/ml for E. coli and 5 μg/ml for L. lactis.
Cloning and overexpression of transcriptional regulators.
All of the transcriptional regulator genes were amplified by PCR using L. lactis subsp. lactis F44 genomic DNA as a template. Primers of the relevant genes used in this study were designed by Primer Premier 5 (Premier Biosoft, Canada) and are listed in Table 5.
TABLE 5.
Primers used in PCR amplifications
| Primer | Oligonucleotide sequence (5′ → 3′) | Length (bp)a |
|---|---|---|
| argR-F | CGCGGATCCGTCAATATATAGTCCATTATATGGT | 495 |
| argR-R | TCCCCCGGGTTATCCTAATAAAATATACTTTATTTCC | |
| yjdC-F | CGCGGATCCTACCGTTAGGGAGGAGTG | 486 |
| yjdC-R | TCCCCCGGGTTAGGCATTTTTTTCCTCCT | |
| ythA-F | CGCGGATCCCACAAGGTTAATTCAACGAT | 489 |
| ythA-R | TCCCCCGGGTTAGAAATCTGACCAGTCATCTT | |
| glnR-F | CGCGGATCCCTATAATGAAAGGCATGGAGG | 438 |
| glnR-R | TCCCCCGGGTTACATCCGCGGTTGGGTA | |
| ycfA-F | CGGGATCCCTGGAAATGGGGTGAAAA | 678 |
| ycfA-R | TCCCCCGGGGTTAGTATTTTTTACGTTTACGTGCT | |
| yfjG-F | CGGGATCCTTTGATTTATTGGTATTAGCTAGAG | 657 |
| yfjG-R | TCCCCCGGGTTAGGTGTTTTTTATATAAATTGGC | |
| rlrD-F | CGCGGATCCATTTTTGTATTTTAGGGAGAAA | 843 |
| rlrD-R | TCCCCCGGGTTAATGAATTTCGGTTTCTTTTT | |
| rcfA-F | CGCGGATCCTACTCAAATAAGGAGAATAAAAT | 702 |
| rcfA-R | TCCCCCGGGTTAAGCAAAATCATCTTCTAATTTC |
That is, the length of the product of PCR.
All of the products were restricted with BamHI and SmaI and subsequently ligated into plasmid pLEB124, which was treated with the same restriction enzymes to yield the expression vector. Vectors were introduced into E. coli TG1 by heat shock transformation. Finally, after antibiotic selection and enrichment, the vectors carrying target gene fragments were transformed into L. lactis F44 to obtain recombinant strains.
Acid resistance assay.
L. lactis cells were harvested by centrifugation after culture for 8 h and subsequently resuspended in equal volumes of fermentation medium that had been acidified to pH 3.0 with hydrochloric acid. After incubation at 30°C for 2.5 h, the number of viable strains was determined by plating on agar plates and was used as the final cell density. The number of cells before acid challenge was set as the initial cell density. Plates were incubated at 37°C for 24 h before colony counting. Survival rates were calculated by dividing the number of final cell density after being challenged at pH 3.0 by the number of the initial cell density immediately after resuspension.
Nisin yield assay.
Plate diffusion method was performed to determine the nisin yield (69). Standard nisin diluted to 200, 100, 50, and 25 IU/ml with 0.02 M HCl was used to draw the standard curve. Accordingly, the fermentation broth was diluted 2-fold by 0.02 mol/liter HCl, and the mixture was heated at 100°C for 5 min. After cooling at room temperature, the samples were centrifuged at 5,000 × g for 5 min to remove the cell pellets. The assay medium (26 ml with 1.5% agar) was pretreated by autoclaving, and 390 μl of Tween 20 (JiangTian, Tianjin, China) was added at approximately 70°C. When the assay medium was cooled to approximately 50°C, the indicator M. flavus ATCC 10240 was added at a ratio of 1% (vol/vol), and the total broth was poured into a sterile plate. Eight wells were drilled using a 7-mm-diameter hole punch on each assay agar plate. Portions (100 μl) of each fermentation sample and standard nisin solution were injected into the corresponding wells. After incubation at 37°C for 24 h, the inhibition zones were measured by using a Vernier caliper. A regression equation was calculated based on the measured data. Each sample was performed in triplicate.
Determination of pHi.
The intracellular pH (pHi) was measured by using a fluorescence method as described previously, and the 5-(and-6)-carboxyfluorescein succimidyl ester (cFDASE) was used as the fluorescent probe (7). Harvested cells were washed three times and resuspended in 50 mM HEPES buffer (pH 8.0), followed by incubation for 10 min at 30°C in the presence of 1.0 μM cFDASE. Subsequently, the cells were washed three times and resuspended in 50 mM potassium phosphate buffer (pH 7.0), and nonconjugated fluorochrome was eliminated by incubation with 10 mM lactose for 30 min at 37°C. The cells were then washed twice and resuspended in 50 mM HEPES buffer (pH 8.0), followed by an additional 10 min at 30°C with 1 μM valinomycin and nigericin. Samples were preserved on ice until required. Calibration curves and pHi values were determined from the ratio of the fluorescence signal measurements using a Fluorescence Spectrophotometer F-2700 (Hitachi High-Tech, Japan) at excitation wavelengths of 490 nm (pH-sensitive wavelength) and 440 nm (pH-insensitive wavelength) with a 5-nm slit width. The emission wavelength was 525 nm with a 5-nm slit width. All values were taken from three independent experiments.
RNA extraction and RNA-seq.
Total RNA of the strains was isolated by using a Quick-RNA MicroPrepkit (ZYMO Research, Irvine, CA) and treated with DNase I (NEB) according to the manufacturer's instructions. The quality of the total RNA samples, such as concentration, RNA integrity number (RIN), 28S/18S, and size, were analyzed by using an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano kit). The purity of the samples was detected by using a NanoDrop apparatus. rRNA depletion was performed using Ribo-Zero magnetic kit (bacteria; Epicentre, Madison, WI), and the sample was purified by using RNAClean XP beads (Agencourt). Afterward, RNA was fragmented into 130 to 170 nucleotides by adding fragment buffer (Ambion) to the samples.
First-strand cDNAs were generated using First-Strand MasterMix and SuperScript II reverse transcription (Invitrogen), and then the second-strand cDNAs were synthesized by adding Second-Strand MasterMix. Subsequently, the purified fragmented cDNAs were combined with End Repair Mix, and an adenine base was added. cDNA fragments were amplified with several rounds of PCR and subsequently purified. Library quantification was performed using an Agilent 2100 bioanalyzer instrument (Agilent DNA 1000 reagents). Subsequently, the qualified cDNA libraries were prepared to generate the cluster on the flow cell (TruSeq PE V3-cBot-HS cluster kit; Illumina) and sequenced on a HiSeq 4000 system (TruSeq SBS KIT-HS V3; Illumina).
Read preprocessing and transcriptome analysis.
Raw reads primarily produced by Illumina HiSeq 4000 were subjected to perform quality control by analyzing the base composition of raw reads and quality distribution of the bases along the reads (Q20 > 95% and Q30 > 90%) to ensure data accuracy. After quality control, raw reads were filtered into clean reads, which were then aligned to the reference sequences by using SOAPaligner/SOAP2 (70). The number of perfect clean reads corresponding to each gene was calculated and normalized to the number of reads per kilobase of exon model per million mapped reads. The false discovery rate (FDR) was used to determine the threshold of the P value. We identified the genes significantly differential expressed at an FDR of <0.001, a P value of <0.05, and a log2 ratio (pYthA/control) ≥ 1.5-fold normalized change. Furthermore, the COG annotation was determined by the BLAST software against the COG protein database (www.ncbi.nlm.nih.gov/COG/) (71).
Transcriptional verification by quantitative real-time PCR.
To determine mRNA levels, reverse transcription was performed using total RNA (0.5 μg) as a template by RevertAid First-Strand cDNA synthesis kit (Thermo Scientific, Waltham, MA). Quantitative real-time PCRs were performed with cDNA (∼100 ng), mix (LightCycler 480 SYBR green I Master), and gene-specific primers on a LightCycler 480 real-time PCR system (Roche, Switzerland). The PCR program was run as follows: preincubation at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 20 s. Specific primers were designed using Primer Premier 5, as listed in Table 6. The 16S rRNA gene in L. lactis F44 was used as an internal control. The value of the relative gene quantification was calculated based on the 2−ΔΔCT analysis method (72).
TABLE 6.
Primers used in qRT-PCR
| Primer | Oligonucleotide sequence (5′ → 3′) | Lengtha (bp) |
|---|---|---|
| argG-F | AGTGGCATTGGGCTCG | 127 |
| argG-R | ACACCAGCTTCAATCGCA | |
| carA-F | AGAGCATCAAATGAGCCAAC | 112 |
| carA-R | AACGACAACTTTTCTGCCTG | |
| dnaK-F | GACCGTAACACAACTATCCCA | 113 |
| dnaK-R | TCAGCTGCCATTGGACG | |
| uspA-F | GGCACAGAAGTTGATTTTGAG | 113 |
| uspA-R | AGACCAGTTGACCCAATTACA | |
| pyrR-F | GACCGTGGACATCGTGAAT | 119 |
| pyrR-R | AAAATACTGTCATTGCCATCG | |
| purA-F | AGAAATCGCTTATCTCGCTG | 133 |
| purA-R | AATCTTCTTATCGCCTTTGG | |
| mtsB-F | AGGAGAACTATCAGGTGGTCAG | 112 |
| mtsB-R | GCTTATCGAATCAATCCCAA | |
| gltS-F | AGGAGCCGTGATTGAAAAC | 122 |
| gltS-R | TAGGAAGTGCAACCGCTG | |
| tcsK7-F | GAGGAGCTGAAGAATCAGGTT | 125 |
| tcsK7-R | TTCACAAGTGAAGCCATACTCA | |
| nisK-F | TCAATATCTTTGTTAATGCCTGT | 117 |
| nisK-R | AAGGATGACCATTATTCCAGAT | |
| arcA-F | CAGGAATTATGGATGGTGCT | 113 |
| arcA-R | GACTTTCTTGAGTGCTGCTTTA | |
| hrcA-F | TTCTTAATGCTCCTCAACGTG | 105 |
| hrcA-R | TTACTTCACCAGTCCCTAATGTT | |
| epsH-F | TTTTTGGATCATACCCCATAA | 102 |
| epsH-R | TACTTCTTCCGAAAATTTCATG | |
| gapA-F | AATGACCCTTGATGGTCCA | 120 |
| gapA-R | AGTTCAGGCAAAACAAGACC | |
| enoA-F | AATTCCACTCTATCGCTATCTTG | 134 |
| enoA-R | CCCGTTTGACTGGTGTAATC |
That is, the length of the qRT-PCR fragment.
Determination of nisin resistance.
Nisin resistance was determined by the serial dilution assay, as described by Xie et al. (73). L. lactis strains F44, F44-p, and FythA were cultivated approximately to 5 × 108 CFU/ml in the fermentation medium at 30°C without rotation. Afterward, the cultures were serially diluted in 10-fold increments in physiological saline solution. Serial dilutions were plated on fermentation medium agar plates containing 18,000 or 20,000 IU/ml nisin. Plates were incubated at 30°C for 24 h. All experiments were performed in triplicate.
DNA motif mining.
MEME suite tools (http://meme-suite.org/) were used to search YthA binding sites in the promoter region of the proposed target genes in the genome of L. lactis F44 (74). The most significantly changed genes in transcriptome analysis were used as the data source (see Table S2 in the supplemental material). Binding site searches were performed using the putative promoter regions (regions that contained the intergenic region and 100 bp upstream of the genes) of the selected genes. Any number of sites per sequence was allowed, and sites should be on the given strand.
Statistical analysis.
To evaluate the statistical significance of the survival rate under acid stress, a Student t test was performed. The statistical significance values for the OD, pH, and pHi were assessed by using a t test. SPSS software version 19.0 (IBM) was used for statistically analyzing the data.
Accession number(s).
The mRNA-seq data of L. lactis F44-p and FythA obtained in this study and used in the sequencing procedures were deposited in the NCBI Gene Expression Omnibus under accession number GSE101424.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Key R&D Program of China (2017YFD0201405), the National Natural Science Foundation of China (31270142, 31770076, and 31570089), and the Funds for Creative Research Groups of China (21621004). J.Q. was supported by the New Century Outstanding Talent Support Program, Education Ministry of China.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02483-17.
REFERENCES
- 1.Song AA-L, In LL, Lim SHE, Rahim RA. 2017. A review on Lactococcus lactis: from food to factory. Microb Cell Fact 16:55. doi: 10.1186/s12934-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong W, Kapuganti VS, Lu T. 2015. A gene network engineering platform for lactic acid bacteria. Nucleic Acids Res 44:e37–e37. doi: 10.1093/nar/gkv1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong W, Blanchard AE, Liao C, Lu T. 2017. Engineering robust and tunable spatial structures with synthetic gene circuits. Nucleic Acids Res 45:1005–1014. doi: 10.1093/nar/gkw1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delves-Broughton J, Blackburn P, Evans R, Hugenholtz J. 1996. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 5.Kong W, Lu T. 2014. Cloning and optimization of a nisin biosynthesis pathway for bacteriocin harvest. ACS Synth Biol 3:439–445. doi: 10.1021/sb500225r. [DOI] [PubMed] [Google Scholar]
- 6.Papadimitriou K, Alegría Á, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C. 2016. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80:837–890. doi: 10.1128/MMBR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Caiyin Q, Feng W, Zhao X, Qiao B, Zhao G, Qiao J. 2016. Enhance nisin yield via improving acid-tolerant capability of Lactococcus lactis F44. Sci Rep 6:27973. doi: 10.1038/srep27973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van de Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, Maguin E. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187–216. doi: 10.1023/A:1020631532202. [DOI] [PubMed] [Google Scholar]
- 9.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 10.Castanié-Cornet M-P, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res 38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai Z, Douillard FP, An H, Wang G, Guo X, Luo Y, Hao Y. 2014. Proteomic characterization of the acid tolerance response in Lactobacillus delbrueckii subsp. bulgaricus CAUH1 and functional identification of a novel acid stress-related transcriptional regulator Ldb0677. Environ Microbiol 16:1524–1537. doi: 10.1111/1462-2920.12280. [DOI] [PubMed] [Google Scholar]
- 13.Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol 70:5315–5322. doi: 10.1128/AEM.70.9.5315-5322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895–3909. doi: 10.1099/mic.0.28186-0. [DOI] [PubMed] [Google Scholar]
- 15.den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. 2005. The Lactococcus lactis CodY regulon identification of a conserved cis-regulatory element. J Biol Chem 280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 16.Ercan O, Wels M, Smid EJ, Kleerebezem M. 2015. Genome-wide transcriptional responses to carbon starvation in nongrowing Lactococcus lactis. Appl Environ Microbiol 81:2554–2561. doi: 10.1128/AEM.03748-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercan O, Wels M, Smid EJ, Kleerebezem M. 2015. Molecular and metabolic adaptations of Lactococcus lactis at near-zero growth rates. Appl Environ Microbiol 81:320–331. doi: 10.1128/AEM.02484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luesink EJ, Van Herpen RE, Grossiord BP, Kuipers OP, De Vos WM. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol 30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 19.Linares DM, del Río B, Ladero V, Redruello B, Martín MC, Fernández M, Alvarez MA. 2013. The putrescine biosynthesis pathway in Lactococcus lactis is transcriptionally regulated by carbon catabolic repression, mediated by CcpA. Int J Food Microbiol 165:43–50. doi: 10.1016/j.ijfoodmicro.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Aleksandrzak-Piekarczyk T, Stasiak-Różańska L, Cieśla J, Bardowski J. 2015. ClaR: a novel key regulator of cellobiose and lactose metabolism in Lactococcus lactis IL1403. Appl Microbiol Biotechnol 99:337–347. doi: 10.1007/s00253-014-6067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson U, Rådström P. 2002. Physiological function of the maltose operon regulator, MalR, in Lactococcus lactis. BMC Microbiol 2:28. doi: 10.1186/1471-2180-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckhardt TH, Skotnicka D, Kok J, Kuipers OP. 2013. Transcriptional regulation of fatty acid biosynthesis in Lactococcus lactis. J Bacteriol 195:1081–1089. doi: 10.1128/JB.02043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 24.Varmanen P, Ingmer H, Vogensen FK. 2000. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology 146:1447–1455. doi: 10.1099/00221287-146-6-1447. [DOI] [PubMed] [Google Scholar]
- 25.Flores-Kim J, Darwin AJ. 2016. The phage shock protein response. Annu Rev Microbiol 70:83–101. doi: 10.1146/annurev-micro-102215-095359. [DOI] [PubMed] [Google Scholar]
- 26.Darwin AJ. 2013. Stress relief during host infection: the phage shock protein response supports bacterial virulence in various ways. PLoS Pathog 9:e1003388. doi: 10.1371/journal.ppat.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toni T, Jovanovic G, Huvet M, Buck M, Stumpf MP. 2011. From qualitative data to quantitative models: analysis of the phage shock protein stress response in Escherichia coli. BMC Syst Biol 5:69. doi: 10.1186/1752-0509-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gueguen E, Savitzky DC, Darwin AJ. 2009. Analysis of the Yersinia enterocolitica PspBC proteins defines functional domains, essential amino acids and new roles within the phage-shock-protein response. Mol Microbiol 74:619–633. doi: 10.1111/j.1365-2958.2009.06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovee ML, Champagne KS, Demeler B, Francklyn CS. 2002. The quaternary structure of the HisZ-HisG N-1-(5′-phosphoribosyl)-ATP transferase from Lactococcus lactis. Biochemistry 41:11838–11846. doi: 10.1021/bi020243z. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, Zhang J, Kang Z, Du G, Yu X, Wang T, Chen J. 2013. Enhanced production of l-phenylalanine in Corynebacterium glutamicum due to the introduction of Escherichia coli wild-type gene aroH. J Ind Microbiol Biotechnol 40:643–651. doi: 10.1007/s10295-013-1262-x. [DOI] [PubMed] [Google Scholar]
- 31.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Wang Y, Song Y, Wang T, Xu S, Peng Z, Lin X, Zhang L, Shen X. 2013. A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ Microbiol 15:557–569. doi: 10.1111/1462-2920.12005. [DOI] [PubMed] [Google Scholar]
- 33.Fernández M, Zúñiga M. 2006. Amino acid catabolic pathways of lactic acid bacteria. Crit Rev Microbiol 32:155–183. doi: 10.1080/10408410600880643. [DOI] [PubMed] [Google Scholar]
- 34.Wu C, Zhang J, Du G, Chen J. 2013. Aspartate protects Lactobacillus casei against acid stress. Appl Microbiol Biotechnol 97:4083–4093. doi: 10.1007/s00253-012-4647-2. [DOI] [PubMed] [Google Scholar]
- 35.Wu C, Zhang J, Chen W, Wang M, Du G, Chen J. 2012. A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl Microbiol Biotechnol 93:707–722. doi: 10.1007/s00253-011-3757-6. [DOI] [PubMed] [Google Scholar]
- 36.Baker-Austin C, Dopson M. 2007. Life in acid: pH homeostasis in acidophiles. Trends Microbiol 15:165–171. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67:429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laws A, Gu Y, Marshall V. 2001. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol Adv 19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 39.Dertli E, Mayer MJ, Colquhoun IJ, Narbad A. 2015. EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb Biotechnol 9:496–501. doi: 10.1111/1751-7915.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolly L, Stingele F. 2001. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int Dairy J 11:733–745. doi: 10.1016/S0958-6946(01)00117-0. [DOI] [Google Scholar]
- 41.Ayabe-Chujo Y, Usami Y, Yoshida T, Omori T, Nojiri H. 2012. Membrane topology and functional analysis of Methylobacillus sp. 12S genes epsF and epsG, encoding polysaccharide chain-length determining proteins. Biosci Biotechnol Biochem 76:608–612. doi: 10.1271/bbb.110831. [DOI] [PubMed] [Google Scholar]
- 42.Ge X, Wang W, Han Y, Wang J, Xiong X, Zhang W. 2013. Methylovorus sp. MP688 exopolysaccharides contribute to oxidative defense and bacterial survival under adverse condition. World J Microbiol Biotechnol 29:2249–2258. doi: 10.1007/s11274-013-1391-4. [DOI] [PubMed] [Google Scholar]
- 43.Agrawal R, Saini DK. 2014. Rv1027c–Rv1028c encode functional KdpDE two-component system in Mycobacterium tuberculosis. Biochem Biophys Res Commun 446:1172–1178. doi: 10.1016/j.bbrc.2014.03.066. [DOI] [PubMed] [Google Scholar]
- 44.Freeman ZN, Dorus S, Waterfield NR. 2013. The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog 9:e1003201. doi: 10.1371/journal.ppat.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janulczyk R, Ricci S, Björck L. 2003. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun 71:2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun X, Baker HM, Ge R, Sun H, He QY, Baker EN. 2009. Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes. Biochemistry 48:6184–6190. doi: 10.1021/bi900552c. [DOI] [PubMed] [Google Scholar]
- 47.Bai G, Schaak DD, Smith EA, McDonough KA. 2011. Dysregulation of serine biosynthesis contributes to the growth defect of a Mycobacterium tuberculosis crp mutant. Mol Microbiol 82:180–198. doi: 10.1111/j.1365-2958.2011.07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leduc D, Gallaud J, Stingl K, de Reuse H. 2010. Coupled amino acid deamidase-transport systems essential for Helicobacter pylori colonization. Infect Immun 78:2782–2792. doi: 10.1128/IAI.00149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trötschel C, Kandirali S, Diaz-Achirica P, Meinhardt A, Morbach S, Krämer R, Burkovski A. 2003. GltS, the sodium-coupled l-glutamate uptake system of Corynebacterium glutamicum: identification of the corresponding gene and impact on l-glutamate production. Appl Microbiol Biotechnol 60:738–742. doi: 10.1007/s00253-002-1170-x. [DOI] [PubMed] [Google Scholar]
- 50.Martinussen J, Schallert J, Andersen B, Hammer K. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J Bacteriol 183:2785–2794. doi: 10.1128/JB.183.9.2785-2794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunin R, Glansdorff N, Pierard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji X. 2016. Structural insights into cell cycle control by essential GTPase Era. Postepy Biochem 62:335. [PMC free article] [PubMed] [Google Scholar]
- 53.Verstraeten N, Fauvart M, Versées W, Michiels J. 2011. The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev 75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karimova G, Dautin N, Ladant D. 2008. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 190:8248. doi: 10.1128/JB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Meer J, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, De Vos W. 1993. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol 175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol 60:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, Fu S, Liu M, Xu Q, Bei W, Chen H, Tan C. 2014. The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. Microbiol Res 169:541–546. doi: 10.1016/j.micres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Kingston AW, Liao X, Helmann JD. 2013. Contributions of the σW, σM, and σX regulons to the lantibiotic resistome of Bacillus subtilis. Mol Microbiol 90:502–518. doi: 10.1111/mmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martínez B, Zomer AL, Rodríguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol Microbiol 64:473–486. doi: 10.1111/j.1365-2958.2007.05668.x. [DOI] [PubMed] [Google Scholar]
- 61.Eldholm V, Gutt B, Johnsborg O, Brückner R, Maurer P, Hakenbeck R, Mascher T, Håvarstein LS. 2010. The pneumococcal cell envelope stress-sensing system LiaFSR is activated by murein hydrolases and lipid II-interacting antibiotics. J Bacteriol 192:1761–1773. doi: 10.1128/JB.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ni Z-J, Zhang X-y, Liu F, Wang M, Hao R-h, Ling P-x, Zhu X-Q. 2017. Effect of co-overexpression of nisin key genes on nisin production improvement in Lactococcus lactis LS01. Probiotics Antimicrob Proteins 9:204–212. doi: 10.1007/s12602-017-9268-8. [DOI] [PubMed] [Google Scholar]
- 63.AlKhatib Z, Lagedroste M, Fey I, Kleinschrodt D, Abts A, Smits SH. 2014. Lantibiotic immunity: inhibition of nisin-mediated pore formation by NisI. PLoS One 9:e102246. doi: 10.1371/journal.pone.0102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.AlKhatib Z, Lagedroste M, Zaschke J, Wagner M, Abts A, Fey I, Kleinschrodt D, Smits SH. 2014. The C terminus of nisin is important for the ABC transporter NisFEG to confer immunity in Lactococcus lactis. Microbiologyopen 3:752–763. doi: 10.1002/mbo3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hiron A, Falord M, Valle J, Débarbouillé M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol 81:602–622. doi: 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 66.Ohki R, Tateno K, Masuyama W, Moriya S, Kobayashi K, Ogasawara N. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol 49:1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Liu S, Du Y, Feng W, Liu J, Qiao J. 2014. Genome shuffling of Lactococcus lactis subspecies lactis YF11 for improving nisin Z production and comparative analysis. J Dairy Sci 97:2528–2541. doi: 10.3168/jds.2013-7238. [DOI] [PubMed] [Google Scholar]
- 68.Lin Z, Zhang Y, Wang J. 2013. Engineering of transcriptional regulators enhances microbial stress tolerance. Biotechnol Adv 31:986–991. doi: 10.1016/j.biotechadv.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 69.Tramer J, Fowler G. 1964. Estimation of nisin in foods. J Sci Food Agric 15:522–528. doi: 10.1002/jsfa.2740150802. [DOI] [Google Scholar]
- 70.Cherney LT, Cherney MM, Garen CR, James MN. 2010. Crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression. J Mol Biol 399:240–254. doi: 10.1016/j.jmb.2010.03.065. [DOI] [PubMed] [Google Scholar]
- 71.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Xie ZX, Li BZ, Mitchell LA, Wu Y, Qi X, Jin Z, Jia B, Wang X, Zeng BX, Liu HM. 2017. “Perfect” designer chromosome V and behavior of a ring derivative. Science 355:eaaf4704. doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 74.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.