Abstract
A keloid is the process of skin healing, collagen synthesis and metabolism of the loss of normal control in a sustained hyperactive state, resulting in excessive proliferation of collagen fibers. A large-scale genome-wide association study (GWAS) has identified multiple single nucleotide polymorphisms (SNPs) in the 3q22.3 loci that are associated with keloids in a Japanese population. However, the associations of SNPs in 3q22.3 with keloids were not confirmed in a selected Chinese population by a replication study. Thus, in the present study, the relationships between keloids and 3q22.3 were assessed in another independent Chinese Han population, including 309 keloid patients and 1080 control subjects. The results displayed that rs940187 was associated with keloids (OR=1.88, 95% CI 1.27-2.78, P=1.35E-3) and remained significant after Bonferroni’s correction for multiple testing, while rs1511412 showed only a trend association (OR=2.23, 95% CI 1.09-4.55, P=0.02) with keloids. In addition, we subsequently checked the annotation datasets for rs940187 with eQTLs and obtained two hits, trans-proteins SLC7A9 and LEMD3, with significant P values less than 1e-4. In summary, genomic risk variants at 3q22.3 are associated with keloids in a Chinese Han population and contribute to the development and deterioration of the keloids, together with environmental factors.
Keywords: Keloids, SNP, association
Introduction
Keloids are benign proliferations of scar tissue that develop as a response to trauma to the dermis and are caused by aberrations in the wound healing process in predisposed individuals. Keloids are characterized by growth beyond the borders of the original scar and lack spontaneous regression [1]. Keloids were first described in 1806 and called “Cheloide” from the Greek word “Chele”, meaning crab’s claw, because of the sideways growth of the scar into normal skin [2]. Apart from esthetic concerns, they may also give significant subjective symptoms such aspain, pruritus and objective symptoms, such as deformations and contractures, and may significantly decrease the quality of life [3,4]. The frequency of keloids rises with the level of skin pigmentation; the reported incidence of keloids in the general population ranges from 0.09% in England to 16% among adults in Zaire [5]. Pathogenesis of keloids is complex and not fully understood. A number of hypotheses, such as high skin tension [6,7], hypoxia [8], endocrine dysfunction [9], fatty acids [10], autoimmunity [11] and genetic [12,13] hypotheses have been proposed in the past to explain the keloid phenomenon. Genetic factors and skin trauma in typical localization seem to play the crucial role [14]. The treatment of keloids is challenging, and an optimal method of treatment has not yet been established. Since the etiopathology of keloid formation remains unclear, and the treatment protocols do not seem to be effective, there is still a need for further research concerning keloids. Recently, a large-scale genome-wide association study (GWAS) identified multiple single nucleotide polymorphisms (SNPs) in the 3q22.3 loci that are associated with keloids in a Japanese population [15]. However, the associations of SNPs in 3q22.3 with keloids were not confirmed in a selected Chinese population by a replication study [16]. Thus, in the present study, the relationships between keloids and 3q22.3 were assessed in another independent Chinese Han population.
Material and methods
Patients
309 keloid patients and 1080 control subjects were recruited from the Department of Dermatology, Anhui provincial hospital. All keloid patients were diagnosed by at least two experienced dermatologists, and all controls were healthy individuals without keloids, immune diseases, neoplastic diseases or other family history of genetic diseases. All the cases and controls were collected using uniform criteria, and their clinical information was collected using the structured questionnaire. This prospective study was approved by the Ethical Committee of the Anhui Provincial Hospital. The tenets of the Declaration of Helsinki were performed, and written informed consent was provided by all patients and healthy control subjects.
Genomic DNA extraction and SNP genotyping
By using a Qiagen kit (Hilden, Germany), the genomic DNA of patients and healthy controls was extracted from peripheral blood samples, which were stored in a -80°C freezer. Two SNPs selected from a previous reported GWAS analysis of the 3q22.3 region, rs1511412 and rs940187, were genotyped using the Sequenom iPlex platform (Sequenom, Inc, San Diego, CA, USA) at the State Key Laboratory Incubation of Dermatology, Ministry of Science and Technology, Hefei, Anhui, China.
Statistical analysis
The exact test statistics for Hardy Weinberg equilibriums were calculated by PLINK (version 1.07). Power for detection of a positive association was estimated using the Power and Sample (PS) size calculation program (PS version 3.1.2). An association test was performed between an SNP and keloids by using Chi-squared tests, while Pearson’s 2*2 contingency tables were implemented in PLINK. Given that the known distance between the two SNPs is too close, we checked the linkage disequilibrium (LD) pattern of the 3q22.3 region based on a two-marker EM estimate that the maximum-likelihood values of the four gamete frequencies in a population of Chinese and Japanese ancestry with Haploview software (version 4.2), and the squared correlation (R squared value) based on the genotypic allele counts between these two SNPs was calculated within our own data by using the PLINK program. While there is some evidence of LD between rs1511412 and rs940187, we implemented a Bayesian statistical method for reconstructing haplotypes from these two SNPs by using the PHASE (version 2.1) program, then using local script to reform the data and perform association analysis.
Bioinformatics analysis
Variants were functionally annotated by an open source program, ANNOVA, with three types of databases. Expression quantitative trait loci (eQTLs) research was performed in molQTL (http://preview.ncbi.nlm.nih.gov/gap/eqtl/studies/), and the regulome database score was calculated using the RegulomeDB online website (http://www.regulomedb.org/index). Interaction between protein and long non-coding RNA was predicted by starBase v2.0 (http://starbase.sysu.edu.cn/). We found all proteins that were predicted to relate with each one of the SNPs and subsequently, performed GO annotation and pathway analysis using KOBAS V3.0 (http://kobas.cbi.pku.edu.cn/index.php) and clusterProfiler (version 3.4.4).
Results
The comparison of the patients with keloids and the controls revealed that rs940187 was found to be associated with keloids (OR=1.88, 95% CI 1.27-2.78, P=1.35E-3), and this finding remained significant after Bonferroni’s correction for multiple comparisons, while rs1511412 showed only a trend association (OR=2.23, 95% CI 1.09-4.55, P=0.02) with keloids, which is likely due to the detective power not being large enough (power =31.8) (Table 1). An association plot of region 3q22.3 (Figure 1) is constructed using the Locus Zoom website (http://locuszoom.org/).
Table 1.
Association between single-nucleotide polymorphisms and keloids susceptibility
| SNP | Chr | Gene | Allele (risk) | Oddsratio (95% CI) | P value | Power |
|---|---|---|---|---|---|---|
| rs1511412 | 3q22.3 | FOXL2 | G/A (A) | 2.23 (1.09-4.55) | 2.37E-2 | 31.8 |
| rs940187 | 3q22.3 | BPESC1 | G/A (A) | 1.88 (1.27-2.78) | 1.35E-3 | 80 |
Abbreviations: CI, confidence interval; SNP, single-nucleotide polymorphism.
Figure 1.
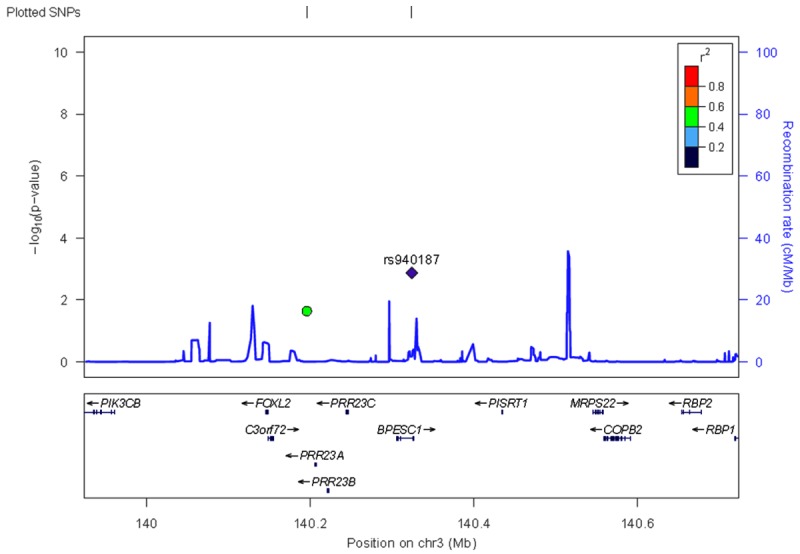
Regional association plots showing signals in replication samples for 3q22.3. The P-values of SNPs (shown as -log10 values in y-axis, from the genome-wide single-marker association analysis) were plotted against their map positions (x-axis). Gene annotations were adapted from the University of California at SantaCruz Genome Browser (http://genome.ucsc.edu/).
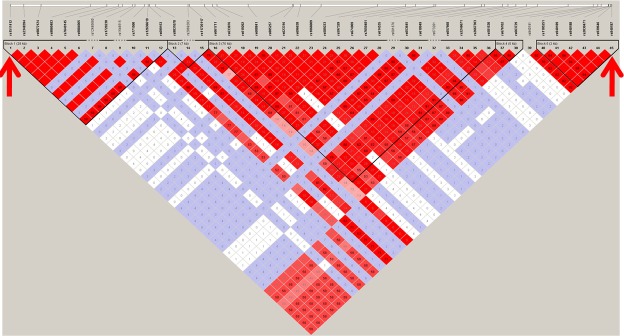
While the positions of the two variants are too close, we checked the LD pattern of the 3q22.3 region by using a HapMap data of Chinese and Japanese populations. The r2 calculated between these two SNPs is 0.54 (Figure 2). Then, we also checked the LD information between the two SNPs of the Asian race from the data of 1000 genomes, where the r2 value is close to 0.5 (HaploReg version 4.1). In addition, in our own data, the calculated r2 value is 0.2013. These results imply that the two polymorphisms might be in LD in a Chinese Han population. Therefore, we followed up with a haplotype analysis to analyze the association between haplotypes constructed by these two SNPs and keloids; the protective haplotype of GG demonstrated a more significant association than each SNP alone (Table 2). These findings supported our hypothesis that genetic variants at the 3q22.3 loci may contribute to keloid susceptibility in a Chinese Han population.
Figure 2.
The LD pattern of the 3q22.3 region in populations of Chinese and Japanese ancestry. The LD pattern (r2) was created in the Haploview by using the genotyping data (only SNPs with MAF > 0.01) from the HapMap project. The positions of the two SNPs (rs1511412, rs940187) identified in this study are indicated by red arrows. It shows there were several independent LD blocks in the entire region, and the high r2 value (0.58) indicates these two SNPs were in LD.
Table 2.
Haplotype association analysis
| Haplotype | A1 | F_A | F_U | A2 | P | Oddsratio |
|---|---|---|---|---|---|---|
| H1 | GA | 0.0356 | 0.0204 | - | 1.25E-02 | 1.77 (1.12-2.79) |
| H2 | - | 0.0502 | 0.0270 | GG | 9.22E-04 | 1.90 (1.29-2.80) |
| H3 | AA | 0.0129 | 0.0059 | - | 3.68E-02 | 2.19 (1.03-4.66) |
| H4 | AG | 0.0016 | 0.0006 | - | 3.86E-01 | 2.51 (0.29-21.48) |
Haplotype constructed by rs1511412 and rs940187, H1 is haplotype GA, while the other haplotype “-” means not GA, and H2 is GG, H3 is AA, H4 is AG. Abbreviations: A1, minor haplotype; F_A, minor haplotype frequence in case; F_U, minor haplotype frequence in control; A2, major haplotype.
Variants were functionally annotated using ANNOVAR [17]; rs1511412 is an intergenic variant and has two nearby genes, FOXL2 and PRR23A, while rs940187 is located in a long non-coding RNA (lncRNA) gene, BPESC1. BPESC1 is expressed in many tissues, such as human skin tissue. Using the starBase website, version 2.0 (http://starbase.sysu.edu.cn/index.php), we found that it can bind with the DGCR8 protein, which can play a complex role in controlling the fate of several classes of RNAs.
To check the relationship between these SNPs and the development of keloids, we subsequently checked the annotation results for two SNPs with one large expression quantitative trait loci (eQTLs) dataset [18] and obtained five hits of trans-proteins with significant P values less than 1e-4 (Table 3). Furthermore, GO annotation and pathway analysis of these proteins were performed using the KOBAS website (version 3.0), which showed that these genes did not have a common pathway and a common gene set function had not been constructed. Next, while using an R package, clusterProfiler [19], we found 4 genes within one predicted GO class with an adjusted P value less than 1.6E-4 (Table 4).
Table 3.
Check the Expression quantitative trait loci (eQTLs) annotation in one large dataset for rs940187 and rs1511412
| Rs_ID | Trans_Chr | Trans_Gene_Symbol | log10(P) | Effect | r2 |
|---|---|---|---|---|---|
| rs940187 | 12 | LEMD3 | -4.4866 | -0.009 | 0.0033 |
| rs940187 | 6 | SLC7A9 | -4.0502 | 0.0082 | 0.0029 |
| rs1511412 | 11 | HARBI1 | -4.8035 | -0.0266 | 0.0036 |
| rs1511412 | 5 | ZRSR2 | -4.2142 | 0.04 | 0.0031 |
| rs1511412 | 12 | GTF2H3 | -4.033 | -0.0258 | 0.0029 |
Rs_ID, Identifier of the marker in the current version of dbSNP; Transcript_Chr, The chromosome on which the gene encoding the transcript (whose expression is regulated) is located; Transcript_Gene_Symbol, The alphanumeric gene symbol representing the gene whose expression varies; log10(P), the log base 10 of the P value for the T test statistic; Effect, the fraction of variance explained by the SNP; r2, the correlation of the gene expression fit.
Table 4.
The GO enrichment analysis result by using ClusterProfiler
| ID | Description | Gene Ratio | B g Ratio | P value | p. adjust | Gene ID |
|---|---|---|---|---|---|---|
| GO:0090305 | Nucleic acid phosphodiester bond hydrolysis | 4/6 | 273/16672* | 1.03E-06 | 1.58E-04 | DGCR8/HARBI1/GTF2H3/FOXL2 |
| GO:0005675 | Holo TFIIH complex | 1/6 | 11/17942 | 0.00367 | 0.02934 | GTF2H3 |
| GO:0005639 | Integral component of nuclear inner membrane | 1/6 | 14/17942 | 0.00467 | 0.02934 | LEMD3 |
| GO:0031229 | Intrinsic component of nuclear inner membrane | 1/6 | 14/17942 | 0.00467 | 0.02934 | LEMD3 |
| GO:0044453 | Nuclear membrane part | 1/6 | 15/17942 | 0.00501 | 0.02934 | LEMD3 |
| GO:0032806 | Carboxy-terminal domain protein kinase complex | 1/6 | 21/17942 | 0.00700 | 0.02934 | GTF2H3 |
| GO:1902555 | Endoribonuclease complex | 1/6 | 22/17942 | 0.00734 | 0.02934 | DGCR8 |
| GO:0005689 | U12-type spliceosomal complex | 1/6 | 26/17942 | 0.00866 | 0.02971 | ZRSR2 |
| GO:0005637 | Nuclear inner membrane | 1/6 | 54/17942 | 0.01793 | 0.05378 | LEMD3 |
| GO:1902554 | Serine/threonine protein kinase complex | 1/6 | 78/17942 | 0.02581 | 0.06260 | GTF2H3 |
| GO:1902911 | Protein kinase complex | 1/6 | 90/17942 | 0.02973 | 0.06260 | GTF2H3 |
| GO:0016591 | DNA-directed RNA polymerase II, holoenzyme | 1/6 | 97/17942 | 0.03201 | 0.06260 | GTF2H3 |
| GO:0090575 | RNA polymerase II transcription factor complex | 1/6 | 102/17942 | 0.03363 | 0.06260 | GTF2H3 |
| GO:0000428 | DNA-directed RNA polymerase complex | 1/6 | 120/17942 | 0.03947 | 0.06260 | GTF2H3 |
| GO:0055029 | Nuclear DNA-directed RNA polymerase complex | 1/6 | 120/17942 | 0.03947 | 0.06260 | GTF2H3 |
| GO:0030880 | RNA polymerase complex | 1/6 | 122/17942 | 0.04012 | 0.06260 | GTF2H3 |
| GO:0044798 | Nuclear transcription factor complex | 1/6 | 127/17942 | 0.04173 | 0.06260 | GTF2H3 |
| GO:0031301 | Integral component of organelle membrane | 1/6 | 153/17942 | 0.05009 | 0.06978 | LEMD3 |
| GO:0031300 | Intrinsic component of organelle membrane | 1/6 | 160/17942 | 0.05233 | 0.06978 | LEMD3 |
| GO:0005681 | Spliceosomal complex | 1/6 | 174/17942 | 0.05680 | 0.07175 | ZRSR2 |
Gene Ratio, related genes with this GO term compare to input gene set; B g Ratio, related genes with this GO term in background compare to all background genes;
using GO term BP as background, the others are using GO term CC as background.
Discussion
In previous research, keloids were found to be associated with single SNPs or multiple susceptible SNPs. Recently, a large-scale genome-wide association study (GWAS) identified multiple single nucleotide polymorphisms (SNPs) in the 3q22.3 loci that are associated with keloids in a Japanese population. In addition, in the present study, we analyzed the associations of SNPs in 3q22.3 with keloids in an independent Chinese Han population. We found that rs940187 was associated with keloids even after Bonferroni’s correction for multiple comparisons, while rs1511412 showed only a trend associated with keloids, which may because by a limited sample size or low minor allele frequency. The results with rs940187 are consistent with those of the previously described GWAS study [15]. However, the rs1511412 SNP did not associate significantly with keloid susceptibility. Power calculation demonstrated that the power of rs1511412 is poor (31.8), whereas the power of rs940187 is higher (80). This may explain why only rs940187 is associated strongly with keloid susceptibility in our study.
A previous study showed that the two SNPs, rs1511412 and rs940187, were in different blocks. However, considering that rs1511412 has a huge difference in minor allele frequency between Chinese and Japanese groups, we checked the LD information between these two SNPs of the Asian race in data of 1000 genomes and HapMap data. In addition, in our own data, by using the Plink program, the r2 value that we calculated is 0.2013. These results imply that there could be some LD in this region. As multiple SNPs may act in combination to increase the risk of keloids, the haplotypes of this region were constructed, and their frequencies were compared between the case and control groups. Significant association evidence for one protective haplotype of GG has been found. These findings supported our hypothesis that genetic variants at the 3q22.3 loci may contribute to keloid susceptibility in a Chinese Han population.
Annotated using the ANNOVA program, SNP rs940187 is located inside a long intergenic non-coding RNA (lncRNA) gene, BPESC1, while rs1151412 is co-localized with the FOXL2 gene, which encodes a fork-head transcription factor that can bind to DNA and stimulate the expression of gonadotropin-releasing hormone (GnRH) [20]. It has been reported that tamoxifen and nonsteroidal anti-estrogens inhibit keloid fibroblast proliferation and reduce collagen production via the downregulation of TGF-β signaling [21,22]. Thus, it was speculated that either severely decreased levels of FOXL2 protein or the existence of an abnormal FOXL2 protein might contribute to keloid susceptibility via its effects on the levels of GnRH and/or steroid hormones and that gonadal hormones and pregnancy estrogens might affect keloid formation.
To further explore whether these variants were regulatory sites, we examined these SNPs in the RegulomeDB database [23]; the result provides evidence that SNP rs940187 can be a TF binding site to perform as a regulatory role. In addition, while this SNP is involved with an lncRNA gene, the protein-lncRNA interaction has been investigated, and the result demonstrates that it can bind with the DGCR8 protein, which can play a complex role in controlling the fate of several classes of RNAs.
We subsequently checked the annotation datasets for two SNPs in a large eQTLs database, and obtained two hits for trans-proteins, SLC7A9 and LEMD3, for rs940187 and another three hits for the trans-proteins, HARBI1, ZRSR2 and GTF2H3, for rs1511412 with significant P values less than 1e-4.
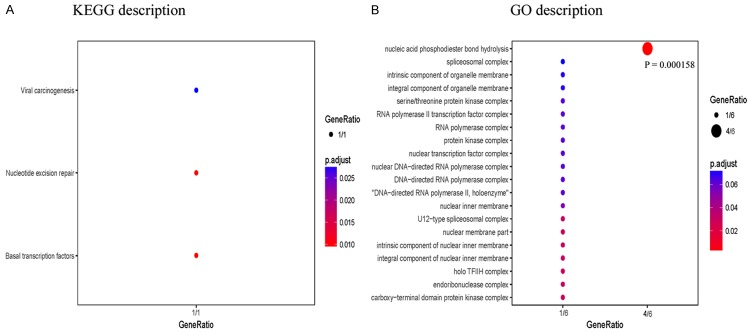
After that, investigation was accomplished, GO annotation and pathway analysis of those proteins were performed. These analyses showed that these genes did not have a common pathway, but four genes, DGCR8, HARBI1, GTF2H3 and FOXL2, had a constructed common gene set function that included nucleic acid phosphodiester bond hydrolysis (Figure 3), which was consistent with the result of the functional annotation clustering analysis (Figure 4). Phosphodiester bonds are central to all life on Earth; hydrolysis of these bonds means that the cleavage of nucleic acid, which always leads to several subcategories, such as UV-damage excision repair (GO:1990731), a process that results in the endonucleolytic cleavage of the damaged strand of DNA immediately 5’ of a UV-induced damage site [24] and is involved in the repair of a wide variety of DNA lesions. As expected, one nationwide scale indicated that the incidence rate of keloids in Asians is slightly higher than American Caucasians, but much less than that of black and Hispanic populations [25-27]. While the degree of ultraviolet damage for skin DNA is low for the white race, moderate for the yellow race, and serious for the black race, if the function of DNA repair is reduced, it may lead to hyperplasia of scars.
Figure 3.
Pathway and GO enrichment analysis using ClusterProfiler method. Pathway analysis based on KEGG database (A). GO enrichment analysis based on Gene Ontology database (B). Gene Ratio corresponds to the number of genes from a specific category. Enrichment term is represented by coloured dots (red indicates high enrichment and blue indicates low enrichment).
Figure 4.
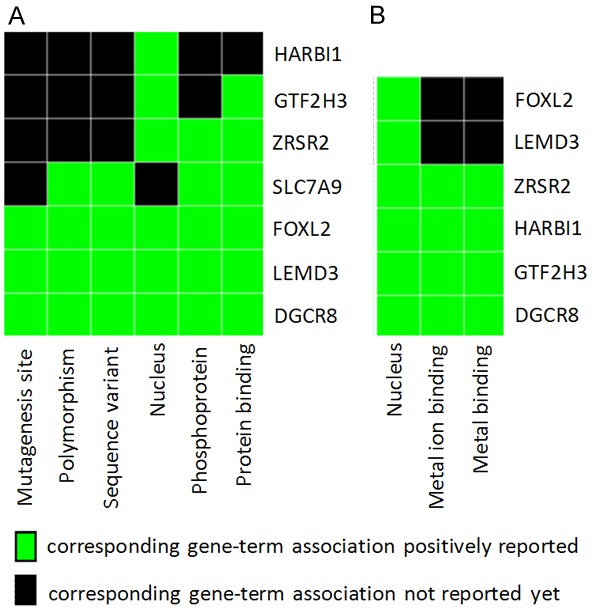
Functional Annotation Clustering analysis by using DAVID online method (version 6.8). A. Functional cluster 1, enrichment score 0.45; B. Functional cluster 2, enrichment score 1.08; all show evidence of nucleus function for input gene set.
In conclusion, we first report the association between the 3q22.3 loci and the risk of keloids in a Chinese Han population. Further large-scale association studies and functional investigations will be useful to show pathogenic mechanisms and to build predictive markers for keloids. The analysis of the polymorphism and LD structure at the 3q22.3 loci in various populations will significantly narrow the region carrying the causative SNPs and identify the real causative SNP.
Acknowledgements
We would like to thank the patients for participating in the study. This study was funded by the AnHui Provincial Natural Science Foundation (1608085MH175) and the Youth Program of the Natural Science Foundation of Anhui Province (1708085QH211).
Disclosure of conflict of interest
None.
References
- 1.Thompson LD. Skin keloid. Ear Nose Throat J. 2004;83:519. [PubMed] [Google Scholar]
- 2.Rockwell WB, Cohen IK, Ehrlich HP. Keloids and hypertrophic scars: a comprehensive review. Plast Reconstr Surg. 1989;84:827–837. doi: 10.1097/00006534-198911000-00021. [DOI] [PubMed] [Google Scholar]
- 3.Sobanko JF, Sarwer DB, Zvargulis Z, Miller CJ. Importance of physical appearance in patients with skin cancer. Dermatol Surg. 2015;41:183–188. doi: 10.1097/DSS.0000000000000253. [DOI] [PubMed] [Google Scholar]
- 4.Halim AS, Emami A, Salahshourifar I, Kannan TP. Keloid scarring: understanding the genetic basis, advances, and prospects. Arch Plast Surg. 2012;39:184–189. doi: 10.5999/aps.2012.39.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu WS, Zheng XD, Yao XH, Zhang LF. Clinical and epidemiological analysis of keloids in Chinese patients. Arch Dermatol Res. 2015;307:109–114. doi: 10.1007/s00403-014-1507-1. [DOI] [PubMed] [Google Scholar]
- 6.Chipev CC, Simon M. Phenotypic differences between dermal fibroblasts from different body sites determine their responses to tension and TGFbeta1. BMC Dermatol. 2002;2:13. doi: 10.1186/1471-5945-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brissett AE, Sherris DA. Scar contractures, hypertrophic scars, and keloids. Facial Plast Surg. 2001;17:263–272. doi: 10.1055/s-2001-18827. [DOI] [PubMed] [Google Scholar]
- 8.Alster TS, Tanzi EL. Hypertrophic scars and keloids: etiology and management. Am J Clin Dermatol. 2003;4:235–243. doi: 10.2165/00128071-200304040-00003. [DOI] [PubMed] [Google Scholar]
- 9.Kelly AP. Keloids. Dermatol Clin. 1988;6:413–424. [PubMed] [Google Scholar]
- 10.Louw L. Keloids in rural black South Africans. Part 1: general overview and essential fatty acid hypotheses for keloid formation and prevention. Prostaglandins Leukot Essent Fatty Acids. 2000;63:237–245. doi: 10.1054/plef.2000.0207. [DOI] [PubMed] [Google Scholar]
- 11.Placik OJ, Lewis VL Jr. Immunologic associations of keloids. Surg Gynecol Obstet. 1992;175:185–193. [PubMed] [Google Scholar]
- 12.Marneros AG, Norris JE, Olsen BR, Reichenberger E. Clinical genetics of familial keloids. Arch Dermatol. 2001;137:1429–1434. doi: 10.1001/archderm.137.11.1429. [DOI] [PubMed] [Google Scholar]
- 13.Marneros AG, Norris JE, Watanabe S, Reichenberger E, Olsen BR. Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. J Invest Dermatol. 2004;122:1126–1132. doi: 10.1111/j.0022-202X.2004.22327.x. [DOI] [PubMed] [Google Scholar]
- 14.Velez Edwards DR, Tsosie KS, Williams SM, Edwards TL, Russell SB. Admixture mapping identifies a locus at 15q21.2-22.3 associated with keloid formation in African Americans. Hum Genet. 2014;133:1513–1523. doi: 10.1007/s00439-014-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, Hosono N, Kubo M, Nakamura Y, Zembutsu H. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–771. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 16.Zhu F, Wu B, Li P, Wang J, Tang H, Liu Y, Zuo X, Cheng H, Ding Y, Wang W, Zhai Y, Qian F, Yuan X, Ha W, Hou J, Zhou F, Wang Y, Gao J, Sheng Y, Sun L, Liu J, Yang S, Zhang X. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS One. 2013;8:e62377. doi: 10.1371/journal.pone.0062377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joehanes R, Zhang X, Huan T, Yao C, Ying SX, Nguyen QT, Demirkale CY, Feolo ML, Sharopova NR, Sturcke A, Schaffer AA, Heard-Costa N, Chen H, Liu PC, Wang R, Woodhouse KA, Tanriverdi K, Freedman JE, Raghavachari N, Dupuis J, Johnson AD, O’Donnell CJ, Levy D, Munson PJ. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 2017;18:16. doi: 10.1186/s13059-016-1142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol. 2003;206:93–111. doi: 10.1016/s0303-7207(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 21.Chau D, Mancoll JS, Lee S, Zhao J, Phillips LG, Gittes GK, Longaker MT. Tamoxifen downregulates TGF-beta production in keloid fibroblasts. Ann Plast Surg. 1998;40:490–493. doi: 10.1097/00000637-199805000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Hu D, Hughes MA, Cherry GW. Topical tamoxifen--a potential therapeutic regime in treating excessive dermal scarring? Br J Plast Surg. 1998;51:462–469. doi: 10.1054/bjps.1997.0100. [DOI] [PubMed] [Google Scholar]
- 23.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alleva JL, Zuo S, Hurwitz J, Doetsch PW. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry. 2000;39:2659–2666. doi: 10.1021/bi992751n. [DOI] [PubMed] [Google Scholar]
- 25.Sun LM, Wang KH, Lee YC. Keloid incidence in Asian people and its comorbidity with other fibrosis-related diseases: a nationwide population-based study. Arch Dermatol Res. 2014;306:803–808. doi: 10.1007/s00403-014-1491-5. [DOI] [PubMed] [Google Scholar]
- 26.Barrett J. Birth Defect Compendium. 1973. Keloid. [Google Scholar]
- 27.Cosman B, Crikelair GF, Ju DM, Gaulin JC, Lattes R. The surgical treatment of keloids. Plast Reconstr Surg. 1961;27:335–358. [Google Scholar]