Abstract
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), as a long chain non-coding RNA (lncRNA), has been reported to be upregulated in Parkinson’s disease (PD). However, the mechanisms underlying this process remain unknown. Hence, to investigate the role of MALAT1 in PD, N-methyl-4-phenylpyridinium (MPP+) was used to induce PD in vitro in the MN9D dopaminergic neuronal cell line and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) was used to induce PD in vivo in C57BL/6 mice. Quantitative Real-Time PCR (qRT-PCR) and western blot assay showed that the expression levels of MALAT1 and leucine-rich repeat kinase (LRRK2) were increased, and that of miR-205-5p was decreased in the midbrains of mice in which PD was induced by MPTP. MALAT1 suppressed the expression of miR-205-5p in MN9D cells. The results of luciferase reporter assay indicated that LRRK2 was a direct target of miR-205-5p. Transfection with the miR-205-5p mimics decreased, whereas transfection with miR-205-5p inhibitor increased the expression levels of LRRK2 mRNA and protein. The cell counting kit-8 (CCK-8) and flow cytometry assays showed that overexpression of LRRK2 reduced the viability and promoted apoptosis in MN9D cells treated with MPP+. MALAT1 knockdown exerted a protective effect on the viability and apoptosis of MN9D cells treated with MPP+, which was abrogated by LRRK2 overexpression and miR-205-5p inhibition. Our study demonstrates that the MALAT1/miR-205-5p axis regulates MPP+-induced apoptosis in MN9D cells by targeting LRRK2, thereby improving our understanding of the molecular pathogenesis of PD.
Keywords: Parkinson’s disease, long non-coding RNA, MALAT1, miR-205-5p, LRRK2, apoptosis
Introduction
Parkinson’s disease (PD) is the second-most common neurodegenerative disease, after Alzheimer’s disease. Epidemiological studies have shown that the prevalence of PD in Chinese populations over 65 years old is about 1.7%, showing an increasing trend year by year, which brings a heavy burden to patients’ families and society, in general [1]. The clinical symptoms of PD are static tremors, myotonia, bradykinesia, anxiety, depression, cognitive dysfunction, among others [2]. Previous studies have shown that genetic, environmental, and age-related factors are closely associated with the onset of PD [3]. The degeneration and loss of substantia nigra dopaminergic neurons, and the decreased level of dopamine in the brain tissue are considered to be the main causes of PD, and are also the focus of recent studies on the pathological mechanism of PD [4].
Long non-coding RNA (lncRNA) is a class of non-protein coding RNA longer than 200 nucleotides. Studies have shown that lncRNAs play important roles in a wide range of biological processes, such as transcriptional regulation, tumor progression, and cell apoptosis [5]. Metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1) is abnormally expressed in many cancers. MALAT1 interacts with the serine/arginine splicing factor to regulate its distribution in splice sites and the phosphorylation levels, thereby modulating the alternative splicing patterns of mRNA precursors [6]. In breast cancer, MALAT1 promotes the migration and invasion of human breast cancer cells by competitively binding with miR-1 to upregulate the expression of cell division cycle 42 (cdc42) [7]. Several studies have shown that MALAT1 plays an important role in the occurrence and progression of cancer [8], but its function in PD remains unknown.
Kraus et al. revealed that the expression levels of lincRNA-p21, MALAT1, SNHG1, and TncRNA were significantly upregulated in 30 brain specimens derived from 20 patients with PD [9]. Microarray analysis showed that approximately 756 lncRNAs were aberrantly expressed in the substantia nigra pars compacta of pre-symptomatic mice over-expressing human A30P*A53T α-synuclein [10]. Bernard et al. found that MALAT1 can interact with the serine/arginine splicing factor to regulate the expression of synaptic-related genes in cultured hippocampal neurons, thus affecting the synaptic density [11]. Liu et al. found that MALAT1 expression levels were increased, and miR-124 was decreased in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced PD mice and MPP+-treated SH-SY5Y cells. MALAT1 promotes apoptosis by sponging miR-124 in a MPTP-induced PD mouse model and in MPP+-treated SH-SY5Y cells [12]. However, further research should be conducted to elucidate the role of MALAT1 and its underlying mechanism in the progression of PD.
In this study, we explored the role of MALAT1 in N-methyl-4-phenylpyridinium (MPP+)-induced cell injury in MN9D cells, which may aid a further understanding about the pathogenesis of PD, and may provide a theoretical basis for the development of novel targeted drugs against PD.
Materials and methods
Animals and reagents
Male C57BL/6 mice (aged 8-10 weeks) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Mice were randomly divided into two groups with 8 mice in each group: the control group and the MPTP group. MPTP hydrochloride (Sigma, St. Louis, MO, USA) was intraperitoneally injected four times at 2 h intervals, at a dose of 20 mg/kg body weight. The vehicle control was injected with an equivalent volume of sterile saline solution. All mice were sacrificed 7 days after the last MPTP injection, and their brains were harvested for tissue analysis, without perfusing the mice. All experimental procedures were performed in compliance with the Guide for Care and Use of Laboratory Animals and were approved by the institutional animal care and use committee of Tongji Hospital, Tongji Medical College of Huazhong University of Science and Technology.
The Lipofectamine 2000 transfection reagent was purchased from Invitrogen (Life Technologies, Carlsbad, CA, USA). The anti-LRRK2 and anti-β-actin antibody were purchased from Abcam (Cambridge, MA, USA). The horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz (Santa Cruz Biotechnology, CA, USA). The primers for qRT-PCR were synthesized by Shanghai Sangon Biotech Co., Ltd (Shanghai, China). The miR-205-5p mimics, MALAT1 and LRRK2 specific siRNAs (si-MALAT1/si-LRRK2), were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). The luciferase reporter plasmid fused with the 3’UTR of human LRRK2, pcDNA-MALAT1 and pcDNA-MUT-MALAT1 plasmids were constructed professionally and purchased from Wuhan Viraltherapy Technologies Co., Ltd (Wuhan, China).
Cell culture and treatment
The MN9D dopaminergic neuronal cell line (American Type Culture Collection, Manassas, Va., USA) was cultured in DMEM high glucose medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., Hangzhou, China) and 100 U/mL of penicillin (Invitrogen) and 100 μg/mL of streptomycin (Invitrogen). The cells were incubated at 37°C in an incubator with a humidified atmosphere of 95% air and 5% CO2. MN9D cells were pretreated with 100 μM MPP+ (Sigma) for 24 h to establish the in vitro PD model.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Cells were collected and the total RNA was extracted using the Trizol kit (Invitrogen). The first strand of cDNA was synthesized by using One Step PrimeScript cDNA synthesis kit (Takara, Dalian, China), according to the manufacturer’s instructions. qRT-PCR was performed using the SYBR®Premix Ex TaqTM II (Takaba) in an ABI 7500 real time PCR system (Applied Biosystems). The gene-specific primers were as follows: MALAT1 (forward: 5’-AGCGGAAGAACGAATGTAAC-3’; reverse: 5’-GAACAGAAGGAAGAGCCAAG-3’); miR-205-5p (forward: 5’-GCGGCGGTGTAGTGTTTCCTA-3’; reverse: 5’-GTGCAGGGTCCGAGGT-3’); U6 (forward: 5’-TGCTTCGGCAGCACATATAC-3’; reverse: 5’-ATGGAACGCTTCACGAATTT-3’); β-actin (forward: 5’-TGAGCGCGGCTACAGCTT-3’; reverse 5’-TCCTTAATGTCACGCACGATTT-3’). For the detection of LRRK2 mRNA and MALAT1 mRNA, β-actin was used as an internal reference gene. The internal reference gene of miR-205-5p was U6. The expression of miR-205-5p was normalized using U6-snRNA as internal control. The expression level was analyzed by using the 2-ΔΔCt method.
Western blot
An equal amount of total protein (10 μg) was separated on a 12% gel using the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) system, and the protein bands were transferred onto a polyvinylidinefluoride (PVDF) membrane using transfer buffer. The membrane was blocked with 5% milk in Tris-buffered saline with 0.05% Tween 20 (TBST), to prevent the non-specific binding of the primary antibody. Membranes were incubated with the primary antibody overnight at 4°C and then washed three times with TBST at room temperature. The blots were labeled with the corresponding HRP-conjugated secondary antibodies for 1 h at room temperature, followed by three washes with 1 × TBST. An enhanced chemilumniscent substrate (Thermo Scientific) was used for detection of the HRP, and the chemiluminescent western blots were exposed onto an X-ray film in the dark. The optical density values were measured with the Image J software (NIH Image, Bethesda, MD, USA).
Dual-luciferase activity assay
MN9D cells were seeded onto a 24-well plate, such that the cell density was 1 × 105 cells/mL, 24 h before transfection. MN9D cells were transfected with 0.4 µg of wild type pGL3-LRRK2-3’UTR or mutant pGL3-LRRK2-3’UTR plasmid, along with 50 nM miR-205-5p mimic or scramble mimic using Lipofectamine 2000 transfection regent (Invitrogen). At 48 h after transfection, cells were harvested and the luciferase activity was determined using the dual-luciferase kit (Promega, Madison, WI, USA) in accordance with the manufacturer’s instruction.
Cell viability assays
The cell viability was assessed with a Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). MN9D cells, at the concentration of 2 × 105 cells/mL, were seeded onto a 96-well plate. A total of 10 μL of CCK-8 reagent was added to each well, and then incubated at 37°C for 2 h. The absorbance in different wells was measured at 450 nm using a microplate reader, according to the manufacturer’s instructions.
Flow cytometry
The apoptotic cells were examined using an Annexin V-FITC/propidium iodide apoptosis detection kit (Invitrogen), according to the manufacturer’s instructions. The MN9D cells in the six-well plate were harvested, and then rinsed twice with PBS at 4°C. Cells (5 × 105) were resuspended with 500 µL of 1 × binding buffer, and then incubated with 5 µL Annexin V-FITC and 5 µL propidium iodide (PI) for 15 min at room temperature in the dark. Stained cells were identified by a Fluorescence-activated cell sorter (FACS; Becton Dickinson, CA, USA), and the cell apoptotic rate was analyzed by the FACStation software (Becton Dickinson).
Statistical analysis
The experimental data were presented as the means ± standard deviation and processed by SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The difference between the two groups (Figure 1) was analyzed by Student’s t-test. The difference among more than two groups (Figures 2, 3, 4 and 5) was analyzed by one-way ANOVA followed by a post hoc Tukey’s test. The difference was considered to be statistically significant when *P < 0.05.
Figure 1.

Changes in MALAT1, miR-205-5p and LRRK2 expression in both in vivo and in vitro models of PD. Seven days after the last MPTP injection, mice were sacrificed and brain tissues were harvested. MN9D cells were treated with 100 μM MPP+ for 24 h. A-C. qRT-PCR was performed to determine the expression of MALAT1, miR-205-5p, and LRRK2 mRNA in mice in which PD was induced by MPTP, and in cells treated with MPP+. D. The result of western blot showed that the LRRK2 protein was overexpressed in the midbrains of MPTP-induced PD mice and in cells treated with MPP+. Data are expressed as the mean ± SD (n = 3). **P < 0.01, ***P < 0.001.
Figure 2.
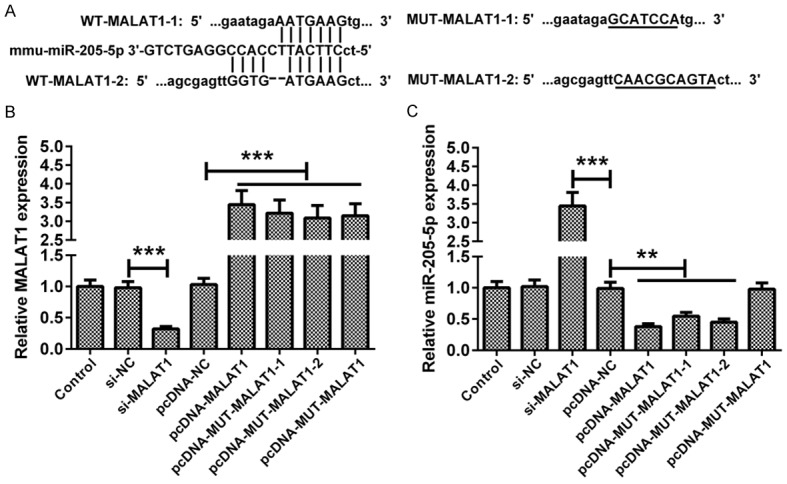
MALAT1 inhibits miR-205-5p expression. A. Bioinformatics analysis showed the combination of MALAT1 and miR-205-5p. WT-MALAT1-1 and WT-MALAT1-2 were the two target locations. B. pcDNA-MALAT1: WT-MALAT1-1 + WT-MALAT1-2. pcDNA-MUT-MALAT1-1: MUT-MALAT1-1 + WT-MALAT1-2. pcDNA-MUT-MALAT1-2: WT-MALAT1-1 + MUT-MALAT1-2. After transfection with different molecules, MN9D cells were cultured for 48 h. The regulatory effects of si-MALAT1, pcDNA-MALAT1, pcDNA-MUT-MALAT1-1, pcDNA-MUT-MALAT1-2 or pcDNA-MUT-MALAT1 were validated by qRT-PCR. C. The effects of MALAT1 knockdown and overexpression on the expression of miR-205-5p were assessed in MN9D cells. Data are expressed as the mean ± SD (n = 3). **P < 0.01, ***P < 0.001.
Figure 3.

LRRK2 was a target gene of miR-205-5p. A. The putative miR-205-5p binding sites on LRRK2 WT 3’UTR and alignment of the seed sequence with MUT LRRK2 3’UTR. B. MN9D cells were co-transfected with a miR-205-5p mimic or inhibitor and WT-LRRK2 or MUT-LRRK2 luciferase reporter plasmids. 48 h after transfection, MN9D cells were harvested, and the luciferase activity was determined. The results showed that miR-205-5p suppressed the luciferase activity of WT LRRK2 3’UTR. C, D. At 24 h after transfection with a miR-205-5p mimic or inhibitor, MN9D cells were harvested and then subjected to qRT-PCR and a western blot analysis. Transient transfection with a miR-205-5p mimic in MN9D cells attenuated LRRK2 mRNA and protein expression, and transient transfection with miR-205-5p inhibitor increased LRRK2 mRNA and protein expression. Data are expressed as the mean ± SD (n = 3). **P < 0.01, ***P < 0.001.
Figure 4.

LRRK2 promoted MPP+-induced apoptosis of MN9D cells. After transfection with si-LRRK2, pcDNA-LRRK2, or their corresponding controls, MN9D cells were treated with 100 μM MPP+ for 24 h. A. LRRK2 protein expression in MN9D cells was downregulated and upregulated by transfection with si-LRRK2 and pcDNA-LRRK2, respectively. B, D. The effects of LRRK2 overexpression and knockdown on the cell viability and apoptosis in MN9D cells, in the presence of MPP+. C. The representative images of Flow cytometry were shown. Data are expressed as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 5.
The effects of LRRK2 overexpression and miR-205-5p downregulation on the cell viability and apoptosis induced by si-MALAT1. MN9D cells were transfected with the indicated molecules and then treated with 100 μM MPP+ for 24 h. A, B. qRT-PCR analysis of MALAT1 and miR-205-5p expression in MN9D cells with different treatments. C. CCK-8 assay was performed to assess the viability of MN9D cells transfected with indicated molecules. D. Flow cytometry assay was conducted to evaluate the apoptosis of MN9D cells transfected with indicated molecules. Data are expressed as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Expression of MALAT1, miR-205-5p, and LRRK2 in both in vivo and in vitro models of PD
Compared to those in the control group, the expression levels of MALAT1, LRRK2 mRNA and protein in the MPTP group were increased in both the in vivo and in vitro models of PD (Figure 1A, 1C and 1D). The expression level of miR-205-5p was decreased in the midbrains of mice in which PD was induced by MPTP, and in MN9D cells treated with MPP+ (Figure 1B).
MALAT1 suppressed the expression of miR-205-5p
As shown in Figure 2A, the prediction of bioinformatics analysis showed that MALAT1 could bind to miR-205-5p. MUT-MALAT1-1 and MUT-MALAT1-2 were the mutant sequences of pcDNA-MUT-MALAT1. MALAT1 expression was downregulated or upregulated by transfection with si-MALAT1, pcDNA-MALAT1 or pcDNA-MUT-MALAT1 (Figure 2B). The expression level of miR-205-5p in MN9D cells transfected with si-MALAT1 was markedly higher than that in the si-NC group. Transfection with pcDNA-MALAT1, MUT-MALAT1-1 or MUT-MALAT1-2 plasmid decreased the expression of miR-205-5p. Most importantly, no changes were found in miR-205-5p expression in MN9D cells transfected with pcDNA-MUT-MALAT1 (Figure 2C).
LRRK2 was a target gene of miR-205-5p
By using the online software TargetScan (www.targetscan.org), we found that LRRK2 was a putative target gene of miR-205-5p. The miR-205-5p binding sites in the LRRK2 3’UTR and the modified sequence in the MUT LRRK2 3’UTR are shown in Figure 3A. In order to confirm the prediction that LRRK2 was a target gene of miR-205-5p, MN9D cells were transfected with luciferase reporter plasmids containing the WT or mutant LRRK2 3’UTR, together with a miR-205-5p mimic or inhibitor. We found that transfection with miR-205-5p decreased the luciferase activity, whereas transfection with miR-205-5p inhibitor increased the luciferase activity. However, no changes were observed in the MUT-LRRK2 group (Figure 3B). To further confirm the regulation of LRRK2 by miR-205-5p, the expression levels of LRRK2 mRNA and protein in MN9D cells transfected with the miR-205-5p mimic or inhibitor were detected. The results showed that the expression levels of LRRK2 mRNA and protein in the miR-205-5p mimic group were lower than those in the miR-NC group, while the LRRK2 mRNA and protein levels in miR-205-5p inhibitor group were higher than those in the miR-NC group (Figure 3C and 3D). These results suggested that LRRK2 was a target gene of miR205-5p.
LRRK2 promoted MPP+-induced apoptosis in MN9D cells
To investigate the effect of LRRK2 on MPP+-induced apoptosis, we regulated LRRK2 expression by using si-LRRK2 and pcDNA-LRRK2. The expression level of LRRK2 protein was decreased in MN9D cells transfected with si-LRRK2, and increased in MN9D cells transfected with pcDNA-LRRK2 (Figure 4A). The scaxtter plots of Flow cytometry were shown in Figure 4C. Compared with the blank in the control group, the cell viability in the MPP+ group was decreased, and the apoptosis rate was increased. Overexpression of LRRK2 reduced cell viability and promoted apoptosis in MN9D cells treated with MPP+. On the other hand, MPP+-induced apoptosis and reduction of cell viability were attenuated in MN9D cells transfected with si-LRRK2, suggesting that LRRK2 is a critical mediator in MPP+-induced cytotoxicity (Figure 4B and 4D).
MALAT1/miR-205-5p axis is involved in the regulation of MPP+-induced apoptosis in MN9D cells
To further study the mechanism of MALAT1 involved in PD, the expression of MALAT1 was downregulated and upregulated by transfection of si-MALAT1 and pcDNA-MALAT1, respectively. Compared with the blank in the control group, the expression of MALAT1 was upregulated in MN9D cells treated with MPP+. Transfection of si-MALAT1 could effectively downregulate MALAT1 expression in MN9D cells (Figure 5A). The expression of miR-205-5p was downregulated in MN9D cells treated with MPP+, but upregulated when MALAT1 was knocked down (Figure 5B). As shown in Figure 5C, the cell viability was decreased in the MPP+ group compared with the blank in the control group. In addition, the effect of si-MALAT1 on the cell viability was reversed by LRRK2 overexpression, as well as the miR-205-5p inhibitor (Figure 5C). Inhibition of miR-205-5p completely abrogated the inhibitory effects of si-MALAT1 on MPP+-induced apoptosis in MN9D cells, suggesting that the pro-apoptotic role of MALAT1 was dependent on the activity of miR-205-5p (Figure 5D).
Discussion
With recent advancements in the knowledge of lncRNAs, it has been demonstrated that some lncRNAs participate in the regulation of gene expression via multiple ways that are not yet fully understood [13]. lncRNAs were found to act as mediators of mRNA degradation, host genes of miRNAs, and regulatory factors in protein function and chromatin remodeling [14]. MPTP is a kind of neurotoxin, and has been used to establish PD animal models in a large number of researches. MPP+ is a toxic metabolite of MPTP and is used to construct an in vitro PD cell model [15]. Consistent with the report by Zhang and colleagues [16], in this study, our results showed that the expression level of MALAT1 was increased in MPTP-induced PD mice and MN9D cells treated with MPP+, suggesting that MALAT1 might play an important role in the apoptosis of neurons in PD.
miRNA is a kind of non-coding single-stranded RNA with a length of about 22 nucleotides. After transcription, mature miRNAs target the 3’UTR of mRNA to degrade mRNA or inhibit the following translation process, thereby inhibiting the expression of the target gene [17]. In recent years, many researches have shown that miRNAs play a vital role in the pathogenesis of PD. The miRNA-based molecular drug, which targets neuronal loss, may be a novel therapeutic strategy for the treatment of PD [18]. miR-205-5p was involved in the regulation of multiple biological processes in SH-SY5Y cells [19]. with MPP+. Furthermore, MA-expression was downregulated, and LRRK2 expression was abnormally upregulated in the brains of patients with sporadic PD [20]. In order to further study the mechanism by which MALAT1 is involved in neuronal apoptosis, we identified miR-205-5p as an inhibitory target of MALAT1 by sequence complementarity analysis, using bioinformatics software. The results showed that MALAT1 was upregulated, and miR-205-5p was downregulated in MN9D cells treated with MPP+. Furthermore, MALAT1 could negatively regulate the expression of miR-205-5p in MN9D cell lines. Therefore, we speculated that MALAT1 may be involved in the neuronal degeneration associated with PD, by regulating the expression of its downstream target gene, miR-205-5p.
LRRK2, also known as PARK8, is located on chromosome 12q12, and has a total length of 144 kb. Consisting of 2527 amino acids, the LRRK2 protein is a serine-threonine kinase, which can catalyze the phosphorylation of cellular proteins involved in the MAPK signaling pathway [21]. Numerous studies have shown that the LRRK2 gene is associated with autosomal dominant PD [22]. In vivo and in vitro experiments demonstrated that LRRK2 modulates the eIF4E/4E-BP signaling pathway to stimulate eIF4E-mediated protein translation. In Drosophila, the overexpression of LRRK2 aggravated oxidative stress damage and induced the apoptosis of dopaminergic neurons [23]. LRRK2 facilitates the efficient endocytosis of the synaptic membrane, and modulates EndoA-dependent membrane deformation by phosphorylating EndoA at S75 [24]. In the mammalian brain, LRRK2 maintains the complexity of neurites through an interaction with p21-activated kinase (PAK6) [25]. Lee et al. have found that LRRK2 kinase inhibitors have a neuroprotective effect, and can prevent neuronal degeneration in LRRK2 cells and mouse models of PD [26]. In this study, we found that the mRNA and protein expressions of LRRK2 were increased in MPP+-treated MN9D cells, and that the overexpression of LRRK2 protein decreased the viability of MN9D cells. In addition, we demonstrated that miR-205-5p downregulated LRRK2 mRNA and protein levels by targeting its 3’UTR, and MALAT1 upregulated LRRK2 expression by competitively binding to miR-205-5p. Knockdown of MALAT1 was shown to upregulate miR-205-5p expression, which improved cell viability and decreased the rate of apoptosis after MPP+ treatment. However, the simultaneous overexpression of LRRK2 could abrogate this benefit, corroborating the assertion that LRRK2 functions downstream of miR-205-5p and MALAT1. We clarified the mechanism by which MALAT1 contributed to the progression of PD.
In summary, our study demonstrated that MALAT1 promoted the apoptosis of MN9D cells induced by MPP+ via the downregulation of miR-205-5p, thereby upregulating the expression level of the LRRK2 protein, suggesting that MALAT1 might serve as a therapeutic target for PD.
Disclosure of conflict of interest
None.
References
- 1.Ma CL, Su L, Xie JJ, Long JX, Wu P, Gu L. The prevalence and incidence of Parkinson’s disease in China: a systematic review and meta-analysis. J Neural Transm (Vienna) 2014;121:123–134. doi: 10.1007/s00702-013-1092-z. [DOI] [PubMed] [Google Scholar]
- 2.Manza P, Zhang S, Li CS, Leung HC. Resting-state functional connectivity of the striatum in early-stage Parkinson’s disease: cognitive decline and motor symptomatology. Hum Brain Mapp. 2016;37:648–662. doi: 10.1002/hbm.23056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 4.Lu C, Zhang J, Shi X, Miao S, Bi L, Zhang S, Yang Q, Zhou X, Zhang M, Xie Y, Miao Q, Wang S. Neuroprotective effects of tetramethylpyrazine against dopaminergic neuron injury in a rat model of Parkinson’s disease induced by MPTP. Int J Biol Sci. 2014;10:350–357. doi: 10.7150/ijbs.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 8.Gutschner T, Hammerle M, Diederichs S. MALAT1--a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 9.Kraus TFJ, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA. Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol Neurobiol. 2017;54:2869–2877. doi: 10.1007/s12035-016-9854-x. [DOI] [PubMed] [Google Scholar]
- 10.Jiao F, Wang Q, Zhang P, Bu L, Yan J, Tian B. Expression signatures of long non-coding RNA in the substantia nigra of pre-symptomatic mouse model of Parkinson’s disease. Behav Brain Res. 2017;331:123–130. doi: 10.1016/j.bbr.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 11.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Zhang Q, Zhang J, Pan W, Zhao J, Xu Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017;7:19. doi: 10.1186/s13578-017-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci U S A. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yazdani U, German DC, Liang CL, Manzino L, Sonsalla PK, Zeevalk GD. Rat model of Parkinson’s disease: chronic central delivery of 1-methyl-4-phenylpyridinium (MPP+) Exp Neurol. 2006;200:172–183. doi: 10.1016/j.expneurol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Zhang QS, Wang ZH, Zhang JL, Duan YL, Li GF, Zheng DL. Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting alpha-synuclein protein expression. Biomed Pharmacother. 2016;83:153–159. doi: 10.1016/j.biopha.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Wei L, Wu F, Hu Z, Liu Z, Yuan W. Advances with microRNAs in Parkinson’s disease research. Drug Des Devel Ther. 2013;7:1103–1113. doi: 10.2147/DDDT.S48500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patil KS, Basak I, Pal R, Ho HP, Alves G, Chang EJ, Larsen JP, Moller SG. A proteomics approach to investigate miR-153-3p and miR-205-5p targets in neuroblastoma cells. PLoS One. 2015;10:e0143969. doi: 10.1371/journal.pone.0143969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, Sun L, Ma B, Ding J, Vancraenenbroeck R, Lobbestael E, Baekelandt V, Taymans JM, He P, Troncoso JC, Shen Y, Cai H. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Esteves AR, Swerdlow RH, Cardoso SM. LRRK2, a puzzling protein: insights into Parkinson’s disease pathogenesis. Exp Neurol. 2014;261:206–216. doi: 10.1016/j.expneurol.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in drosophila. Embo J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, Delbroek L, Swerts J, Chavez-Gutierrez L, Esposito G, Daneels G, Karran E, Holt M, Gevaert K, Moechars DW, De Strooper B, Verstreken P. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 25.Civiero L, Cirnaru MD, Beilina A, Rodella U, Russo I, Belluzzi E, Lobbestael E, Reyniers L, Hondhamuni G, Lewis PA, Van den Haute C, Baekelandt V, Bandopadhyay R, Bubacco L, Piccoli G, Cookson MR, Taymans JM, Greggio E. Leucine-rich repeat kinase 2 interacts with p21-activated kinase 6 to control neurite complexity in mammalian brain. J Neurochem. 2015;135:1242–1256. doi: 10.1111/jnc.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, Dawson VL, Dawson TM. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]



