Abstract
Circular RNAs (circRNAs) are key regulators in the development and progression of human cancers, however its role in cervical cancer tumorigenesis is not well understood. The present study aims to investigate the expression profiles and potential modulation of circRNA on cervical cancer carcinogenesis. Human circRNA microarray was performed to screen for abnormally expressed circRNA in cervical cancer cells and circRNA-000284 was identified as one circRNA significantly upregulated in cervical cancer cells. Subsequent mechanistic investigations suggested that knockdown of circRNA-000284 suppressed cell proliferation and invasion, and caused G0/G1 phase cell cycle arrest. By performing anti-AGO2 RNA precipitation and luciferase reporter assay, we identified miR-506 as the circRNA-000284-associated miRNA. Furthermore, Snail-2 was identified as a direct target of miR-506, and circRNA-000284 could positively regulate the expression of Snail-2. Finally, the tumor promoting effect of circRNA-000284 was abolished by co-expression of miR-506 mimics or Snail-2 silencing vector. In conclusion, circRNA-000284 promotes cell proliferation and invasion in cervical cancer, and may serve as a promising therapeutic target for cervical cancer patients. Therefore, silence of circRNA-000284 could be a future direction to develop a novel treatment strategy.
Keywords: Cervical cancer, circRNA-000284, miR-506, Snail-2
Introduction
Cervical cancer is the second commonest cancer among women in the worldwide, and the majority cause of death in developing countries as well [1]. At the time of diagnosis, most of the patients have developed invasive cancer. Despite many advances in the diagnosis and treatment of this disease, the prognosis of patients with cervical cancer remains poor, with a 5-year overall survival of less than 30% in most countries [2]. Therefore, it is of great significance to find novel diagnostic and prognostic biomarkers and to investigate the mechanism of cervical cancer progression and metastasis.
Circular RNAs (circRNAs) from back-spliced exons have been recently identified as a naturally occurring family of noncoding RNAs (ncRNAs) that is highly prevalent in the eukaryotic transcriptome [3-5]. With the advent of high-throughput sequencing and bioinformatic analysis, thousands of circRNAs have been successfully identified in multiple cell lines and across various species [6,7]. Recently, they have been found to play an important role in the regulation of cancer initiation and progression [8,9]. Certain kinds of circRNAs have been found to be significantly deregulated in gastric cancer, esophageal squamous cancer, and breast cancer, and these deregulated circRNAs is suggested to participate in cancer development [10]. However, the specific role of crcRNAs in cervical cancer progression is still not well known.
These circRNAs mainly arise from exons or introns, and are differentially generated by back splicing or lariat introns [11]. Interestingly, they are found to be enormously abundant, evolutionally conserved and relatively stable in cytoplasm [12]. These features confer numerous potential functions to circRNAs, such as acting as microRNA (miRNA) sponges. For example, some circRNAs associate with cancer-related miRNAs and the circRNA-miRNA axes are involved in cancer-related pathways [13].
MiRNAs are 19-25 nucleotides noncoding RNAs that directly regulate the expression of most mRNAs in various ranges of biological functions. More recently, miRNAs are found to interact with other noncoding RNAs, such as long noncoding RNAs (lncRNAs) and circRNAs, by the way of complementary base pairing [14]. Specifically, the circRNA ciRS-7 (also termed CDR1as), which harbours more than 70 conventional miR-7-binding sites, has been identified as a miRNA inhibitor. However, only a few such circRNAs contain multiple binding sites to trap one particular miRNA, and the function of circRNA remains largely unknown.
In this study, we identified thousands of distinct circRNAs from human cervical cancer cells and cervical normal epithelial cells by using microarray profiling. We further characterize one specific circRNA-000284, which originated from exon 2 of HIPK3 gene [15] and is frequently upregulated cancer patients. Our intergrated investigations reveales that circRNA-000284 promotes cell growth and invasion via sponging miR-506 to suppress the expression of Snail-2.
Materials and methods
Cell culture
Human cervical cancer cell lines HeLa, CaSki, SiHa, C-33A, SW756 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human normal cervical epithelial cells were bought from CHI Scientific, Inc (Maynard, MA, USA). All cervical cancer cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml penicillin and 100 g/ml streptomycin (Life Technologies, Grand Island, NY, USA) at 37°C in 5% CO2 and 95% air. Human normal cervical epithelial cells were grown in DMEM/F12 1:1 medium with 10% FBS, 2.5 mM L-glutamine and 0.3 mg/ml G418 at 37°C in 5% CO2 and 95% air.
Expression profile analysis of circRNAs
The circRNAs chip (Arraystar Human circRNAs chip, ArrayStar) containing 5682 probes specific for human circular RNAs splicing sites was used. After hybridization and washing with samples, five cervical cancer cell lines (HeLa, CaSki, SiHa, C-33A, SW756) and one normal cervical epithelial cell line were analyzed on the circRNAs chips. Exogenous RNAs developed by ERCC (External RNA Controls Consortium) were used as controls. The GEO Accession number is GSE96964.
RNA oligoribonucleotides and cell transfection
The miR-506 mimics and small interfering RNAs (siRNAs) that specifically target Snail-2 (si-Snail-2) were synthesized by GenePharma (Shanghai, China). The circular transcript expression vector possesses two elements termed as the front circular and the back circular frame which were specially designed containing inverted repeat sequences flank. The full-length cDNA of circRNA-000284 was amplified in cervical cells, and was cloned into the specific vector between two frames. To knockdown circRNA-000284, three siRNA against circRNA-000284 and siRNA-NC were synthesized, and the efficiency was examined using RT-qPCR in HeLa and SiHa cells. The most effective siRNA was determined for synthesizing siRNA. si-circRNA: CUACAGGUAUGGCCUCACA. Cervical cancer cells were maintained in a 6-well plate in DMEM supplemented with 10% FBS and cultured until 50-70% confluent. RNA oligoribonucleotides were mixed with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) in reduced serum medium (Opti-MEM, Gibco, USA) according to the manufacturer’s instructions and final concentration of RNA oligoribonucleotides was 100 nM. Knockdown or overexpression effect was examined by RT-qPCR using RNA extracted 48 hours after transfection.
Quantitative real-time PCR (RT-qPCR)
Total RNA was isolated from primary cervical cancer cell lines using TRIzol reagent (Invitrogen). And then, the cDNA was synthesized from 200 ng extracted total RNA using the SuperScript III® (Invitrogen) and amplified by RT-qPCR based on the TaqMan method on an ABI PRISM 7500 Sequence Detection System (Life Technologies, Grand Island, NY, USA), with the housekeeping gene GAPDH or U6 as an internal control. The 2-ΔΔCt method was used to determine the relative quantification of gene expression levels. All the premier sequences were synthesized by RiboBio (Guangzhou, China), and their sequences are shown as follows: circRNA-000284 (Forward) 5’-TATGTTGGTGGATCCTGTTCGGCA-3’, (Reverse) 5’-TGGTGGGTAGACCAAGACTTGTGA-3’; Snail-2 (Forward) 5’-GGACTCTTGGTGCTTGTGGA-3’, (Reverse) 5’-GGACTCTTGGTGCTTGTGGA-3’; GAPDH (Forward) 5’-GAAGGTGAAGGTCGGAGTC-3’; (Reverse) 5’-GAAGATGGTGATGGGATTTC-3’. miR-506 (Forward), 5’-TAAGGCACCCTTCTGAGTAGA-3’, (Reverse), 5’-GCGAGCACAGAATTAATACGAC-3’; U6 (Forward), 5’-AGAGCCTGTGGTGTCCG-3’, (Reverse), 5’-CATCTTCAAAGCACTTCCCT-3’.
RNA FISH
In situ hybridization was performed using specific probes to circRNA-000284 sequence. PCR fragments with T7 promoter were amplified with specific primers for the back-splice region of circRNA-000284. Digoxin or Biotin-labelled RNA probes were transcribed from PCR fragments using the DIG or Biotin RNA labelling mix and T7 RNA polymerase (Roche, Shanghai, China) according to the manufacturers’ instructions. Cervical cells were grown to the exponential phase and were 80-95% confluent at the time of fixation. After prehybridization (1 × PBS/0.5% Triton X-100), cells were hybridized in hybridization buffer (40% formamide, 10% Dextran sulfate, 1 × Denhardt’s solution, 4 × SSC, 10 mM DDT, 1 mg/ml yeast transfer RNA, 1 mg/ml 1 sheared salmon sperm DNA) with DIG-labelled probes specific to circRNA-000284 at 60°C overnight. Signals were detected using tyramide-conjugated Alexa 488 fluorochrome TSA kit (Life Technologies). FISH probes are as follows: FISH-dig-F GCTTTCAGCACCGTAACCA; FISH-dig-T7-R TAATACGACTCACTATAGGGAGACTTGCGCTTCAATCCACAT.
Dual-luciferase reporter assay
The circRNA-000284 sequence in cervical cells was subcloned into the luciferase reporter psiCHECK2 (Promega, Madison, WI) and designated as psiCHECK2-circRNA-000284-WT. The circRNA-000284 sequence with mutation of miR-506 binding site was synthesized using overlap extension PCR and cloned into psiCHECK2 vector designated as psiCHECK2-circRNA-000284-Mut. The Snail-2 3’ UTR cDNA was amplified and cloned to psiCHECK2 and termed as psiCHECK2-Snail-2-3’ UTR-WT. The mutant vector for miR-506 binding site was constructed and termed as psiCHECK2-Snail-2-3’ UTR-Mut. 3 × 104 cervical cancer cells were seeded in 24-well plates in triplicate. Forty-eight h after cotransfection with corresponding plasmids and miRNA mimics, luciferase reporter assays were conducted using Dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s instructions. Relative luciferase activity was normalized to the Renilla luciferase internal control.
Cell proliferation assay
Cell proliferation was quantified by using the Cell Counting Kit-8 (Sigma). Briefly, 100 μl of cells from the different transfection group were seeded onto a 96-well plate at a concentration of 2000 cells per well and were incubated at 37°C. At 72 h, the optical density was measured at 450 nm using a microtiter plate reader, and the rate of cell survival was expressed as the absorbance. The results represent the mean of three replicates under the same conditions.
Cell invasion assay
Cell invasive ability was detected by using Transwell permeable supports (Corning, USA) according to manufacturer’s protocol. Briefly, the transfected/treated cells were plated onto a Matrigel-coated membrane in the upper chamber of a 24-well insert containing medium free of serum. The bottom chamber contained DMEM with 10% FBS. Cells were incubated at 37°C with 5% CO2 for 48 h after plating. Then, the bottom of the chamber insert was fixed with methanol and stained with DAPI for 5 minutes. The number of cells that invaded through the membrane was determined from digital images captured on an inverted microscope and calculated with Image J 1.47 software.
CircRNAs immunoprecipitation (circRIP)
Biotin-labeled circRNA-000284 probe (5’-TAATACGACTCACTATAGGGAGACAACTGCTTGGCTCTACTT-3’-biotin) was synthesized by Sangon Biotech and the circRIP assay was performed as previously described with minor modification [16]. Cervical cancer cells were fixed by 1% formaldehyde for 10 minutes, lysed and sonicated. After centrifugation, 50 µl of the supernatant was retained as input and the remaining part was incubated with circRNA-000284 specific probes-streptavidin dynabeads (M-280, Invitrogen) mixture over night at 30°C. Next day, M-280 dynabeads-probes-circRNAs mixture was washed and incubated with 200 µl lysis buffer and proteinase K to reverse the formaldehyde crosslinking. Finally, the mixture was added with TRIzol for RNA extraction and RT-qPCR detection.
Western blot and antibodies
Cervical cancer cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Sigma). Protein quantification was done using a BCA protein assay kit (Promega, USA). The primary antibodies used for western blotting were rabbit anti-human Snail-2 antibody (1:1000, Cell Signaling Technology, Beverly, MA, USA) and rabbit anti-human β-actin antibody (1:1000, Cell Signaling Technology). Horseradish peroxidase-conjugated (HRP) anti-rabbit antibodies (1:5000; Santa Cruz Biotechnology) were used as the secondary antibodies. A total of 25 μg protein from each sample was separated on 10% Bis-Tris polyacrylamide gel through electrophoresis and then blotted onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare, Piscataway, NJ, USA). Then, the membrane was blocked with 5% (5 g/100 mL) nonfat dry milk (Bio-Rad, CA, USA) in tri-buffered saline plus Tween (TBS-T) buffer for 2 h. Blots were immunostained with primary antibody at 4°C overnight and with secondary antibody at room temperature for 1 h. Immunoblots were visualized using ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore). Protein levels were normalized to β-actin.
Statistical analysis
Kolmogorov-Smirnov test was used to determine the normality of the distribution of data in each group. For cervical cancer vs. normal cell lines, differences were shown in median expression and were determined using the Mann-Whitney U test or Kruskal-Wallis test. Count dates were described as frequency and examined using Fisher’s exact test. The results were considered statistically significant at P<0.05. Error bars in figures represent SD (Standard Deviation). Quantile normalization and subsequent date processing were performed using the R software. The package gplots and function heatmap in R software were used for mapping. Other statistical analyses were performed with GraphPad Prism software (version 5.01, La Jolla, CA, USA) *P<0.05; **P<0.01; ***P<0.001.
Results
CircRNAs expression profiles analysis
To identify specific circRNAs that are differentially expressed between cervical cancer and normal subjects, five cervical cancer cell lines and one normal cervical epithelial cell line were subjected for Arraystar Human circRNA Array. A total of 4760 human circRNAs were detected. There were 512 circRNAs that were differently expressed more than 2.0-fold change between the two groups were identified. Box plots show the normalized intensities from the two groups (Figure 1A). Volcano plots show the different expression of circRNAs (Figure 1B). Subsequently, we narrowed the scope of the study to the 20 most aberrant expressed circRNAs, including 10 upregulated circRNAs and 10 downregulated circRNAs (shown in the heat map, Figure 1C).
Figure 1.
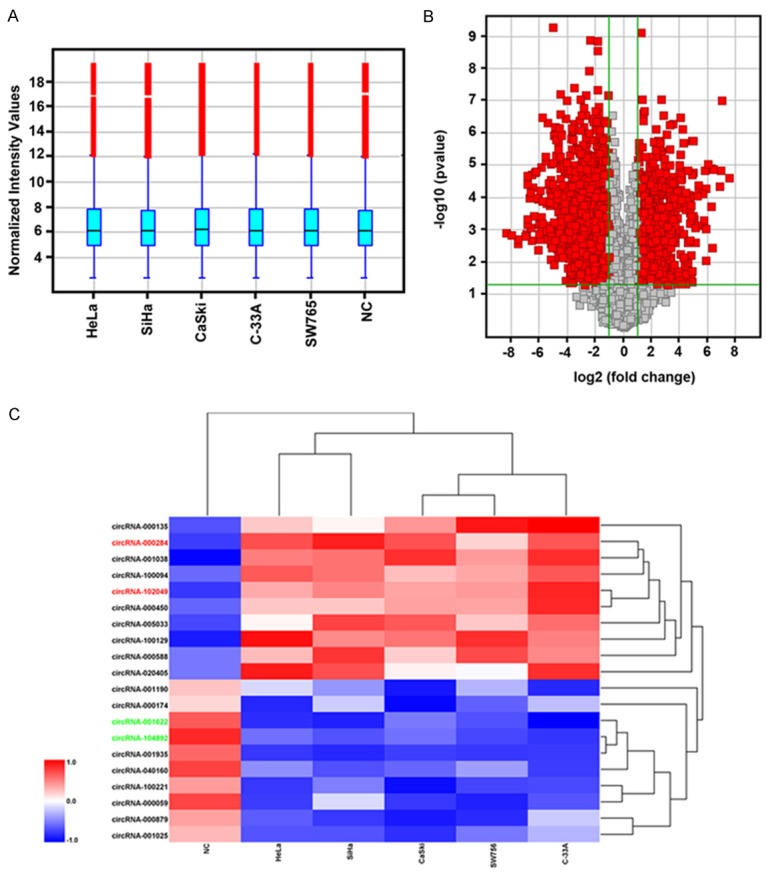
CircRNA profile expression in cervical cancer cells. A: Box plot showed the normalized intensities from five cervical cancer cell lines and one normal cervical epithelial cells (NC). B: Volcano plot of the differentially expressed circRNAs. The vertical lines correspond to 2.0-fold up and down, respectively, and the horizontal line represents a p-value of 0.05. The red point in the plot represents the differentially expressed circRNAs with statistical significance. C: Heat map showed the selected 10 upregulated and 10 downregulated circRNAs. Red indicated the upregulated expression with high fold-change and blue indicated the downregulated expression with low fold-change.
CircRNA-000284 is upregulated in cervical cancer cells
We then detected the expression of these 20 mostly changed circRNAs in cervical cells via RT-qPCR. As shown in Figure 2A-D, four circRNAs were found to be significantly deregulated between cervical cancer cells and normal cells (P<0.05), including two upregulated circRNAs, circRNA-000284 and circRNA-102049, and two downregulated circRNAs, circRNA-001622 and circRNA-104892. More importantly, only circRNA-000284 was significantly dysregulated in all of the 5 cervical cancer cell lines in contrast to normal cervical epithelial cells, and ectopic expression of circRNA102049, circRNA-001622 and circRNA-104892 had no effect on cell viability (data not shown). This inspired us to focus on the clinical and biological significance of circRNA-000284 in cervical cancer progression.
Figure 2.
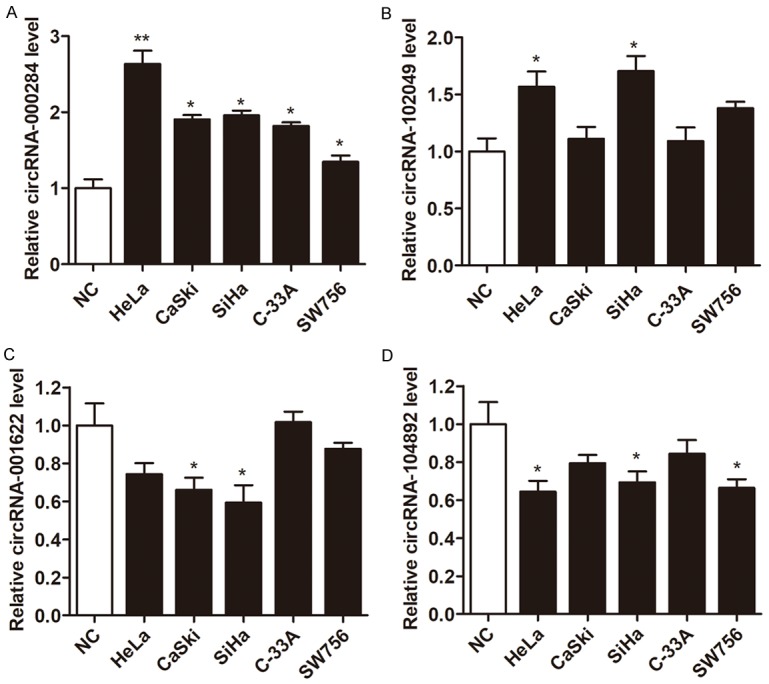
A-D: Relative expression of the four indicated circRNAs from five cervical cancer cell lines and one normal cervical epithelial cells (NC) by RT-qPCR. *P<0.05 vs. NC group.
Knockdown of circRNA-000284 suppresses cell proliferation and invasion
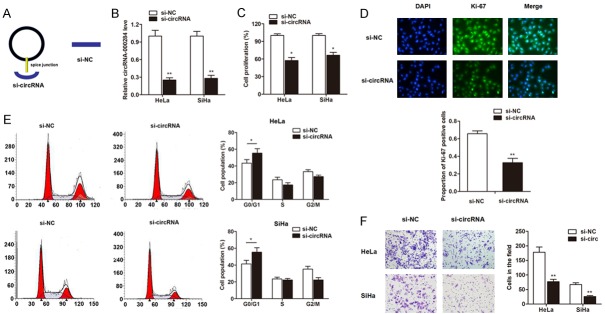
After having validated the upregulation of circRNA-000284 in cervical cancer, we then determined its functional role. The short interfering RNA (siRNA) which covers back-splicing region of circRNA-000284 was constructed (Figure 3A). RT-qPCR clearly showed that transfection of si-circRNA vector significantly silenced the expression of circRNA-000284 in HeLa and SiHa cell lines when compared with si-NC vectors (Figure 3B). Then CCK8 assay was performed to evaluate the function of circRNA-000284 on cell proliferation after transfection for 72 h. As shown in Figure 3C, knockdown of circRNA-000284 dramatically inhibited the cell growth in contrast to control cells. Moreover, the cell proliferation marker Ki-67 was evaluated by immunofluorescence assay. As shown in Figure 3D, the Ki-67 expression level was also downregulated by transfection of si-circRNA vectors in HeLa cells. Furthermore, cell-cycle assays indicated that knockdown of circRNA-000284 caused cell-cycle arrest in G0/G1 phase (Figure 3E). Matrigel invasion assay suggested that the invasive capacity was significantly inhibited by transfection of si-circRNA (Figure 3F).
Figure 3.
Knockdown of circRNA-000284 suppresses cell proliferation and invasion. A: The sketch of structures of si-circRNA, si-NC vector is shown. B: CircRNA-000284 was silenced by transfection of specific silencing vectors in both cervical cell lines. C: CircRNA-000284 was knocked down by specific silencing vectors for 48 h, then, CCK8 assay was performed in cervical cell lines. D: CircRNA-000284 was knocked down by specific silencing vectors for 48 h, then, Immunofluorescence assay was performed to detect the expression change of Ki-67. Blue, DAPI; Green, Ki-67. Merge, the merge of Ki-67 and DAPI. E: CircRNA-000284 was knocked down by specific silencing vectors for 48 h, then, FACS cell-cycle assays were performed and results suggested that knockdown of circRNA-000284 caused cell-cycle arrest in G0/G1 phase. F: Transwell permeable supports were used for the detection of cell invasive, and the results showed that knockdown of circRNA-000284 suppressed the invasive ability of cervical cancer cells. *P<0.05 vs. si-NC group; **P<0.01 vs. si-NC group.
CircRNA-000284 directly binds to miR-506 and suppresses miR-506 activity
Since circRNAs can function as sponges or inhibitors of their interacting miRNAs, circRNAs interacting with miRNAs were predicted. We wondered whether circRNA-000284 exerted its effect through sponging miRNAs. Fluorescence in situ hybridization (FISH) against circRNA-000284 demonstrated that the circular form of circRNA-000284 preferentially localized in the cytoplasm, indicating that it may interact with miRNAs (Figure 4A). Using the TargetScan and PicTar miRNA prediction programs, we identified two sequences that targeted by miR-506 (Figure 4B). We further mutated the miRNA target site from the luciferase reporter with inclusion of the circRNA-000284 sequence in the 3’-UTR and circRNA-000284 expressing vector, respectively. We found that miR-506 mimics (Figure 4C) significantly inhibited luciferase activity of wild type reporter for cirRNA-000284 more than 40%, however, miR-506 did not inhibit the luciferase activity of reporter vector containing the mutant binding sites of cirRNA-000284 (Figure 4D). It has been known that miRNAs repress translation and degrade mRNA in an AGO2-dependent manner by binding to their targets. We conducted anti-AGO2 immunoprecipitation (RIP) in cervical and cervical cells transiently overexpressing miR-506 to pull down the circRNA-000284 using anti-AGO2 antibodies or control IgG, followed by RT-qPCR analysis for circRNA-000284 levels. The results showed that circRNA-000284 pulled down with anti-Ago2 antibodies was significantly enriched in cells transfected with miR-506 mimics compared to controls. (Figure 4E), suggesting that miR-506 could directly target circRNA-000284 in AGO2-dependent manner. In addition, RTqPCR showed that miR-506 was significantly downregulated in cervical cancer cell lines (Figure 4F). Knockdown of circRNA-000284 significantly pr-omoted miR-506 expression in cervical cancer cells (Figure 4G).
Figure 4.
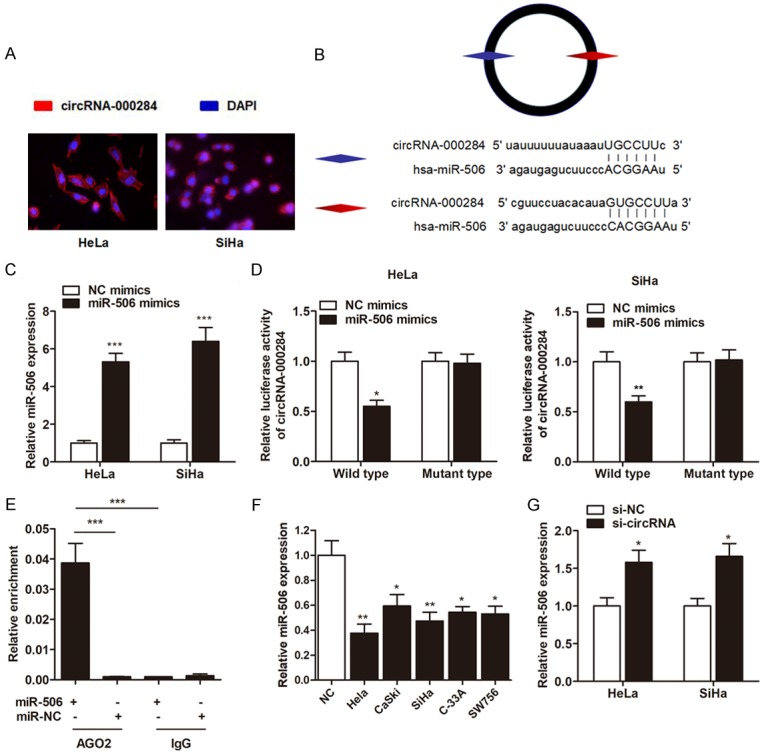
CircRNA-000284 directly binds to miR-506 and suppresses miR-506 activity. A: RNA fluorescence in situ hybridization for circRNA-000284. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). B: The putative sequences of miR-506 and circRNA-000284 with two binding sites. C: MiR-506 was upregulated by transfection of miR-506 mimics in cervical cells. D: Luciferase reporter assay was performed to detect the interaction between miR-506 and circRNA-000284. MiR-506 significantly inhibited luciferase activity of wild type reporter for cirRNA-000284, however, miR-506 did not inhibit the luciferase activity of reporter vector containing the mutant binding sites of cirRNA-000284. E: Anti-AGO2 RIP was performed in HeLa cells transfected with miR-506 mimics or NC, followed by RT-qPCR to detect circRNA-000284. F: RT-qPCR experiment suggested that miR-506 was downregulated in cervical cell lines in contrast to normal epithelial cells. G: Cervical cells were transfected by si-circRNAs, then miR-506 expression was detected via RT-qPCR. *P<0.05 vs. NC group; **P<0.01 vs. NC group; ***P<0.001 vs. Input group.
Snail-2 is identified as a direct target of miR-506 in cervical cancer
According to miRBase prediction, we identified several mRNA targets of miR-506. Specifically, we found that Snail-2 was targeted by miR-506 with high score (Figure 5A), moreover, this gene is essential for the cell proliferation and invasion induced by miR-506 [17]. To further identify whether Snail-2 in cervical cancer cells responded to miR-506 through direct interactions with its 3’-UTR, we cloned the wild type or mutant type 3’-UTR of the putative miR-506 target into reporter plasmid containing the luciferase gene of cervical cells. The Dual-Luciferase reporter experiment revealed that miR-506 attenuated the fluorescence driven by the wild type 3’-UTR by more than 1.5-fold compared with the negative control, whereas the mutant 3’-UTR was not affected by miR-506 (Figure 5B). Furthermore, the gain and loss functional assays clearly showed that overexpression of miR-506 significantly inhibited the expression of Snail-2 at both transcript and protein levels (Figure 5C). Thus, we concluded that Snail-2 was a direct target of miR-506 in cervical cancer.
Figure 5.
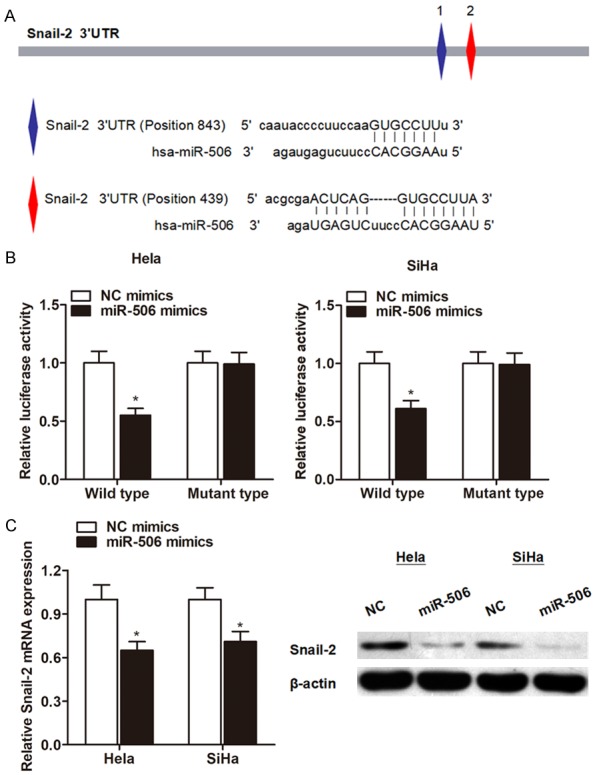
Snail-2 is a direct downstream target of miR-506. A: The putative sequences of miR-506 and Snail-2 with two binding sites. B: Luciferase reporter assay was performed to detect the potential binding between miR-506 and Snail-2. MiR-506 significantly inhibited luciferase activity of wild type reporter for Snail-2, however, miR-506 did not inhibit the luciferase activity of reporter vector containing the mutant binding sites of Snail-2 in both cell lines. C: RT-qPCR and Western blot experiments showed that miR-506 suppressed the expression level of Snail-2 at both transcript and protein levels, respectively. *P<0.05 vs. NC group.
CircRNA-000284 suppresses cell proliferation and invasion via miR-506/Snail-2 pathway
Based on the above observations, we further detected the correlationship between circRNA-000284 and the expression of Snail-2 gene. The expression of Snail-2 was significantly decreased after knockdown of circRNA-000284 (Figure 6A). Then, the full-length cDNA of circRNA-000284 from cervical cells was amplified and cloned into the specific vector (Figure 6B). RT-qPCR showed that circRNA-000284 vector significantly elevated the level of circRNA-000284 in both cells (Figure 6C). Furthermore, RIP assay showed that the enrichment of Snail-2 and miR-506 was signifi-cantly decreased in HeLa cells transfected with circRNA-000284 vector (Figure 6D), suggesting that circRNA-000284 sponges miR-506, which subsequently releases the activity of Snail-2.
Figure 6.
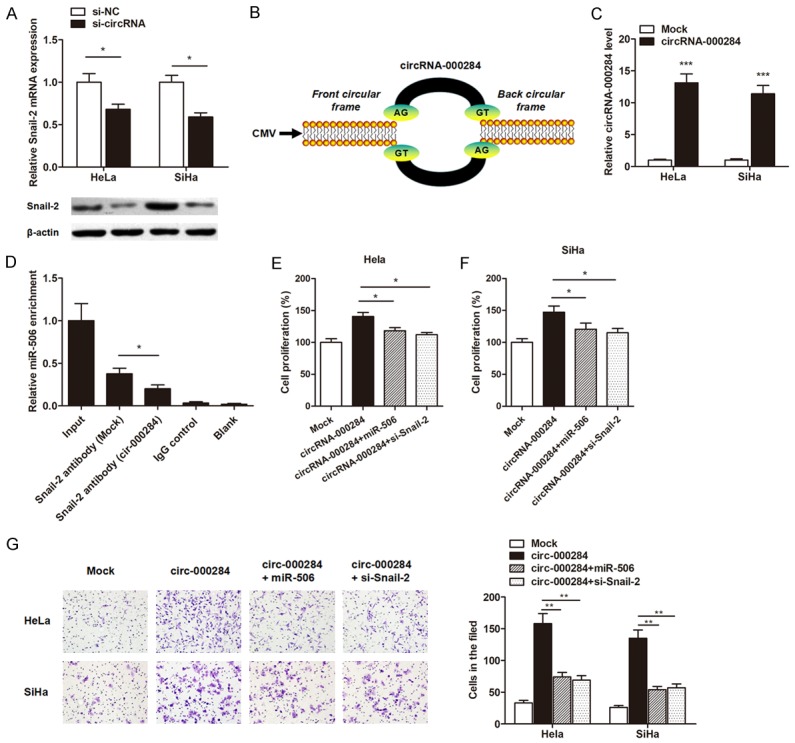
CircRNA-000284 suppresses cell proliferation and invasion via miR-506/Snail-2 pathway. A: RT-qPCR and Western blot assays suggested that knockdown of circRNA-000284 suppressed Snail-2 expression level at both transcript and protein levels, respectively. B: The sketch of structures of circRNA-000284 is shown. C: RT-qPCR showed that circRNA-000284 expression level was dramatically elevated by the transfection of circRNA-000284 overexpressing vector. D: RIP experiments were performed using the anti-Snail-2 antibody to immunoprecipitate RNA and a primer to detect miR-506, and a significantly decreased enrichment of miR-506 was identified in cells transfected with circRNA-000284 overexpressing vector. E, F: CCK8 assay showed that enhanced expression of circRNA-000284 promoted the cell proliferation rate of both cell lines, however, cotransfection of miR-506 mimics or si-Snail-2 significantly reversed this effect. G: Matrigel invasion assay suggested that the increased cell invasion of cervical cancer cells induced by circRNA-000284 was abrogated by cotransfection with miR-506 mimics or si-Snail-2 vector. *P<0.05; **P<0.01; ***P<0.001 vs. respective control groups.
We then conducted the gain and loss functional assays to verify whether circRNA-000284 regulates cervical cancer cell growth and invasion via miR-506/Snail-2 pathway. Enhanced expression of circRNA-000284 promoted the cell proliferation rate, however, cotransfection of miR-506 mimics or si-Snail-2 significantly reversed this effect (Figure 6E, 6F). Similarly, the increased cell invasive capacity of cervical cancer cells induced by circRNA-000284 was abrogated via cotransfection with miR-506 mimics or si-Snail-2 vector (Figure 6G).
Discussion
For more than 30 years, circular RNAs were reported sporadically and long considered to be molecular flukes [18]. Recently, with the advent of next-generation sequencing, numerous of circRNAs were identified from various animal genomes, and many of them were highly stable and abundantly expressed, thus largely reshaped the conventional perspective on circRNAs. In this study, we identified a substantial fraction of circRNAs that were differentially expressed in cancer tissues compared with normal tissues, which suggests that these RNAs may be regulated and may exert a potential function. We further characterized one of the mostly differentially expressed circRNA, circRNA-000284, and provided the first evidence that it may play an important role as a prognostic marker in cases of cervical cancer.
We firstly performed a comprehensive microarray analysis of the global changes in the expression pattern of circRNAs in cervical cancer. A total of 4760 human circRNAs were detected. There were 512 differentially expressed circRNAs, consisting of 231 up-regulated and 281 down-regulated circRNAs, and showed circRNA-000284’s expressive feature in cervical cancer for the first time. Compared with cervical epithelial cells, the expression level of circRNA-000284 significantly increased in cervical cancer cells. CircRNA-000284 is derived from exon2 of the HIPK3 gene, and also identified in previous reports via the deep sequencing of several human cell lines [15,19]. CircRNA-000284 consists solely of a large second exon (1099 bp) from the HIPK3 gene flanked on either side by long introns, which include many complementary Alu repeats. All these features indicate that circRNA-000284 is formed by “direct splicing” and stably expressed in different cell lines and tissues [18,20].
The two most important properties of circRNAs are highly conserved sequences and a high degree of stability in mammalian cells [21]. Compared with other noncoding RNA, such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), these properties provide circRNAs with the potential to become ideal biomarkers in the diagnosis and prognosis of cancers [22]. To date, only a few circRNAs have been explored. They may serve as oncogenes, tumor suppressor genes, or both, depending on circumstances. For example, Fu et al reported that circRNA-0004018 is differentially expressed in HCC and may serve as prognostic marker for HCC patients [23]; Chen et al identified circRNA-PVT1 as a proliferative factor and prognostic marker in gastric cancer [24]; Li et al demonstrated that circRNA-HIPK3 could be a new therapeutic target for the treatment of bladder cancer [19]. To get a closer insight into the role of circRNA-000284 in cervical cancer, we investigated the functional role of in cervical cancer. Many literatures have showed that some specific circRNAs play an important role during cancer initiation and progression. For example, circRNA-MYLK, circRNA-ABCB10 and circRNA-0067934 are reported to promote cell migration and growth [25-27]; while circRNA-MTO1 and circRNA-LARP4 suppress tumor progression by multiple signaling pathways [28,29]. Our in vitro studies clearly demonstrated that circRNA-000284 functions as tumor oncogene through promoting cervical cancer cell proliferation and invasion, which is consistent with its expression feature of clinical materials.
We then sought to define how circRNA-000284 exerts its function. It is known that circRNAs are novel RNA molecules with different biological functions and pathological implications. Among these multiple functions, “miRNA sponge” represents the most conspicuous function. MiRNAs, an abundant class of small noncoding RNAs (~22 nt), posttranscriptionally modulate the translation of target mRNAs via corresponding miRNA response elements (MRE) [30]. Computational searches for miRNA target sites in circRNAs identified a portion of circRNA molecules that contain MREs, which might act as miRNA sponge, reducing miRNA binding to its target genes, thereby releasing the expression of the miRNA targets indirectly [31]. Since the first report of circRNA functioning as a miRNA sponge, the potential of circRNAs in regulating cancer-related genes through fine-tuning miRNAs has recently been recognized. For example, ciRS7 contains more than sixty miR-7-binding sites, thereby acting as an endogenous miRNA sponge to adsorb and subsequently quench normal miR-7 functions [32]. More recently, more and more circRNAs were recognized as miRNA sponges in different cancers, however, this model reported in cervical cancer is very limited. In this study, characterization of circRNA-000284 indicated that the circular form of circRNA-000284 preferentially localized in the cytoplasm, and miR-506 showed a complementary sequence to circRNA-000284 based on the bioinformatic analysis. Eventually, this miRNA was finally identified as the endogenous competing RNA by luciferase reporter assay and RIP assay.
After having validated the interaction between circRNA-000284 and miR-506, we also identified the downstream genes that may account for the functional role of circRNA-000284. Snail-2 gene was found to be the direct target of miR-506 and regulated by circRNA-000284. Moreover, the gain and loss functional assays suggest that circRNA-000284 regulates cell proliferation and invasion through sponging miR-506 and subsequently targeting Snail-2. In conclusion, our study revealed the circular RNA profile of cervical cancer tissues and characterized circRNA-000284 as a differentially expressed circRNA. We found that circRNA-000284 is a new diagnostic factor and prognostic marker in cervical cancer. Our findings suggest that circRNA-000284 may serve as a novel biomarker that may have potential functions and clinical significance in cervical cancer.
Acknowledgements
This study was supported by Shanghai Science and Technology Commission Medical Guide Project (09411963700).
Disclosure of conflict of interest
None.
References
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, Lozano R, Lopez AD, Murray CJ, Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Tewari KS, Sill MW, Long HJ 3rd, Penson RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao MM, Michael HE, Monk BJ. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370:734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Li C, Gao X, Wang J, Liang X. Preparation of small RNAs using rolling circle transcription and site-specific RNA disconnection. Mol Ther Nucleic Acids. 2015;4:e215. doi: 10.1038/mtna.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y, Ao Y. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223–230. doi: 10.1016/j.omtn.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, Lin H, Liu F, Dai Y. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 10.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 11.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond SM. Soaking up small RNAs. Nat Methods. 2007;4:694–695. doi: 10.1038/nmeth0907-694. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su X, Wang H, Ge W, Yang M, Hou J, Chen T, Li N, Cao X. An in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancer. Cancer Res. 2015;75:2875–2885. doi: 10.1158/0008-5472.CAN-14-2218. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Ju J, Ni B, Wang H. The emerging role of miR-506 in cancer. Oncotarget. 2016;7:62778–62788. doi: 10.18632/oncotarget.11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Fu L, Yao T, Chen Q, Mo X, Hu Y, Guo J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7:1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, Li N, Zhou W, Yu Y, Cao X. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]