Abstract
Long noncoding RNA HOXA11 antisense RNA (HOXA11-AS) is involved in tumorigenesis and development of some human cancers. However, the role of HOXA11-AS in human laryngeal squamous cell cancer (LSCC) is yet unclear. In this study, we firstly investigated the expression of HOXA11-AS in LSCC. Microarray and qRT-PCR showed that the level of HOXA11-AS was significantly higher in LSCC than that in the corresponding adjacent non-neoplastic tissues. ISH revealed that HOXA11-AS was strongly expressed in the nucleus and closely related to the T grade, neck nodal metastasis, and clinical stage. Patients with T3-4 grade, neck nodal metastasis, or advanced clinical stage presented a high HOXA11-AS expression. Kaplan-Meier analysis showed that high HOXA11-AS expression could predict a poor prognosis in LSCC patients. Furthermore, HOXA11-AS knockdown significantly inhibited the growth, migration, and invasion of LSCC cells. Taken together, the current data indicated that HOXA11-AS plays an oncogenic role in the cellular processes of LSCC and serve as a novel marker and a potential therapeutic target in LSCC patients.
Keywords: Laryngeal squamous cell cancer (LSCC), long noncoding RNA, HOXA11-AS, prognosis
Introduction
Laryngeal squamous cell carcinoma (LSCC) is a common malignancy of the upper respiratory tract [1]. Current treatments including surgical intervention, radiation therapy, and chemotherapy have a moderate effect on early-stage cases; however, the prognosis of advanced-stage LSCC patients remains poor [2,3]. Therefore, understanding the molecular mechanisms underlying LSCC carcinogenesis or progression is crucial for developing effective therapeutic strategies. In addition to the classical long protein-coding mRNAs, a wide variety of noncoding RNA transcripts also occur in mammalian genomes. The increasing information on long non-coding RNA (lncRNA) has been revealed in the mammalian transcript [4]. Similar to miRNA, lncRNA can also promote cellular pathways that lead to the development and progression of cancer [5]. HOXA11 antisense RNA (HOXA11-AS) has been shown to be involved in tumorigenesis and development of different cancers [6]. However, the physiological roles of HOXA11-AS in LSCC are yet poorly understood. Therefore, herein, we found a significantly upregulated expression of HOXA11-AS in LSCC cancer tissues than that in non-neoplastic tissues. Moreover, the potential association of HOXA11-AS with clinicopathological features was evaluated. The findings suggested that HOXA11-AS plays an oncogenic role and may serve as a therapeutic target in LSCC.
Materials and methods
Collection of patient samples
A total of 25 patients subjected to LSCC surgical resection at the Second Affiliated Hospital of Harbin Medical University were recruited in this study. Among them, 5 patients were chosen for profile analysis, and another 20 for qRT-PCR validation. The fresh paired cancerous and adjacent noncancerous tissues were collected and frozen in liquid nitrogen after surgical resection. Overall, 92 paraffin samples of LSCC patients, who were treated between 2008 and 2011, were recruited in this study, and their paired cancerous and noncancerous tissue blocks were collected from the Department of Pathology of the Second Affiliated Hospital of Harbin Medical University. All patients provided written informed consent in accordance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Harbin Medical University.
RNA extraction, RNA labeling and array hybridization
Five human samples (containing paired neoplastic and non-neoplastic margins) were used for the microarray test performed as described previously [7]. Briefly, the total RNA was isolated from laryngeal carcinoma and corresponding adjacent non-neoplastic tissues (100 mg) by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and quantified using the NanoDrop 1000 (NanoDrop Technologies, Rockland, DE, USA). The human lncRNA array was manufactured by Arraystar (USA). The lncRNAs of HOX cluster were collected from NCBI RefSeq, UCSC, NRED, and RNAdb. About 5 μg total RNA from each of the 5 matched LSCC cancerous and noncancerous tissue samples was used for labeling and array hybridization. The differences in the expression of HOX lncRNAs between neoplastic and non-neoplastic samples was calculated by the ratio of the mean normalized fluorescence values. Genes with >2-fold upregulation were selected for downstream analysis.
qRT-PCR
Quantitative real-time RT-PCR (qPCR) was performed as described previously [8]. The sequences of primers were the primers for HOXA11-AS detection were as follows: 5’-CGGCTAACAAGGAGATTTGG-3’ (forward) and 5’-AGGCTCAGGGATGGTAGTCC-3’ (reverse) [9]. To estimate the expression of HOXA11-AS, the Ct values were normalised using 18S rRNA as internal control. The relative HOXA11-AS expression was calculated using the 2-ΔΔCt.
In situ hybridization (ISH)
Paraffin sections were analyzed according to the protocol developed by Advanced Cell Diagnostics. ISH was performed using the RNAscope® 2.5 Assay (ACD, Inc. Catalog No. 3222335) and HybEZTM Hybridization System (ACD, Inc. 110 VAC, Cat. no. 322000). HOXA11-AS target and positive and negative control probes were purchased from ACD. Technical and slide quality control was certified using a positive control probe targeting the common housekeeping protein, peptidyl prolyl isomerase B (PPIB), and a negative control probe targeting the bacterial protein dihydrodipicolinate, reductase (DapB). Finally, the slides were imaged using an Olympus Dual-CCD microscope digital camera and semi-quantitative scores obtained by estimating the punctate staining.
Cell culture and virus transfection
AMC-HN-8 and Hep-2 cells of human LSCC were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Gibco; Life Technologies, Carls-bad, CA, USA) in a humidified incubator (37°C, 5% CO2). The cells were plated in 24-well plates (2 × 104 cells/well) overnight. The siRNA of human HOXA11-AS lentivirus transfer vector harboring the green florescent protein (GFP) and the control lentivirus (GFP-lentivirus) were constructed by Genechem (Shanghai, China). The lentiviruses were diluted in 0.2 mL [107 transduction units (TU)/mL] complete medium containing hexadimethrine bromide (Polybrene; 8 mg/mL) and incubated with the cells for 1 h at 37°C. Next, the cells were incubated with 0.3 mL freshly prepared Polybrene-DMEM for an additional 24 h. The medium was replaced with fresh DMEM and the cells cultured for another 48 h.
Wound-healing migration assays
Cells were seeded in 6-well plates and grown to about 90% confluency before being wounded using a 200-mL plastic tip across the monolayer. The debris was removed by washing three times with phosphate-buffered saline (PBS), followed by culturing in fresh DMEM containing 5% FBS. The wound healing was observed at different time points within the scrape line, and such images for each cell line were captured. The migration ability of the cells was evaluated measuring the width of the wounds (× 400 magnification). The images were captured after 24 h post-wounding. Each assay was performed in triplicate in at least two independent experiments.
Transwell assays
To evaluate the invasion ability of the AMC-HN-8 cells, a modified version of the standard transfilter assay was conducted. The transwell filters (pore size, 8 μm; Falcon; BD Biosciences) were coated on the lower side with 8 μg/μL Matrigel and placed on a 24-well plate containing DMEM. The cells (1 × 105) were added to the upper compartment of a transwell chamber and allowed to migrate for 24 h at 37°C. Then, the cells were harvested and suspended in DMEM with 1% bovine serum albumin (BSA). After 24 h, Matrigel and cells remaining on the upper side of the membrane were wiped off with PBS-rinsed cotton swabs, and the invading cells that had migrated to the bottom surface of the membrane were fixed in 3.7% paraformaldehyde in PBS. Subsequently, the cells were stained with crystal violet for 10 min at room temperature. The cell migration was quantified by counting the number of cells in three inserts. Data are expressed as the average number of cells per insert.
Cell proliferation assay
CCK-8 cell proliferation kit was used to evaluate the proliferation of AMC-HN-8 and Hep-2 cells according to the manufacturer’s instructions. Briefly, the cells were seeded into 96-well plates at a density of 5 × 103 cells/well and cultured for 48 h. Then, 10 μL CCK-8 solution was added to each well and incubated for 1 h, and the absorbance measured at 450 nm using a microplate reader. Results were representative of three independent experiments in triplicate.
Statistical analysis
All statistical analyses were performed using Statistical Package SPSS 17.0. In addition, comparison between two samples was analyzed by Student’s t-test. The association of HOXA11-AS with clinicopathological features was assayed by X2 test. The overall survival of patients was calculated using the Kaplan-Meier survival curve. P values less than 0.05 were considered statistically significant.
Results
Microarray analysis of HOX cluster lncRNAs’ expressions in LSCC tissues
To identify the potential HOX lncRNAs targets involved in the molecular mechanisms of LSCC, microarray analysis was carried out. Figure 1 showed 18 differently expressing lncRNAs in cancerous and noncancerous tissues. As shown in the heatmap, 8 lncRNAs (HOXA4-66, HOXA10-82, HOXA11-AS, HOXA13-100, HOXB9-206, HOXB9-205, HOXD12-3, and HOTAIR) were upregulated and 10 lncRNAs (HOXA11-92, HOXA13-97, HOXB4-171, HOXB4-173, HOXB9-187, HOXC5-254, HOXC9-131, HOXC11-114, HOXD1-49, and HOXD9-16) were downregulated (≥2-fold). Herein, we first studied HOXA11-AS (P<0.01) with 17.75-fold change; also, it was overexpressed in some tumors.
Figure 1.
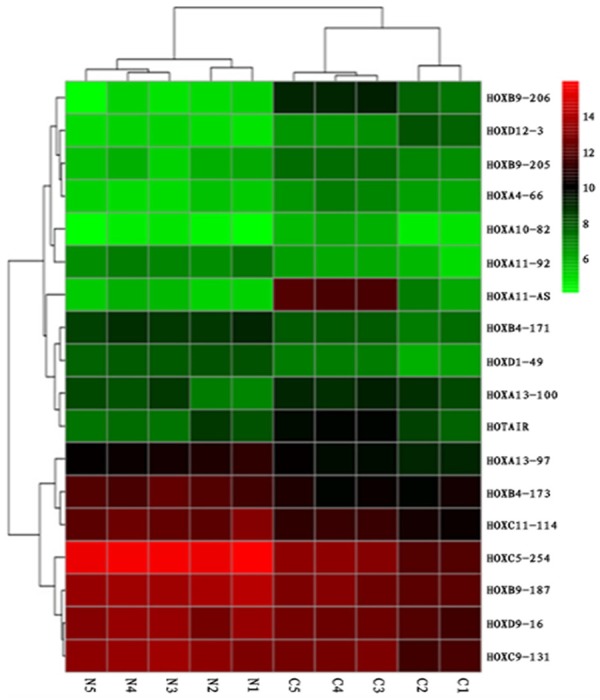
Microarray analysis of HOX cluster lncRNAs in LSCC. Hierarchical clustering is shown as a heat map, and relative lncRNA expression levels are shown in color scales (green, below the mean; red, above the mean; black, median expression). Columns C1-C5 are 5 different LSCC samples, and Columns N1-N5 are the 5 corresponding non-cancerous tissues (n=5).
HOXA11-AS is overexpressed and correlates with malignant clinicopathological features in LSCC
LncRNA HOXA11-AS was selected for further qRT-PCR validation in a large cohort of patients (n=20). As shown in Figure 2, HOXA11-AS levels were significantly higher (8.33-fold) in fresh LSCC tumor tissues than the adjacent non-neoplastic tissues (P<0.01). In order to further evaluate the potential role of HOXA11-AS in LSCC, the expression of HOXA11-AS in 92 paraffin-embedded tissue samples of LSCC and adjacent non-neoplastic tissues was semi-quantitatively examined by ISH. ISH showed that HOXA11-AS was expressed in the nucleus (Figure 3). LSCC tissues showed a significant increase in HOXA11-AS expression as compared to that observed in adjacent non-neoplastic tissues. The HOXA11-AS expression was statistically related to T grade, pathological grade, neck nodal metastasis, and clinical stage of LSCC (Table 1). Tumors with grade T3-4, poor differentiation, lymph node metastasis, or advanced clinical stages expressed high levels of HOXA11-AS.
Figure 2.
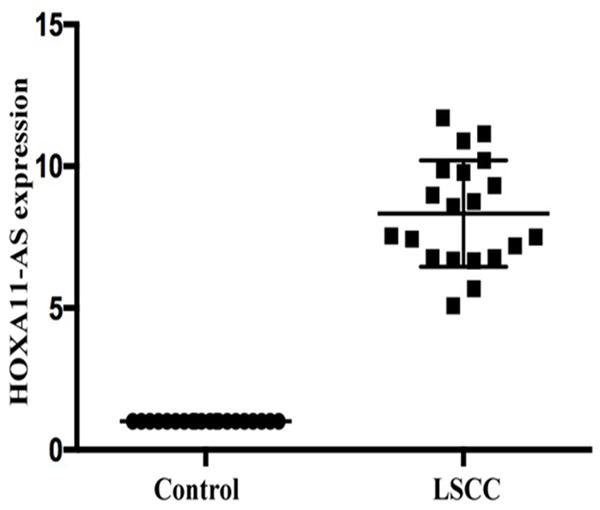
Expression of HOXA11-AS as tested by RT-PCR in a panel of non-cancerous and LSCC tissues. HOXA11-AS showed a significant increase in the expression in LSCC as compared to non-cancerous tissues (n=20).
Figure 3.
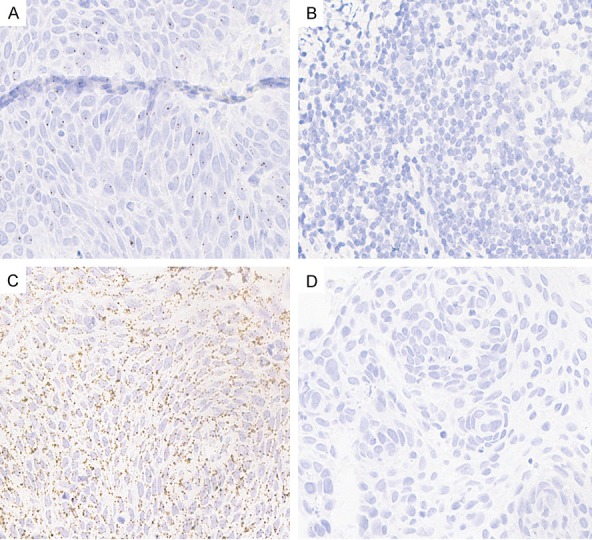
ISH analysis of HOXA11-AS expression. Representative images of ISH for HOXA11-AS in patient samples. A: LSCC tissue; B: Adjacent non-neoplastic tissues; C: Positive control; D: Negative control.
Table 1.
Relationship between HOXA11-AS expression level and clinicopathologic parameters of LSCC
| Characteristics (n) | HOXA11-AS expression | P | |
|---|---|---|---|
|
| |||
| High | Low | ||
| Sex | 0.936 | ||
| Male (64) | 36 | 28 | |
| Female (28) | 16 | 12 | |
| Age | 0.400 | ||
| ≥58 (46) | 28 | 18 | |
| <58 (46) | 24 | 22 | |
| T classification | 0.011 | ||
| T1-2 (53) | 24 | 29 | |
| T3-4 (39) | 28 | 11 | |
| Differentiation | 0.014 | ||
| G1 (51) | 23 | 28 | |
| G2 (41) | 29 | 12 | |
| Lymph node metastasis | 0.026 | ||
| Negative (50) | 23 | 27 | |
| Positive (42) | 29 | 13 | |
| Primary location | 0.137 | ||
| Supraglottic (38) | 18 | 20 | |
| Glottic (54) | 34 | 20 | |
| Clinical stage | 0.001 | ||
| I-II (47) | 19 | 28 | |
| III-IV (45) | 33 | 12 | |
Relationship between HOXA11-AS expression and survival of LSCC patients
To further evaluate the clinical significance of high HOXA11-AS expression in LSCC, the survival curve compared the difference in the survival rate between high and low expression levels of HOXA11-AS patients (n=92, 4 patients were lost to follow-up). The results showed that the overexpression of HOXA11-AS was significantly associated with poor prognosis at 60 months (p=0.019, Figure 4).
Figure 4.
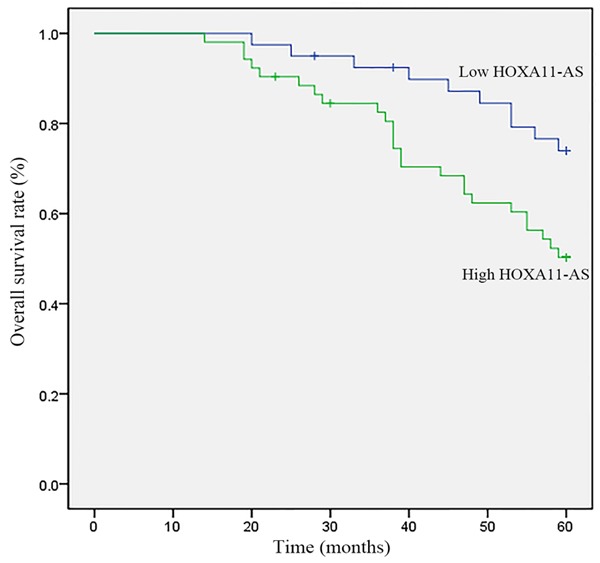
Survival analysis of the correlation of high/low HOXA11-AS expression with overall survival in patients with LSCC. The overall survival rate was estimated by the Kaplan-Meier method and analyzed by the log-rank test (n=92).
HOXA11-AS knockdown inhibits the migration and invasion of LSCC cells
The wound healing assay found that the downregulation of HOXA11-AS inhibited the migration of AMC-HN-8 cells 24 h post-transfection (Figure 5). Additionally, the results of in vitro transwell migration and Matrigel invasion assay showed that the downregulation of HOXA11-AS inhibited the migration and invasion of AMC-HN-8 cells (Figure 6). Taken together, these results suggested that HOXA11-AS promotes the invasion and migration of LSCC cells.
Figure 5.
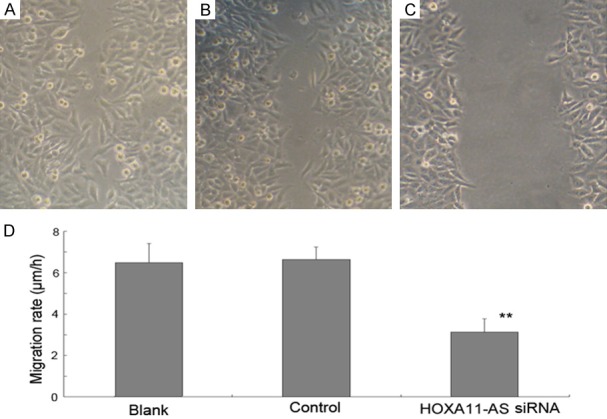
Effect of HOXA11-AS siRNA on the migration of AMC-HN-8 cells. Effect of HOXA11-AS siRNAs on cell migration was determined using scratch-wound healing migration assays at 24 h post-wounding. (A) AMC-HN-8 cells without any treatment, (B) AMC-HN-8 cells transfected with GFP vector control, (C) AMC-HN-8 cells transfected with HOXA11-AS siRNAs, (D) Cell migration was significantly suppressed in AMC-HN-8 cells by transfection them with HOXA11-AS siRNAs (**P<0.01).
Figure 6.
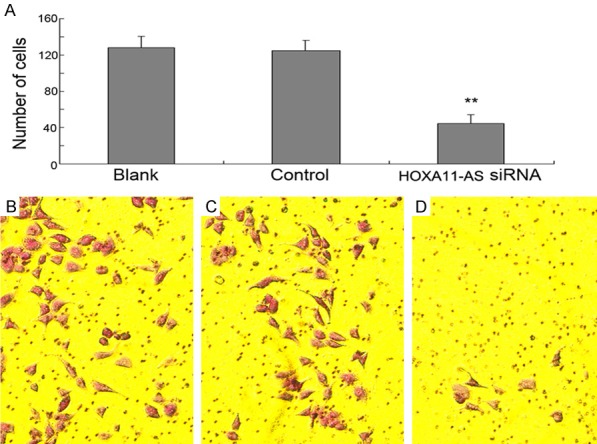
Transwell assay showed that after transfection with HOXA11-AS siRNA for 24 h, the invaded or migrated cells that penetrated the lower surface of the membrane were significantly reduced than that in the controls; **P<0.01.
HOXA11-AS knockdown inhibits the growth and proliferation of LSCC cells
To investigate the growth and proliferation of LSCC cells after HOXA11-AS knockdown, we used the CCK8 method in AMC-HN-8 and Hep-2 cells and found that the downregulation of HOXA11-AS inhibited the proliferation of both AMC-HN-8 (Figure 7A) and Hep-2 cells (Figure 7B) at different time points (24, 48, and 72 h).
Figure 7.
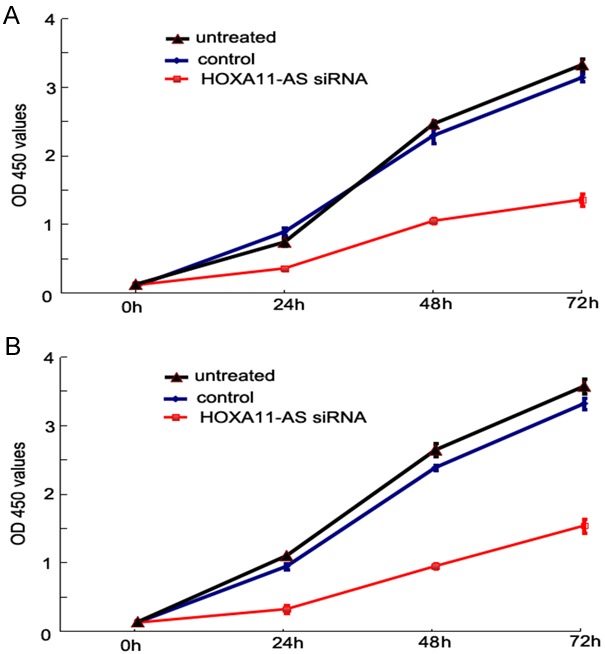
Result of cell proliferation assay. After HOXA11-AS siRNA transfection, the proliferation of LSCC cells was evidently decreased to each time point (24, 48, and 72 h, respectively) as compared to the controls. A: AMC-HN-8 cells; B: Hep-2 cells.
Discussion
LncRNAs are a class of noncoding RNA transcripts with little or no protein-coding capacity. Recent studies suggested that a large number of genomic sequences were transcribed into lncRNAs than the protein-coding RNAs [10]. An increasing number of studies revealed that lncRNAs play important roles in tumorigenesis [11,12]. LncRNAs are involved in several biological functions in carcinogenesis and have been suggested as potential therapeutic targets in many cancers [13]. Therefore, an in-depth understanding of lncRNAs may be useful in the identification of new effective therapeutic strategies for cancers. However, the function of lncRNAs in LSCC is yet not elucidated. Our previous studies showed that some lncRNAs play an oncologic gene function in LSCC [14-16]. Microarray demonstrated that the expression of HOXA11-AS was significantly increased in LSCC tissues. Real-time PCR further showed that lncRNA HOXA11-AS was overexpressed in LSCC. LncRNA HOXA11-AS is one of the Homeobox (HOX) family genes that is localized on 7p15.2 with a 3885 nt length. This gene cluster consists of protein-coding genes and genes for non-coding RNA. Our previous study suggested that the protein-coding gene HOXB9 of HOX family is upregulated and correlated with the prognosis of LSCC [17].
HOXA11-AS, transcribed from the opposite strand of the protein-coding gene HOXA11, is highly conserved across evolution. Recent studies showed that HOXA11-AS is upregulated, and it possesses oncogenic properties in various cancers such as lung,breast, and gastric cancer [9,18,19]. Furthermore, HOXA11-AS may function as a miRNA sponge to promote tumorigenesis via targeting miR-140-5p or miR-124 in glioma, uveal melanoma, and osteosarcoma [20-22]. ISH found a robust expression of HOXA11-AS in the nucleus, where it putatively functions through protein binding. Mechanistically, lncRNAs can regulate the transcription via interactions with protein, DNA, or other cellular macromolecules. Thus, we speculated that HOXA11-AS might function through binding to proteins, such as transcription factors, in order to regulate the downstream genes.
Moreover, our data indicated that the HOXA11-AS RNA is involved in LSCC aggressiveness. Patients with grade T3-4, lymph node metastasis, poor differentiation, or advanced clinical stages expressed high levels of HOXA11-AS. Furthermore, the overexpression of HOXA11-AS was significantly associated with poor prognosis of LSCC. In addition to our studies, Liu et al. reported that HOXA11-AS promoted the migration, invasion, and metastasis of gastric cancer cells in vitro [19]. Another study by Xu et al. revealed that HOXA11-ASacted as an oncogenic lncRNA that promoted cell growth and metastasis of glioma cells [23]. In this study, we found that HOXA11-AS knockdown significantly inhibited the growth, migration and invasion of LSCC cells. These results suggested that high levels of HOXA11-AS promotes the progression of LSCC cells and plays an oncogenic role in LSCC.
In conclusion, our study demonstrated the marked upregulation of lncRNA HOXA11-AS in LSCC. A high level of HOXA11-AS correlated with neck nodal metastasis, advanced clinical stage, and poor prognosis in LSCC patients. Furthermore, the downregulation of HOXA11-AS could significantly inhibit the growth, migration and invasion of LSCC cells. Our findings also suggested that HOXA11-AS may serve as an oncogene and as a potential marker in the development of LSCC; however, additional studies are essential.
Acknowledgements
This study was supported by the scientific funds of National Natural Science Foundation of China (Grant no. 81272965 and 81772874 to Yanan Sun), Daqing Science & Technology Bureau (zdy-2016-027) and Fifth Hospital of Harbin medical university.
Disclosure of conflict of interest
None.
References
- 1.Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;14:673–695. doi: 10.1016/j.otc.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Hardisson D. Molecular pathogenesis of head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2003;260:502–508. doi: 10.1007/s00405-003-0581-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang SY, Lu ZM, Luo XN, Chen LS, Ge PJ. Retrospective analysis of prognostic factors in 205 patients with laryngeal squamous cell carcinoma who underwent surgical treatment. PLoS One. 2013;8:95–98. doi: 10.1371/journal.pone.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosseini ES, Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16:107. doi: 10.1186/s12943-017-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, Yang M, Shi K, Li W. Long non-coding RNA HOXA11-AS in human cancer: a meta-analysis. Clin Chim Acta. 2017;17:30371–30376. doi: 10.1016/j.cca.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Severino P, Alvares AM, Michaluart P Jr, Okamoto OK, Nunes FD, Moreira-Filho CA, Tajara EH Head and Neck Genome Project GENCAPO. Global gene expression profiling of oral cavity cancers suggests molecular heterogeneity within anatomic subsites. BMC Res Notes. 2008;1:113–116. doi: 10.1186/1756-0500-1-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren J, Zhu D, Liu M, Sun Y, Tian L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of laryngeal squamous cell carcinoma. Eur J Cancer. 2010;46:3409–3416. doi: 10.1016/j.ejca.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, Luo DZ, Chen G. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: a study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. doi: 10.1038/s41598-017-05856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 11.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 12.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Wu TY, Zhou H, Jin QQ, He GQ, Xuan LJ, Wang X, Tian LL, Sun YN, Liu M, Qu LM. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res. 2016;35:22. doi: 10.1186/s13046-016-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic non-codingRNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Wu T, Qu L, He G, Tian L, Li L, Zhou H, Jin Q, Ren J, Wang Y, Wang J, Kan X, Liu M, Shen J, Guo M, Sun Y. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C, Han C, Wang P, Jin Y, Sun YN, Qu LM. HOXB9 Expression correlates with histological grade and prognosis in LSCC. Biomed Res Int. 2017;2017:3680305. doi: 10.1155/2017/3680305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Jia G, Qu Y, Du Q, Liu B. Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med Sci Monit. 2017;23:3393–3403. doi: 10.12659/MSM.904892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen QN, Nie FQ, Lu KH, Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui M, Wang J, Li Q, Zhang J, Jia J, Zhan X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother. 2017;92:437–444. doi: 10.1016/j.biopha.2017.05.081. [DOI] [PubMed] [Google Scholar]
- 21.Lu Q, Zhao N, Zha G, Wang H, Tong Q, Xin S. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma. DNA Cell Biol. 2017;36:837–844. doi: 10.1089/dna.2017.3808. [DOI] [PubMed] [Google Scholar]
- 22.Cui Y, Yi L, Zhao JZ, Jiang YG. Long noncoding RNA HOXA11-AS functions as miRNA sponge to promote the glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol. 2017;36:822–828. doi: 10.1089/dna.2017.3805. [DOI] [PubMed] [Google Scholar]
- 23.Xu C, He T, Li Z, Liu H, Ding B. Regulation ofHOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. doi: 10.1016/j.biopha.2017.08.097. [DOI] [PubMed] [Google Scholar]


