Abstract
Head and neck squamous cell carcinomas (HNSCC) are a diverse group of tumor types, including neoplasia of the paranasal sinuses, oral cavity, trachea, pharynx and larynx. Laryngeal cancer is the most common type of HNSCC. The proline-rich 4 (PRR4) protein is synthesized in the acinar cells of human lacrimal glands. Previous studies have demonstrated that PRR4 may function as an antimicrobial protein protecting the ocular surface and the oral cavity. In order to determine differentially expressed genes (DEGs) in laryngeal tumors, a GeneFishing Assay was performed; 27 DEGs were identified. The PRR4 gene expression level in laryngeal tissue samples obtained from 90 patients, and the saliva of 25 healthy smokers and 25 non-smokers, was investigated using reverse transcription-quantitative polymerase chain reaction. It was revealed that PRR4 gene expression was decreased in 65/90 tumor tissues (72.2%) compared with normal tissues. No significant difference was identified between the healthy smoker and the non-smoker groups in terms of PRR4 gene expression. The results of the present study indicated that the PRR4 gene may serve an important role in laryngeal carcinogenesis.
Keywords: laryngeal cancer, GeneFishing, differently expressed gene, annealing control primer, expression
Introduction
Head and neck squamous cell carcinoma (HNSCC) includes epithelial malignancies of the oral cavity, oropharynx, hypopharynx and larynx (1). Laryngeal carcinoma originates from the larynx, which is divided into three regions: The supraglottis, glottis and subglottis (2). Smoking is by far the principal risk factor for laryngeal cancer, followed by alcohol consumption (3). However, dietary factors, including vitamin and fiber intake, are reported to be protective factors for laryngeal cancer (3). In addition to these factors, genetic factors contribute to the development of laryngeal cancer (4). Therefore, a more detailed understanding of the complex molecular mechanisms leading to the development of laryngeal cancer is still required.
Lacrimal proline-rich 4 (PRR4), also known as nasopharyngeal carcinoma-associated proline-rich protein, was discovered in 1995 by Dickson and Thiesse as a PRR protein (PRP) synthesized in the acinar cells of the human lacrimal glands (5). In previous studies, a decrease in the PRR4 protein level in tear fluid was associated with pathological conditions, including dry eye syndrome (DES), thyroid-associated orbitopathy and diabetic proliferative rethinopathy (6–8). It has also been demonstrated that the gene encoding the PRR4 protein is highly expressed in the human submucosal glands (9,10).
Submucosal glands occur in the upper respiratory tract, visual and auditory systems, and the throat and intestines of mammals. The majority of airway mucus is produced by the submucosal glands. Salivary PRPs constitute approximately two-thirds of the proteins secreted by the parotid gland (11,12). PRPs are naturally unfolded, with no stable tertiary structure (13,14). Salivary PRPs make up 70% of the proteins in saliva and are well characterized (11,12). They have several functions including the inhibition of calcium phosphate precipitation, binding bacterial pathogens and binding to dietary tannins (15–17).
The development of novel techniques has allowed the identification of novel macromolecules that may have critical functions in the development and progression of cancer. In the present study, a GeneFishing Assay (18) was performed in order to identify novel genes that participate in laryngeal carcinogenesis. It was identified that the PRR4 gene was one of the downregulated genes, which may be due to the functional importance of PRR4 in laryngeal carcinogenesis. PRR4 mRNA expression levels in the tumor and adjacent normal tissues from 90 patients with laryngeal cancer were also investigated.
Materials and methods
Tissue samples
A total of 90 tumor tissues and matched non-cancerous tissue samples were obtained from patients diagnosed with laryngeal cancer undergoing surgery in the Department of Otorhinolaryngology at the Cerrahpasa Medical Faculty, Istanbul University (Istanbul, Turkey). A total of 87 men (96.7%) and 3 women (3.3%) were recruited in the present study. The age range of patients was 39–81 years and the mean age at diagnosis was 60±9 years. The samples were fresh tissues obtained during surgery and processed immediately. Tumor and normal tissue differentiation were confirmed by a pathologist under a microscope. Only samples with a tumor/stroma cell content >70% were included in the study as tumor samples. Oral saliva was collected from 50 healthy volunteers, including 25 smokers and 25 non-smokers. The subjects did not eat or drink for 2 h prior to saliva collection, and their mouth was rinsed with sterile MilliQ water (Merck KGaA, Darmstadt, Germany). Saliva was collected into a DNase- and RNase-free 50-ml Falcon tube. The Falcon tube was kept on ice during the collection procedure. The saliva was processed immediately after collection.
The present study was approved by the Cerrahpasa Medical Faculty Ethics Committee (approval no., 83045809/604.01/02-235918), and was performed in accordance with the 2013 Declaration of Helsinki. Signed informed consent was obtained from all patients prior to the study.
GeneFishing assay
RNA isolation and first-strand cDNA synthesis
Total RNA was extracted from the tumor and adjacent non-cancerous tissues of 4 patients using the miRCURY™ RNA Isolation kit (Exiqon A/S, Vedbaek, Denmark) according to the manufacturer's protocol. A total of 3 µg RNA was used for first strand cDNA synthesis. The reaction conditions were as follows: 1 µM dT-ACP1 (provided in the GeneFishing™ DEG Premix kit; Seegene, Inc., Seoul, South Korea), 1X reverse transcriptase buffer (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 0.5 mM dNTP, 20 U RNase inhibitor (Biomatik Corporation, Ontario, Canada) and 200 U M-MLV reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) in a 20 µl final reaction volume. Reverse transcription was performed at 42°C for 90 min and 94°C for 2 min. First strand cDNA was diluted by adding 80 µl DNase-free water prior to GeneFishing polymerase chain reaction (PCR).
GeneFishing PCR
A total of 20 different arbitrary annealing control primers (ACPs) provided in the GeneFishing DEG Premix kit (Seegene Inc.) were used for GeneFishing PCR. Diluted first-strand cDNA (50 ng) was used as a template in a reaction volume of 20 µl containing 0.5 µM arbitrary ACP (one type per reaction), 0.5 µM dT-ACP2 and 1X SeeAmp™ ACP master mix. The reaction conditions were 94°C for 5 min, 50°C for 3 min, 72°C for 1 min, then 40 cycles of 94°C for 40 sec, 65°C for 40 sec and 72°C for 40 sec, and a final step at 72°C for 5 min. GeneFishing PCR products were analyzed by electrophoresis on 2% agarose gels, and all bands were quantified using BioCapt analysis software (version 11.03; Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany). Bands were purified and cloned if there was >10% difference in the band intensities between the tumor and normal tissue. The differentially expressed bands were extracted from the gels using the Zymoclean™ Gel DNA Recovery kit (Zymo Research Corp., Irvine, CA, USA) according to the manufacturer's protocol.
Cloning and sequencing
The purified PCR products were cloned into a TA cloning vector using the TOPO TA Cloning kit for Sequencing (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The plasmid vector pCR™4-TOPO was used for cloning. Subsequent to performing the cloning reaction, the plasmid vector was transformed into One Shot® TOP10 Chemically Competent E. coli (Thermo Fisher Scientific, Inc.) cells, according to the manufacturer's protocol. E. coli cells were spread onto Luria-Bertani (LB) agar plates containing 50 µg/ml kanamycin and incubated overnight at 37°C. A total of 2–6 colonies were selected and cultured overnight in LB medium containing 50 µg/ml kanamycin. The plasmid DNA was isolated using the PureLink™ Quick Plasmid Miniprep kit (Invitrogen; Thermo Fisher Scientific, Inc.) and sequenced using an ABI Prism 3100-Avant™ Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the ABI Prism BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The DNA sequences were analyzed by evaluating the data using the Basic Local Alignment Search Tool program 2.4.0 (19).
Validation of the GeneFishing assay data by RT-quantitative PCR (RT-qPCR)
Total RNA was extracted using the PureLink RNA Mini kit (Ambion; Thermo Fisher Scientific, Inc.) from the 90 tumor and non-cancerous adjacent tissues. Total RNA (400 ng) from each sample was transcribed in a 20 µl reaction volume using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. PRR4 gene expression levels were analyzed by RT-qPCR using the LightCycler 480-II system (Roche Diagnostics GmbH, Mannheim, Germany). RT-qPCR was performed in a final volume of 15 µl containing 1X master PCR mix (SolGent, Inc., Daejeon, South Korea) with EvaGreen (Biotium, Inc., Freemont, CA, USA), 600 nM gene-specific primers, cDNA and nuclease free water. The β-actin gene was used as a housekeeping gene for normalization of mRNA levels. The sequences of the primers are presented in Table I. The PCR cycling conditions were as follows: 95°C for 15 min, followed by 40 cycles at 95°C for 15 sec, 59°C for 30 sec and 72°C for 30 sec, and a final 10 sec at 50°C. The relative quantification of mRNA levels was calculated using the comparative 2−ΔΔCq method (20). The expression levels of 12 differentially expressed RNAs in HNSCC or laryngeal cancer have been analyzed in Gene Ontology, Serial Analysis of Gene Expression and The Cancer Genome Atlas databases (21–23).
Table I.
Sequences for the primers used quantitative polymerase chain reaction in the present study.
| Gene | Primer | Sequence |
|---|---|---|
| Proline-rich 4 | Forward | 5′-ACGAGGACACCGTCAACTCT-3′ |
| Reverse | 5′-TCAATGTCATGGCTTTCTGAAG-3′ | |
| β-actin | Forward | 5′-CTCGCGCTACTCTCTCTTTCTGG-3′ |
| Reverse | 5′-GCTTACATGTCTCGATCCCACTTAA-3′ |
RNA isolation and cDNA synthesis from saliva
Saliva samples were centrifuged at 11,000 × g for 20 min at 4°C to separate the supernatant from the cellular fraction. The cell pellet was resuspended in 1 ml 1X PBS buffer, and RNA was extracted from the cell pellet using the Hybrid-R Blood RNA kit (GeneAll Biotechnology Co., Ltd., Seoul, South Korea) according to the manufacturer's protocol. A total of 400 ng isolated RNA was reverse-transcripted using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) according to the manufactuer's protocol.
Statistical analysis
Statistical analyses were performed using SPSS version 20 (IBM Corp., Armonk, NY, USA). A paired Student's t-test or Pearson's χ2 test were used to calculate P-values. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of differently expressed genes
In order to identify the genes that were differently expressed in tumor tissues compared with normal tissues, ACP-based GeneFishing PCR was performed using a combination of 20 arbitrary primers and two anchored oligo (dT) primers (dT-ACP1 and dT-ACP2). A total of 4 pairs of tumor and normal tissues were used for the GeneFishing assay.
A total of 27 differently expressed genes (DEGs) were identified; of these, 15 DEGs were downregulated and 12 DEGs were upregulated in tumor tissues compared with normal tissues. Amongst the 27 DEGs, 12 DEGs were isolated, cloned, sequenced and searched in GenBank if there was a difference of >10% in band intensity. The isolated DEGs and their characteristics are summarized in Table II. The PRR4 gene was identified as one of the downregulated genes, and due to the functional importance of PRR4 in the saliva, PRR4 mRNA expression levels were further investigated in the tumor tissues and adjacent normal tissues from 90 patients with laryngeal cancer.
Table II.
Identified DEGs in the present study.
| Annealing control primer | DEG no. | Sequence homology search | GeneBank accession no. | Function |
|---|---|---|---|---|
| 2 | DEG4 | Human DNA sequence from clone RP11-15N12 on chromosome 6, complete sequence (contains solute carrier family 22 member 23 isoform a) | AL160398.27 | Transmembrane protein that transports organic ions across cell membranes |
| 2 | DEG5 | Homo sapiens tetraspanin 1, mRNA | NM_005727.3 | Transmembrane protein that regulates cell adhesion, migration, proliferation and differentiation |
| 3 | DEG7 | Homo sapiens chromosome 1 clone RP5-1014C4, complete sequence | AC104456.2 | |
| 4 | DEG9 | Human DNA sequence from clone RP5-1148A21 on chromosome 6, complete sequence | AL135905.6 | |
| 8 | DEG10 | Homo sapiens mitochondrial ribosomal protein L53, mRNA | NM_053050.4 | Component of the large subunit of the mitochondrial ribosome that is encoded by the nuclear genome |
| 9 | DEG11 | Human DNA sequence from clone RP4-668J24 on chromosome 6p25.1–25.3, complete sequence | AL034346.31 | |
| 10 | DEG14 | Homo sapiens ribosomal protein S26, mRNA | NM_001029.3 | Ribosomal protein that is a component of the 40S subunit |
| 11 | DEG17 | Homo sapiens chromosome 16 clone RP11-488I20, complete sequence | AC007353.5 | |
| 12 | DEG20 | Homo sapiens immunoglobulin heavy constant γ 1 (G1m marker), mRNA (cDNA clone IMAGE:4851063) | BC018747.1 | |
| 13 | DEG21 | Homo sapiens proline-rich 4 (lacrimal), transcript variant 2, mRNA | NM_007244.2 | Secreted protein which may have protective functions in the eye and the mouth |
| 15 | DEG22 | Homo sapiens 1-acylglycerol-3-phosphate O-acyltransferase 3, transcript variant 1, mRNA | NM_020132.4 | Acyltransferase that converts lysophosphatidic acid into phosphatidic acid, which is the second step in the de novo phospholipid biosynthetic pathway |
| 17 | DEG23 | Homo sapiens follistatin-like 1, mRNA | NM_007085.4 | Promotes keratinocyte migration and wound repair |
DEG, differentially expressed gene; ACP, annealing control primer.
Confirmation of ACP data by RT-qPCR for PRR4
In order to confirm the expression pattern of DEG21 (PRR4 gene), its expression levels were analyzed using RT-qPCR in 90 tumor samples and adjacent non-cancerous tissue samples. The PRR4 transcript was detected in all tumor and normal tissue samples except for 3 tumor tissues and 1 normal tissue. However, PRR4 expression was significantly decreased in 65 (72.2%) of the 90 tumor samples when compared with the paired non-cancerous tissue (P<0.001; Table III). Increased expression was observed in 24 tumor samples (26.7%), and no change was detected in 1 sample. The mean ΔCq levels were 9.1±3.7 and 5.7±5.5 for the tumor and the normal tissue samples, respectively, and a significant difference was identified (P<0.001; Fig. 1; Table IV). This indicates an 11-fold decrease in PRR4 expression in the tumor tissues compared with the non-cancerous tissue. No association was identified with any clinicopathological characteristic, including clinical stage, histology, sex, age, histological grade and smoking status (Table III).
Table III.
Distribution of PRR4 gene expression and its association with clinicopathological characteristics of the patients.
| PRR4 gene expression, n (%) | ||||
|---|---|---|---|---|
| Clinicopathological parameter | Decreased | No change | Increased | P-valuea |
| Clinical stage | 0.903 | |||
| Early stage (I+II) | 7 (7.8) | 0 (0) | 3 (3.3) | |
| Advanced stage (III+IV) | 57 (63.3) | 1 (1.1) | 20 (22.2) | |
| Unknown | 1 (1.1) | 0 (0) | 1 (1.1) | NT |
| Histology | NT | |||
| SCC | 64 (71.1) | 1 (1.1) | 21 (23.3) | |
| Non-SCC | 0 (0) | 0 (0) | 2 (2.2) | |
| Unknown | 1 (1.1) | 0 (0) | 1 (1.1) | |
| Sex | 0.951 | |||
| Female | 2 (2.2) | 0 (0) | 1 (1.1) | |
| Male | 63 (70) | 1 (1.1) | 23 (25.6) | |
| Age | 0.5 | |||
| ≤50 | 8 (8.9) | 0 (0) | 5 (5.6) | |
| >50 | 57 (63.3) | 1 (1.1) | 18 (20) | |
| Unknown | 0 (0) | 0 (0) | 1 (1.1) | NT |
| Histological grade | 0.327 | |||
| Low grade (1+2) | 31 (34.4) | 0 (0) | 8 (8.9) | |
| High grade (3+4) | 30 (33.3) | 1 (1.1) | 14 (15.6) | |
| Unknown | 4 (4.4) | 0 (0) | 2 (2.2) | NT |
| Smoking | 0.815 | |||
| Smoker | 53 (58.9) | 1 (1.1) | 20 (22.2) | |
| Non-smoker | 11 (12.2) | 0 (0) | 3 (3.3) | |
| Unknown | 1 (1.1) | 0 (0) | 1 (1.1) | NT |
| Total | 65 (72.2) | 1 (1.1) | 24 (26.7) | 0.001 |
Statistical analyses were performed using the Pearson's χ2 test. NT, not tested; SCC, squamous cell carcinoma; PRR4, proline-rich 4.
Figure 1.
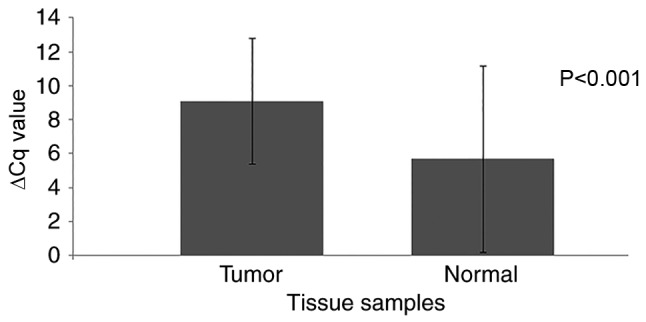
Quantification of the mean expression levels of the proline-rich 4 gene in tumor and normal tissue samples. Cq, quantification cycle.
Table IV.
Mean expression levels of the PRR4 gene in tumor and normal tissues.
| Tissue type | PRR4 Cq (mean ± SD) | β-actin Cq (mean ± SD) | ΔCq (mean ± SD) | ΔΔCq | 2−ΔΔCq | P-value |
|---|---|---|---|---|---|---|
| Tumor | 31.4±4.1 | 22.3±2.5 | 9.1±3.7 | 3.4 | 0.09 | <0.001 |
| Normal | 28.6±5.9 | 22.9±2.6 | 5.7±5.5 | 0 | 1 |
Statistical analysis was performed using a paired Student's t-test. PRR4, proline-rich 4; Cq, quantification cycle; SD, standard deviation; PRR4, proline-rich 4.
Expression of PRR4 in the control group saliva
PRR4 expression was analyzed in the saliva of healthy smoker and non-smoker groups, and the expression levels were compared. The mean ΔCq values were 7.7±1.9 and 7.7±1.7 for the smoker and the non-smoker groups, respectively (data not shown). No significant differences were identified between the studied groups.
Discussion
Cancer may be described as a disease of altered gene expression. As a result of the up- or downregulation of different genes, numerous genes are activated or silenced, which alters the overall activity of the cell and supports tumor development (24). Therefore, identifying differentially expressed genes in tumor cells may help in understanding the molecular mechanisms that underlie the development and progression of cancer (25,26). To date, by using different high-throughput technologies including cDNA arrays and transcriptome analyses, numerous genes differentially expressed in tumor cells have been identified (27–29). However, it remains difficult to determine the genetic events that function in tumorigenesis.
The PCR-based GeneFishing technology is a relatively novel method that can be used to reveal differences in the gene expression levels between two or more samples (18). In the present study, 27 differentially expressed RNAs in laryngeal tumor samples were identified, and 12 of these were characterized by cloning and sequencing. Based on Gene Ontology, Serial Analysis of Gene Expression and The Cancer Genome Atlas database queries, no associations between these genes and HNSCC or laryngeal cancer could be identified. However, the effect of tetraspanin 1 and follistatin-like 1 in the progression of other types of cancer have been reported (30,31).
PRR4 mRNA was discovered in 1995 by Dickinson and Thiesse (5) in the acinar cells of the human lacrimal gland. Transcriptome studies demonstrated that PRR4 is highly expressed at the mRNA level in the submucosal glands, including the parotid gland, and is considered to be a potential biomarker for indicating the functional efficiency of the gland (9,10). PRR4 is one of the salivary PRPs, constituting approximately two-thirds of the proteins secreted by human parotid glands (11,12). PRPs contain repetitive PRR sequences or multiple tandem repeats with minor variations between repeated sequences (32–34). Salivary PRPs have several functions, but most are likely to serve a protective role by binding to tannins via a repetitive domain in the epithelial surfaces (15–17). Tannins are water-soluble polyphenols that are present in a variety of plant-derived foods. It has been reported that tannins are mutagenic and carcinogenic compounds; animal experiments also revealed that the subcutaneous injection of tannins resulted in tumor formation (35). Due to its function in binding to pathogens and tannins, the PRR4 gene was selected for further investigation in the present study.
A number of PRPs additionally function in the regulation of transcription by binding to transcription factors (36,37). However, their exact functions remain unclear. The downregulation of the PRR4 mRNA in tear fluid has been associated with pathological conditions including DES, thyroid associated orbitopathy, Sjögren syndrome, blepharitis and diabetic retinopathy (6–8,38–41). We hypothesized that laryngeal cells may also express PRR4 mRNA depending on the anatomical location in the larynx. Areas of the larynx have small glands, termed the minor salivary glands, which produce mucus and saliva to lubricate and moisten the area. Therefore, the expression of the PRR4 gene was used for further validation. As a result of expression analysis, it was observed that in the majority of tumor tissues, PRR4 expression was downregulated.
PRR4 is a relatively novel protein and there are few studies at present investigating the PRR4 gene in cancer. Zinovyeva et al (42) identified the expression of 80 genes downregulated in the esophageal tumor cells compared with normal tissues using suppression subtractive hybridization. Although one of the identified genes was PRR4, the study did not select this gene for further confirmation analysis. However, another PRP that belonged to the same family (small proline-rich protein 3) was selected for further analysis and its downregulation was associated with esophageal squamous cell carcinoma. As a result of the pilot study CapLC-ESI-Q-TOF, Casado et al (43) reported that PRR4 was the one of the proteins present in the sputum of a non-smoker study group, whilst it was absent from the sputum of the chronic obstructive pulmonary disease group. In the same previous study, the authors demonstrated that PRR4 was downregulated in healthy smokers. In the present study, 82.2% of the patients with laryngeal cancer were smokers. As a result of this, we hypothesized that the downregulation of PRR4 in the tumor samples may be due to smoking. To test the potential effect of smoking on PRR4 mRNA expression, PRR4 mRNA expression levels in the saliva of 25 smokers and 25 non-smoking healthy subjects were investigated. However, no difference was observed between the PRR4 levels of these groups. Therefore, the results of the present study indicate that the expression rate of the PRR4 gene was directly associated with malignancy in laryngeal tumors. As a novel protein, and due to its function in clearing bacterial pathogens and binding tannins, it may be concluded that PRR4 functions in the progression of laryngeal cancer and HNSCC.
Laryngeal cancer is a tumor of the upper aerodigestive tract with a low overall survival rate (44). Therefore, there is an urgent need for the earlier detection of laryngeal cancer and the identification of theurapeutic target molecules. The results of the present study suggest that the function of PRR4 in laryngeal tumor warrants further study.
Acknowledgements
The present study was supported by the Scientific Research Projects Coordination Unit of Istanbul University (grant nos. 49005 and 20452).
Glossary
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- PRR4
proline-rich 4
- PRP
proline rich protein
- DES
dry eye syndrome
- ACP
annealing control primer
- DEG
differently expressed gene
- LB
Luria-Bertani
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- PBS
phosphate buffered saline
- GO
gene ontology
- SAGE
serial analysis of gene expression
- TCGA
the cancer genome atlas
- COPD
chronic obstructive pulmonary disease
References
- 1.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 2.Bray I, Brennan P, Boffetta P. Projections of alcohol- and tobacco-related cancer mortality in Central Europe. Int J Cancer. 2000;87:122–128. doi: 10.1002/1097-0215(20000701)87:1<122::AID-IJC18>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Edefonti V, Bravi F, Garavello W, La Vecchia C, Parpinel M, Franceschi S, Dal Maso L, Bosetti C, Boffetta P, Ferraroni M, Decarli A. Nutrient-based dietary patterns and laryngeal cancer: Evidence from an explatory analysis. Cancer Epidemiol Biomarkers Prev. 2010;19:18–27. doi: 10.1158/1055-9965.EPI-09-0900. [DOI] [PubMed] [Google Scholar]
- 4.Loyo M, Pai S. The molecular genetics of laryngeal cancer. Otolaryngol Clin North Am. 2008;41:657–672. doi: 10.1016/j.otc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Dickinson DP, Thiesse M. A major human lacrimal gland mRNA encodes a new proline-rich protein family member. Invest Ophthalmol Vis Sci. 1995;36:2020–2031. [PubMed] [Google Scholar]
- 6.Grus FH, Podust VN, Bruns K, Lackner K, Fu S, Dalmasso EA, Wirthlin A, Pfeiffer N. SELDI-TOF-MS ProteinChip array profiling of tears from patients with dry eye. Invest Ophthalmol Vis Sci. 2005;46:863–876. doi: 10.1167/iovs.04-0448. [DOI] [PubMed] [Google Scholar]
- 7.Matheis N, Okrojek R, Grus FH, Kahaly GJ. Proteomics of tear fluid in thyroid-associated orbitopathy. Thyroid. 2012;22:1039–1045. doi: 10.1089/thy.2012.0119. [DOI] [PubMed] [Google Scholar]
- 8.Csősz É, Boross P, Csutak A, Berta A, Tóth F, Póliska S, Török Z, Tőzsér J. Quantitative analysis of proteins in the tear fluid of patients with diabetic retinopathy. J Proteomics. 2012;75:2196–2204. doi: 10.1016/j.jprot.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Fischer AJ, Goss KL, Scheetz TE, Wohlford-Lenane CL, Snyder JM, McCray PB., Jr Differential gene expression in human conducting airway surface epithelia and submucosal glands. Am J Respir Cell Mol Biol. 2009;40:189–199. doi: 10.1165/rcmb.2008-0240OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamkin MS, Arancillo AA, Oppenheim FG. Temporal and compositional characteristics of salivary protein adsorption to hydroxyapatite. J Dent Res. 1996;75:803–808. doi: 10.1177/00220345960750021101. [DOI] [PubMed] [Google Scholar]
- 11.Edgar WM. Saliva: Its secretion, composition and functions. Br Dent J. 1992;172:305–312. doi: 10.1038/sj.bdj.4807861. [DOI] [PubMed] [Google Scholar]
- 12.Azen EA, Amberger E, Fisher S, Prakobphol A, Niece RL. PRB1, PRB2, and PRB4 coded polymorphisms among human salivary concanavalin-A binding, II-1, and Po proline-rich proteins. Am J Hum Genet. 1996;58:143–153. [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function; Proc Natl Acad Sci USA; 2009; pp. 19819–19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 15.Hagerman AE. Chemistry of Tannin-Protein Complexation. In: Hemingway RW, Karchesy JJ, Branham SJ, editors. Chemistry and Significance of Condensed Tannins. Plenum Press; New York: 1989. pp. 323–333. [DOI] [Google Scholar]
- 16.Hatton MN, Loomis RE, Levine MJ, Tabak LA. Masticatory lubrication. The role of carbohydrate in the lubricating property of a salivary glycoprotein-albumin complex. Biochem J. 1985;230:817–820. doi: 10.1042/bj2300817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loomis RE, Bergey EJ, Levine MJ, Tabak LA. Circular dichroism and fluorescence spectroscopic analyses of a proline-rich glycoprotein from human parotid saliva. Int J Pept Protein Res. 1985;26:621–629. doi: 10.1111/j.1399-3011.1985.tb03220.x. [DOI] [PubMed] [Google Scholar]
- 18.Hwang IT, Kim YJ, Kim SH, Kwak CI, Gu YY, Chun JY. Annealing control primer system for improving specificity of PCR amplification. Biotechniques. 2003;35:1180–1184. doi: 10.2144/03356st03. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium: Gene ontology consortium: Going forward. Nucleic Acids Res. 2015;43(Database Issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, et al. A public database for gene expression in human cancers. Canc Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 23.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boundless: Altered gene expression in cancer. Boundless Biology. Boundless. 2016. https://www.boundless.com/biology/textbooks/boundless-biology-textbook/gene-expression-16/cancer-and-gene-regulation-118/altered-gene-expression-in-cancer-470-11690/ https://www.boundless.com/biology/textbooks/boundless-biology-textbook/gene-expression-16/cancer-and-gene-regulation-118/altered-gene-expression-in-cancer-470-11690/
- 25.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 27.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 28.van Baal JW, Milana F, Rygiel AM, Sondermeijer CM, Spek CA, Bergman JJ, Peppelenbosch MP, Krishnadath KK. A comparative analysis by SAGE of gene expression profiles of esophageal adenocarcinoma and esophageal squamous cell carcinoma. Cell Oncol. 2008;30:63–75. doi: 10.1155/2008/328529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen X, Ma L, Bi J, Wei G, Fong G, Xue X. TSAPN1 functions as an oncogene in gastric cancer and is downregulated by miR-573. FEBS Lett. 2015;589:1988–1994. doi: 10.1016/j.febslet.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 31.Ni X, Cao X, Wu Y, Wu J. FSTL1 supresses tumor cell proliferation, invasion and survival in non-small cell lung cancer. Oncol Rep. 2018;39:13–20. doi: 10.3892/or.2017.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boze H, Marlin T, Durand D, Pérez J, Vernhet A, Canon F, Sarni-Manchado P, Cheynier V, Cabane B. Proline-rich salivary proteins have extended conformations. Biophys J. 2010;99:656–665. doi: 10.1016/j.bpj.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennick A. Structural and genetic aspects of proline-rich proteins. J Dent Res. 1987;66:457–461. doi: 10.1177/00220345870660021201. [DOI] [PubMed] [Google Scholar]
- 34.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoltz DR. In: Carcinogenes and mutagenes in the Environment. Stich HF, editor. Vol. 3. CRC Press; Boca Raton, Florida: 1982. p. 75. [Google Scholar]
- 36.Mermod N, O'Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 37.Gessler M, Poustka A, Cavenee W, Neve RL, Orkin SH, Bruns GA. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature. 1990;343:774–778. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- 38.Aluru SV, Agarwal S, Srinivasan B, Iyer GK, Rajappa SM, Tatu U, Padmanabhan P, Subramanian N, Narayanasamy A. Lacrimal proline-rich 4 (LPRR4) protein in the tear fluid is a potential biomarker of dry eye syndrome. PLoS One. 2012;7:e51979. doi: 10.1371/journal.pone.0051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perumal N, Funke S, Pfeiffer N, Grus FH. Characterization of lacrimal proline-rich protein 4 (PRR4) in human tear proteome. Proteomics. 2014;14:1698–1709. doi: 10.1002/pmic.201300039. [DOI] [PubMed] [Google Scholar]
- 40.Tsai PS, Evans JE, Green KM, Sullivan RM, Schaumberg DA, Richards SM, Dana MR, Sullivan DA. Proteomic analysis of human meibomian gland secretions. Br J Ophthalmol. 2006;90:372–377. doi: 10.1136/bjo.2005.080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koo BS, Lee DY, Ha HS, Kim JC, Kim CW. Comparative analysis of the tear protein expression in blepharitis patients using two-dimensional electrophoresis. J Proteome Res. 2005;4:719–724. doi: 10.1021/pr0498133. [DOI] [PubMed] [Google Scholar]
- 42.Zinovyeva MV, Monastyrskaya GS, Kopantzev EP, Vinogradova TV, Kostina MB, Sass AV, Filyukova OB, Uspenskaya NY, Sukhikh GT, Sverdlov ED. Identification of some human genes oppositely regulated during esophageal squamous cell carcinoma formation and human embryonic esophagus development. Dis Esophagus. 2010;23:260–270. doi: 10.1111/j.1442-2050.2009.01008.x. [DOI] [PubMed] [Google Scholar]
- 43.Casado B, Iadarola P, Pannell LK, Luisetti M, Corsico A, Ansaldo E, Ferrarotti I, Boschetto P, Baraniuk JN. Protein expression in sputum of smokers and chronic obstructive pulmonary disease patients: A pilot study by CapLC-ESI-Q-TOF. J Proteome Res. 2007;6:4615–4623. doi: 10.1021/pr070440q. [DOI] [PubMed] [Google Scholar]
- 44.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]


