Abstract
Colon cancer is one of common cancer in the world and glycolysis is one of the major problems in colon cancer therapy. MicroRNAs (miRNAs) involve in colon cancer progression. Sushi repeat-containing protein X-linked 2 (SRPX2) is associated with poor prognosis in some cancer patients, however, the role of SRPX2 including glycolytic metabolism regulated by miRNAs is unclear in colon cancer. So, the purpose of the present study is to elucidate the underlying mechanism in colon cancer metabolism mediated by SRPX2. Our results revealed that miR-192-5p (miR-192) and miR-215-5p (miR-215) inhibited glycolysis by regulating SRPX2 expression in colon cancer cells. We also found that miR-192 and miR-215 were both regulated by PI3K-Akt. Our results indicate that SRPX2 facilitates colon cancer cell glycolysis by miR-192 and miR-215, which are down-regulated by PI3K-Akt.
Keywords: SRPX2, miR-192, miR-215, colon cancer, PI3K-Akt
Introduction
Sushi repeat protein X-linked 2 (SRPX2), named as a sushi-repeat protein, was first identified in pro-B leukemia cells [1]. SRPX2 acts as a ligand for the urokinase plasminogen activator surface receptor, which plays a role in angiogenesis by inducing endothelial cell migration and the formation of vascular network [1,2]. SRPX2 is commonly overexpressed in many types of cancer such as glioblastoma [3,4], pancreatic cancer [5], colorectal cancer [6,7] and gastric cancer [8,9]. SRPX2 enhances cellular migration and adhesion in gastric cancer cells and the conditioned-medium obtained from SRPX2-producing cells promotes cell migration and adhesion [1,2]. SRPX2 is also associated with poor prognosis in cancer patients. SRPX2 overexpression promotes interactions between tumor cells and endothelial cells and further accelerates tumor progression and metastasis [2]. All these reported data suggest that SRPX2 functions as an oncogene in cancer.
Colon cancer is a common and aggressive digestive tumor in the world and the major problems for colon cancer therapy include chemotherapy resistance, glycolytic state and metastasis. It is necessary to elucidate the underlying mechanism in colon cancer glycolysis. MicroRNAs (miRNAs) play an important role in regulating colon cancer [10-12]. Based on the background, we hypothesize that SRPX2 regulates colon cancer cell proliferation and glycolysis via miRNAs regulated by PI3K-Akt. In this study, we investigated and confirmed that SRPX2 was regulated by PI3K-Akt in colon cancer cells. Cellular functions including cell growth and glycolysis were performed for verify the role of SRPX2 in colon cancer cells. Further studies verified that SRPX2 was a target gene of miR-192-5p (miR-192)/miR-215-5p (miR-215), which were negatively regulated by PI3K-Akt. It was also confirmed that SRPX2 expression was negatively related with the levels of miR-192/miR-215, which are the down-stream miRNAs of PI3K-Akt.
Materials and methods
Cell lines and cell culture
Human colon cell lines (HCT8 and SW116) were obtained from Shanghai Institute of Biochemistry and Cell Bank at the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in Dulbecco’s modified eagle medium (Invitrogen, USA) with 10% fetal bovine serum (FBS), streptomycin (100 U/mL) and penicillin (100 U/mL). Cells were cultured under a condition of a humidified atmosphere of 5% carbon dioxide at 37°C.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA and miRNA was extracted from the colon cancer cells using Trizol (Invitrogen, CA, USA) based on the protocol. RNA was firstly synthesized to cDNA (Takara, Japan). For mRNA and miRNA expression detection, real time RT-PCR was performed usinga sequence detector (ABI-Prism, Applied Biosystems) following the instructions. The relative expression levels were calculated by the 2-ΔΔCq method. All data were normalized to β-actin for mRNA and U6 for miRNA.
Luciferase assay
The wide-type or mutated type of SRPX2 3’-UTR was cloned into PGL3 firefly luciferase reporter vector. The luciferase activity was assayed using dual luciferase assay system (Promega, USA) according to the protocol.
Cell proliferation
The Cell Counting Kit-8 (CCK-8) was used to determine cell viability according to the manufacturer’s instructions.
Western blot analysis
The cell lysates were separated by SDS-PAGE and transferred to PVDF membranes (Millipore, Bedford, MA). After blocking in 5% non-fat dry milk for 1 h at room temperature, the membranes were incubated with primary antibodies for 2 h at room temperature, and then incubated with the secondary antibodies conjugated for 1 h at room temperature. Protein bands were visualized on X-ray film using an automatic imager.
Glucose consumption and lactate production
The supernatants from the cultured cells with SRPX2 siRNAs or miR-192/miR-215 transfection were used for glucose consumption and lactate production measurement. Metabolite concentrations were quantified according to the protocols of glucose consumption and lactate production (Invitrogen). Glucose consumption and lactate production were normalized to protein content using the Pierce BCA Protein assay (Thermo Scientific).
Patients and tissue specimens
Primary human colon cancer samples were obtained from patients undergoing tumor resection. Informed consent was obtained from Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute (Shenyang, China).
Statistical analysis
All the data were analyzed using the SPSS 15.0 software (Chicago, IL, USA) or Excel. All the in vitro experiments were done for three dependent times. Data are presented as mean and standard error (mean ± SEM). Student’s t-test was used for comparing means between two groups. Statistical significance was analyzed by one-way ANOVA in three or more groups. In all experiments, a p-value of 0.05 or less was considered statistically significant.
Results
PI3K-Akt regulates SRPX2 expression in colon cancer cells
To determine whether SRPX2 is regulated by PI3K-Akt in colon cancer cells, SW116 and HCT8 cells were treated with a PI3K inhibitor, LY294002 and then PI3K down-stream genes were examined by western blotting. It was found that SRPX2 protein levels were decreased in colon cancer cells after inhibiting PI3K (Figure 1A), so did in the cells with Akt1/2 siRNAs transfection (Figure 1B). Next, SRPX2 mRNA levels was examined in the colon cancer cells. SW116 and HCT8 cells were treated with LY294002 or transfected with Akt1/2 siRNAs and it was shown that inhibition of PI3K-Akt decreased SRPX2 mRNA level in colon cancer cells significantly (Figure 1C and 1D). The data indicate that SRPX2 is a down-stream molecule of PI3K-Akt in colon cancer cells.
Figure 1.
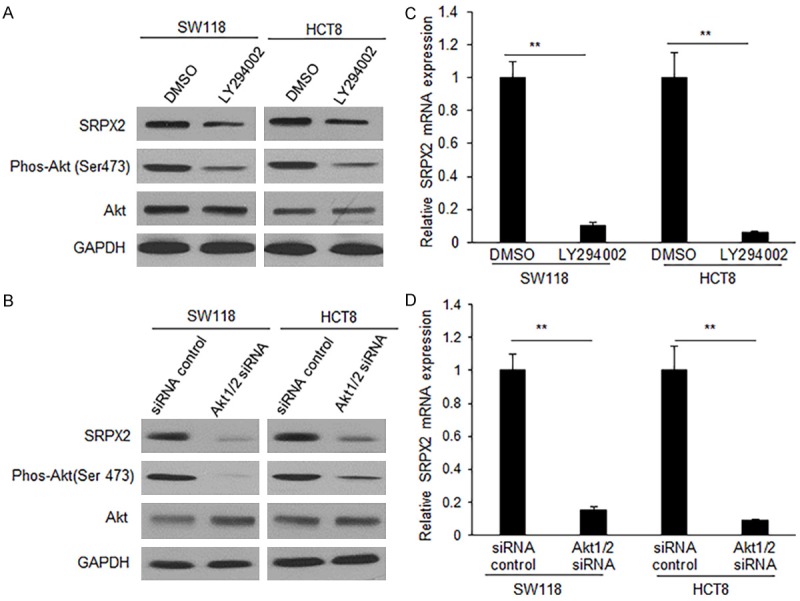
PI3K-Akt up-regulates SRPX2 expression in colon cancer cells. A. SRPX2 expression in SW116 and HCT8 cells. The cells were treated with LY294002 (20 uM) for 24 h and then protein was extracted for western blotting analysis. B. SRPX2 expression in SW116 and HCT8 cells. The cells were transfected with Akt1/2 siRNAs (100 uM) for 48 h and then protein was extracted for western blotting analysis. C. SRPX2 expression in SW116 and HCT8 cells. The cells were treated with LY294002 (20 uM) for 24 h and then total RNA was extracted for real time RT-PCR. D. SRPX2 expression in SW116 and HCT8 cells. The cells were transfected with Akt1/2 siRNAs (100 uM) for 48 h and then total RNA was extracted forreal time RT-PCR. **P<0.01 compared with the control.
MiR-192/miR-215 regulates SRPX2 expression in colon cancer cells
To further explore the molecular mechanism of PI3K-Akt regulating SRPX2 expression in colon cancer cells, we used the online prediction tools to find miRNAs that may regulate SRPX2 expression. MiR-192 and miR-215 were among the predicted miRNAs which may regulate SRPX2 expression (Figure 2A). When SW116 or HCT8 cells were transfected with miR-192 and miR-215 mimics, miR-192 and miR-215 levels were both increased (Figure 2B). When SW116 cells were co-transfected with the SRPX2 3’UTR and miR-192/miR-215, we found that the luciferase activity of SRPX2 3’UTR (wide type) was suppressed significantly comparing to the luciferase activity in the cells with mutated SRPX2 3’UTR transfection (Figure 2C). To further validate the result, we examined whether or not miR-192/miR-215 could influence the expression of SRPX2. It was found that the expression of SRPX2 mRNA and protein decreased in SW116 or HCT8 cells with miR-192/miR-215 transfection compared with the control (Figure 2D and 2E). Collectively, these findings suggested that miR-192 and miR-215 regulated the expression of SRPX2 in colon cancer cells post-transcriptionally.
Figure 2.
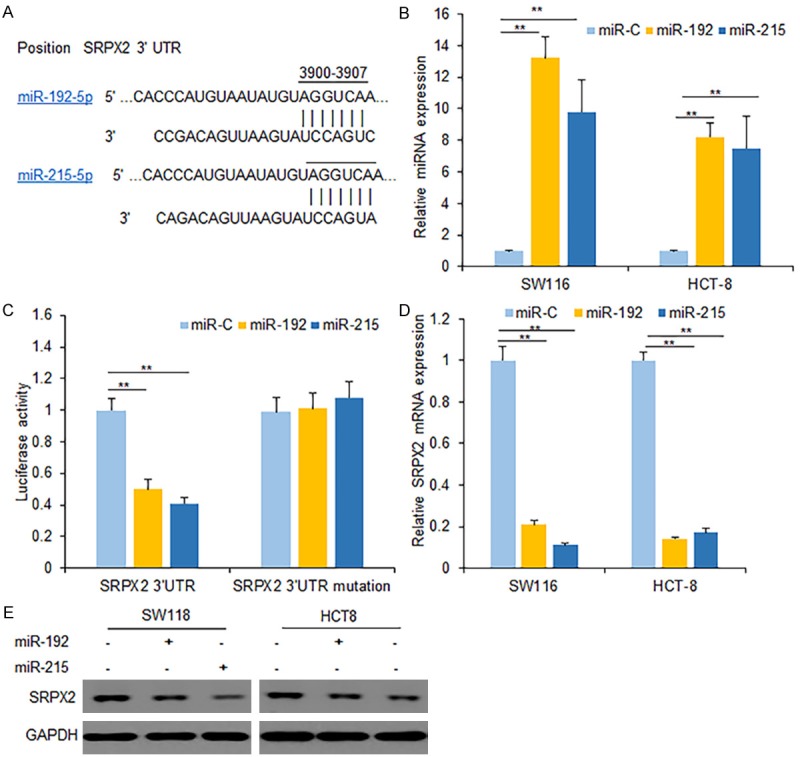
MiR-192/miR-215 regulates SRPX2 expression in colon cancer cells. (A) miR-192 and miR-215 are the predicted miRNAs targeting SPRX2. (B) miR-192 and miR-215 expression increased in colon cancer cells. SW116 and HCT-8 cells were transfected miR-192 and miR-215 for 48 h, and then total RNA was extracted for real time RT-PCR. (C) Relative luciferase activity of the indicated SRPX2 reporter construct in SW116 cells. SW116 cells were co-transfected with SRPX2 promoter plasmid or miR-192 and miR-215 for 24 h. The cells were collected according to the protocol and analyzed. (D) miR-192 and miR-215 expression increased in colon cancer cells. SW116 and HCT8 cells were transfected miR-192 and miR-215 for 48 h, and then total RNA was extracted for SPRX2 mRNA analysis by real time RT-PCR. (E) Western blot assays were performed to detect the expression of SRPX2 upon transfection with miR-192 and miR-215 in SW116 and HCT8 cells. SW116 and HCT8 cells were transfected miR-192 and miR-215 for 48 h, and then total protein was extracted for SPRX2 protein analysis by western blotting. **P < 0.01 compared with the control.
MiR-192 or miR-215 expression is negatively related to SRPX2 expression in colon cancer samples
To know the relationship between miR-192 or miR-215 levels with SRPX2 expression in colon cancer samples, we detected SRPX2 expression in 78 pairs of colon cancer tissues by qRT-PCR. Most of colon cancer tissues exhibited overexpression of SRPX2 (Figure 3A) and down-regulation of both miR-192 and miR-215 comparing with the matched normal tissues (Figure 3B and 3C). It was also shown that SRPX2 mRNA was negatively correlated with miR-192 and miR-215 expression in most of colon cancer samples (Figure 3D and 3E). These data confirmed that SRPX2 might play important roles in colon cancer, which was regulated by miR-192 and miR-215.
Figure 3.
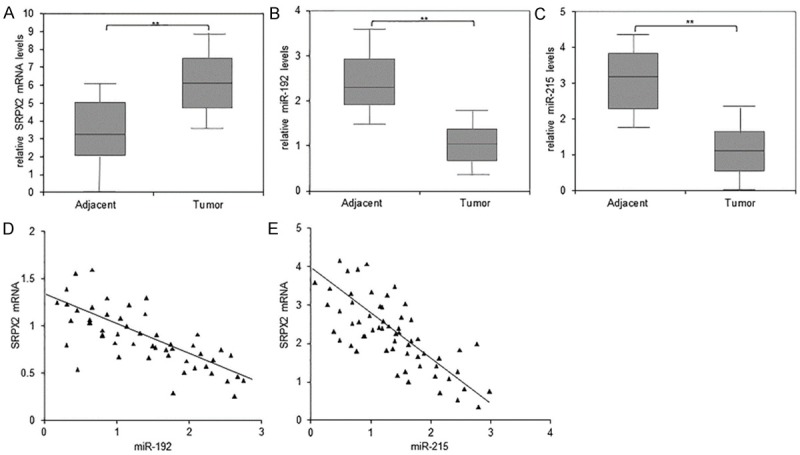
MiR-192 or miR-215 expression is related to SRPX2 mRNA levels in colon cancer samples negatively. A. The expression of SRPX2 mRNA in 49 pairs of colon cancer tissues and adjacent normal tissues. SPRX2 mRNA was examined by real time RT-PCR. B, C. The expression of miR-192 and miR-215 in 49 pairs of colon cancer tissues and adjacent normal tissues. MiR-192 and miR-215 levels was examined by real time RT-PCR. D. Relationship between miR-192 and SRPX2 expression in colon cancer tissues. E. Relationship between miR-215 and SRPX2 expression in colon cancer tissues. **P<0.01 compared with normal tissues.
SRPX2 promotes cell proliferation regulated by miR-192/miR-215
To explore the roles of miR-192 and miR-215 in colon cancer cells, cells were transfected with miR-192 or miR-215 or SRPX2 siRNA to observe cell survival ability and glycolyticmetabolism. As expected, the level of SRPX2 decreased efficiently when SRPX2 siRNA was transfected into SW116 and HCT8 cells (Figure 4A). Inhibiting SRPX2 expression decreased cell proliferation (Figure 4B and 4C) in SW116 and HCT8 cells infected with SRPX2 siRNA. We also observed that miR-192 or miR-215 decreased ATP and lactate production in SW116 and HCT8 cells (Figure 4D and 4E). These results demonstrated that miR-192 or miR-215 suppressed colon cancer cell proliferation and cell metabolism via SRPX2.
Figure 4.
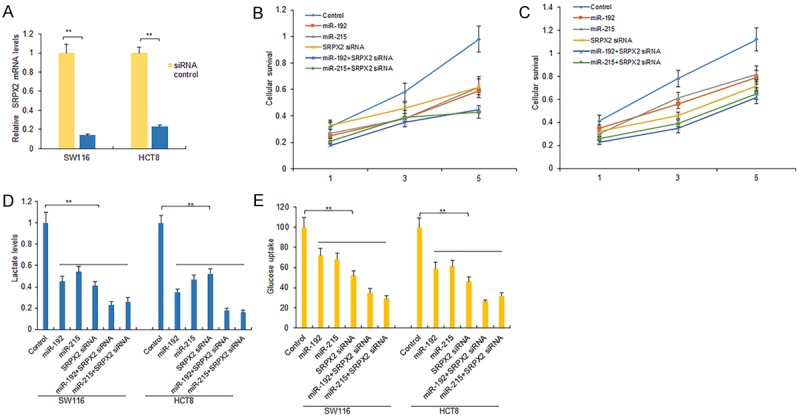
SRPX2 regulates cell proliferation and metabolism via miR-192 or miR-215. A. The expression of SRPX2 mRNA in SW116 and HCT8 cells. SW116 and HCT8 cells were transfected with SRPX2 siRNA or siRNA controls and total RNA was isolated for real time RT-PCR analysis. B, C. CCK-8 assay was used to detect the proliferation. SW116 and HCT8 cells transfected with miR-192 or miR-215 or SRPX2 siRNA for 24 h. Cell survival rates were assayed by CCR8 at the day 1, 3 and 5. D. Glucose uptake in SW116 and HCT8 cells. Cells were transfected with miR-192 or miR-215 or SRPX2 siRNA for 48 h and glucose uptake was assayed using glucose consumption kit. E. Lactate production in SW116 and HCT8 cells. Cells were transfected with miR-192 or miR-215 or SRPX2 siRNA for 48 h. Lactate was measured using lactate production assay kit. **P<0.01 compared with the controls.
PI3K-Akt down-regulates miR-192/215 in colon cancer cells
Previous reports show that various signal pathways such as PI3K-Akt involving in regulating miRNA expression. To know the correlation of PI3K-Akt and miR-192/215 expression in colon cancer cells, colon cancer cells were treated with LY294002 and miR-192 and miR-215 expression was measured using qRT-PCR. MiR-192/215 increased in SW116 and HCT8 cells treated with LY294002 compared with the cells treated with DMSO (Figure 5A and 5B). It was also shown that miR-192/215 increased in the colon cancer cells with Akt1/2 siRNA transfection (Figure 5C and 5D). These data indicated that PI3K-Akt negatively regulated miR-192/215 in colon cancer.
Figure 5.
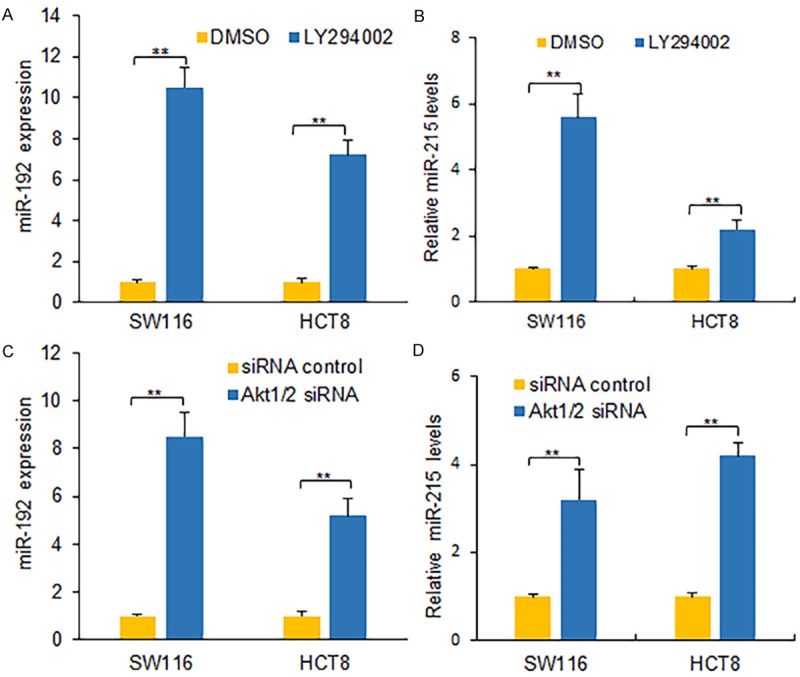
MiR-192 and miR-215 are regulated by PI3K/Akt in colon cancer cells. A, B. miR-192 and miR-215 expression in colon cancer cells. SW116 and HCT8 cells were seeded in 6-well plates and treated with LY294002 (20 uM) for 24 h and then miR-192 or miR-215 expression levels were examined by real time RT-PCR. C, D. miR-192 and miR-215 expression in colon cancer cells. SW116 and HCT8 cells were seeded in 6-well plates and transfected with Akt1/2 siRNAs for 24 h and then miR-192 or miR-215 expression levels were examined by real time RT-PCR. **P<0.01 compared with the controls.
Discussion
The reports showed that SRPX2, an oncogene, is involved in the progression of several types of cancer including glioblastoma [3,4], pancreatic cancer [5] and colorectal cancer [6], gastrointestinal cancer [8,9]. A report shows that SRPX2 is overexpressed in glioblastoma, and can promote epithelial-mesenchymal transition, which suggests SRPX2 involves in glioblastoma metastasis [3]. SRPX2 promotes pancreatic and colorectal cancer metastasis [5,6]. In addition, the published reports show that there are other cancer cellular functions on SRPX2 inducing temozolomide resistance. It is reported that SRPX2 is regulated by MAPK [3], FAK [5], Wnt/β-catenin [6] and miRNAs such as miR-149 [7]. SPRX2 is a potential down-stream gene of the PI3K-Akt pathway based on our previous genome array. Here, we identified that SRPX2 is regulated by PI3K-Akt and further study verified that it was a target gene of both miR-192 and miR-215, which were down-regulated by PI3K-Akt in colon cancer cells.
To verify that PI3K-Akt regulates SRPX2 expression in colon cancer, cells were treated with a PI3K inhibitor or transfected with Akt1/2 siRNAs, and when PI3K-Akt was effectively suppressed, SRPX2 levels decreased in SW116 and HCT8 cells. The experiments demonstrate that SRPX2 is a down-stream gene of PI3K-Akt. MiRNAs are a class of non-coding RNA with short length and involve in many tumor aggressive processes such as cell proliferation, metastasis, metabolism and others. In order to find the new miRNAs targeting SRPX2, potential miRNAs are predicted using TargetScan and miRBase. We found that miR-192 and miR-215 were among the most potential miRNAs that regulated SRPX2 expression in colon cancer cells. The findings identified that both miR-192 and miR-215 regulated SRPX2 expression based on the results from luciferase assay, real time RT-PCR and western blotting. A sequence complementary to miR-192 and miR-215 was identified in the 3’UTR of the SRPX2 mRNA sequence, and introduction of miR-192 and miR-215 to the colon cancer cells led to a significant reduction in SRPX2 expression at mRNA and protein levels. Previous studies show that miR-192 [13-16] and miR-215 [17-21] are tumor suppressors in many kinds of cancer, here, we verified that SRPX2 is new target gene of miR-192 and miR-215.
The relationship between SRPX2 expression and miR-192/miR-215 expression in colon cancer tissue was analyzed, which elucidated that SPRX2 levels was negatively correlated to miR-192 or miR-215 levels. SRPX2 expression is overexpressed in colon cancer tissues in comparison with adjacent normal tissues. In addition, we found that the introduction of miR-192 or miR-215 into the colon cancer cells could inhibit cell proliferation, lactate production and glucose uptake, which were strengthened by down-regulation of SRPX2. The evidence indicates that SRPX2 benefits to colon cancer growth through the negative control of miR-192 and miR-215 expression. Further research elucidates that miR-192 and miR-215 expression are enhanced by inhibition of PI3K-Akt. Our study also confirmed that miR-192 and miR-215 are tumor suppressors in colon cancer.
In a conclusion, our study found a new molecular mechanism of SRPX2 regulation in colon cancer cells. Our study elucidated the following points: (1) SRPX2 is overexpressed in colon cancer including cells and tissues and is a down-stream gene of PI3K-Akt; (2) SRPX2 is regulated by miR-192 and miR-215 in colon cancer; (3) PI3K-Akt decreases miR-192 and miR-215 expression in colon cancer cells; (4) Suppression of SRPX2 inhibits cell proliferation and glycolysis in colon cancer due to lack of miR-192 and miR-215 expression in colon cancer. There needs further investigation to evaluate SRPX2, miR-192 and miR-215 as the potential targets in colon cancer.
Acknowledgements
The study was supported by Clinical Capability Construction Project for Liaoning Provincial Hospitals (China).
Disclosure of conflict of interest
None.
References
- 1.Royer-Zemmour B, Ponsole-Lenfant M, Gara H, Roll P, Lévêque C, Massacrier A, Ferracci G, Cillario J, Robaglia-Schlupp A, Vincentelli R, Cau P, Szepetowski P. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum Mol Genet. 2008;17:3617–3630. doi: 10.1093/hmg/ddn256. [DOI] [PubMed] [Google Scholar]
- 2.Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, Massacrier A, Valenti MP, Roeckel-Trevisiol N, Jamali S, Beclin C, Seegmuller C, Metz-Lutz MN, Lemainque A, Delepine M, Caloustian C, de Saint Martin A, Bruneau N, Depétris D, Mattéi MG, Flori E, Robaglia-Schlupp A, Lévy N, Neubauer BA, Ravid R, Marescaux C, Berkovic SF, Hirsch E, Lathrop M, Cau P, Szepetowski P. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 3.Tang H, Zhao J, Zhang L, Zhao J, Zhuang Y, Liang P. SRPX2 enhances the epithelial-mesenchymal transition and temozolomide resistance in glioblastoma cells. Cell Mol Neurobiol. 2016;36:1067–76. doi: 10.1007/s10571-015-0300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung NA, Eiholzer RA, Kirs S, Zhou J, Ward-Hartstonge K, Wiles AK, Frampton CM, Taha A, Royds JA, Slatter TL. Telomere profiles and tumor-associated macrophages with different immune signatures affect prognosis in glioblastoma. Mod Pathol. 2016;29:212–226. doi: 10.1038/modpathol.2015.156. [DOI] [PubMed] [Google Scholar]
- 5.Gao Z, Zhang J, Bi M, Han X, Han Z, Wang H, Ou Y. SRPX2 promotes cell migration and invasion via FAK dependent pathway in pancreatic cancer. Int J Clin Exp Pathol. 2015;8:4791–4798. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu KL, Wu J, Zhou Y, Fan JH. Increased Sushi repeat-containing protein X-linked 2 is associated with progression of colorectal cancer. Med Oncol. 2015;32:99. doi: 10.1007/s12032-015-0548-4. [DOI] [PubMed] [Google Scholar]
- 7.Øster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET, Sandoval J, Moran S, Esteller M, Hansen TF, Lamy P COLOFOL steering group. Laurberg S, Ørntoft TF, Andersen CL. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer. 2013;132:2303–15. doi: 10.1002/ijc.27921. [DOI] [PubMed] [Google Scholar]
- 8.Yamada T, Oshima T, Yoshihara K, Sato T, Nozaki A, Shiozawa M, Ota M, Yoshikawa T, Akaike M, Numata K, Rino Y, Kunisaki C, Tanaka K, Imada T, Masuda M. Impact of overexpression of Sushi repeat-containing protein X-linked 2 gene on outcomes of gastric cancer. J Surg Oncol. 2014;109:836–840. doi: 10.1002/jso.23602. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka K, Arao T, Maegawa M, Matsumoto K, Kaneda H, Kudo K, Fujita Y, Yokote H, Yanagihara K, Yamada Y, Okamoto I, Nakagawa K, Nishio K. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124:1072–1080. doi: 10.1002/ijc.24065. [DOI] [PubMed] [Google Scholar]
- 10.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–92. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149:1204–1225. e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng W, Feng J, Qin H, Ma Y, Goel A. An update on miRNAs as biological and clinical determinants in colorectal cancer: a bench-to-bedside approach. Future Oncol. 2015;11:1791–808. doi: 10.2217/fon.15.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krattinger R, Boström A, Schiöth HB, Thasler WE, Mwinyi J, Kullak-Ublick GA. microRNA-192 suppresses the expression of the farnesoid X receptor. Am J Physiol Gastrointest Liver Physiol. 2016;310:G1044–1051. doi: 10.1152/ajpgi.00297.2015. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Fan Z, Lu S, Yang J, Hao T, Huo Q. MiR-192 suppresses the tumorigenicity of prostate cancer cells by targeting and inhibiting nin one binding protein. Int J Mol Med. 2016;37:485–492. doi: 10.3892/ijmm.2016.2449. [DOI] [PubMed] [Google Scholar]
- 15.Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, Wang Q, Zhao F, Li J, Yao M, Qin L, Liang L, He X. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7:2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C, Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene. 2014;33:5332–5340. doi: 10.1038/onc.2013.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li N, Zhang QY, Zou JL, Li ZW, Tian TT, Dong B, Liu XJ, Ge S, Zhu Y, Gao J, Shen L. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7:4817–4828. doi: 10.18632/oncotarget.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A, Girgis H, Von Both I, Bjarnason GA, Yousef GM. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis. 2013;34:2231–2239. doi: 10.1093/carcin/bgt184. [DOI] [PubMed] [Google Scholar]
- 19.Deng Y, Huang Z, Xu Y, Jin J, Zhuo W, Zhang C, Zhang X, Shen M, Yan X, Wang L, Wang X, Kang Y, Si J, Zhou T. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342:27–35. doi: 10.1016/j.canlet.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Senanayake U, Das S, Vesely P, Alzoughbi W, Fröhlich LF, Chowdhury P, Leuschner I, Hoefler G, Guertl B. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis. 2012;33:1014–1021. doi: 10.1093/carcin/bgs126. [DOI] [PubMed] [Google Scholar]
- 21.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]


