Abstract
The aim of the present study was to evaluate the clinical importance of melanoma-associated antigen D4 (MAGE-D4) expression in glioma, and to identify it as a valuable prognostic biomarker and therapeutic target. To achieve this, the expression of MAGE-D4 protein in 124 tumor tissues from patients with glioma was measured using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and immunohistochemistry (IHC), and the associations between MAGE-D4expression and clinicopathological factors were evaluated. The survival analysis demonstrated the significant prognostic value of MAGE-D4 in glioma using follow-up data. RT-qPCR and IHC analysis confirmed that MAGE-D4 mRNA and protein expression levels were significantly increased in glioma tissues compared with those in normal brain tissues. The present study demonstrated that the percentage of glioma tissues with high expression of MAGE-D4 mRNA was 67.74%, and the percentage positive for MAGE-D4 protein expression was 78.23%. All patients with high MAGE-D4 expression in cancerous tissues experienced significantly reduced median overall survival (OS; 18.00 vs. 33.29 months; P<0.001) and recurrence-free survival (RFS; 12.7 vs. 28.3 months; P<0.001) times compared with those with low MAGE-D4 expression. In the patients with lower grade glioma [World Health Organization (WHO), I–II], similar results were obtained for the OS (26.11 vs. 57.85 months; P=0.013) and RFS (22.7 vs. 55.3 months; P=0.010) times; however, in patients with high-grade glioma (WHO, III–IV), there were no significant differences between high and low MAGE-D4 expression levels with regard to OS and RFS times (P>0.05). Multivariate analysis indicated that high MAGE-D4 protein expression was an important independent prognostic factor for patients with glioma (hazard ratio, 2.384; P=0.005), and was significantly associated with higher grade glioma (P<0.001). These results indicated that MAGE-D4 may be a potential biomarker for glioma and an important prognostic factor for patients with new or recurring glioma.
Keywords: melanoma-associated antigen-D4, glioma, prognostic, reverse transcription-quantitative polymerase chain reaction, immunohistochemistry
Introduction
Globally, malignant glioma is the most common type of central nervous system tumor in humans, and is characterized by high morbidity and mortality rates (1). Thus far, the standard treatment has included tumor resection with maximal range, adjuvant radiotherapy and concomitant chemotherapy (1,2). Numerous types of treatment methods have been adopted, but the median survival time remains limited to 16–19 months (2). This poor prognosis is primarily due to the invasive potential of malignant glioma (3). To date, the molecular mechanisms underlying the invasion of glioma remain unclear (4); therefore, it is urgent that biological and molecular mechanisms underlying glioma development and progression are elucidated, which can assist in the development of more effective treatments.
The melanoma-associated antigen (MAGE) gene was first discovered in melanoma cells, and melanoma and glial tumors are known to be commonly derived from neural ectoderm. MAGE has also been demonstrated to serve as a glioma-specific antigen, and an increasing amount of evidence has indicated that it is particularly expressed in glioma tumors (5). The MAGE gene was first recorded by van der Bruggen et al in 1991 (6), when it was identified to be a novel gene encoding a tumor-specific antigen presented by human melanoma cells. In total, >50 MAGE genes have been identified and divided into two types, based on differences in the sequence and tissue-specific gene expression patterns (7). Type I MAGE genes, which encode for MAGE-A, -B and -C proteins, are located on the X chromosome; these proteins are expressed in the process of germ cell development, but not in normal, mature somatic cells (8). By contrast, the localization and expression of type II MAGE proteins (MAGE-D, -E, -F, -G and -H) have yet to be clearly elucidated. MAGE-D is divided into four groups, which belong to the type II MAGE protein family (7).
MAGE-D4 is a tumor-specific antigen that serves an important role in invasion and metastasis. Numerous studies have demonstrated that MAGE-D4 can drive cell progression and metastasis, including that of renal cell carcinoma (9), non-small cell lung cancer (10), breast cancer (11), oral squamous cell carcinoma (12), hepatocellular carcinoma (13), squamous cell carcinoma of the esophagus (14) and colorectal cancer (15). In a previous study, it was identified that the MAGE-D4 mRNA and protein expression levels were relatively high in gliomas, but that limited, lower or no expression was present in normal tissue (16); however, an analysis of the function of MAGE-D4 for survival in glioma cells has not been reported. The evaluation of MAGE-D4 expression levels in glioma, independent of other MAGE genes, would be beneficial to glioma treatment options; therefore, in the present study, the association of MAGE-D4 overexpression with clinical indices and outcomes [sex, age, tumor size, World Health Organization (WHO) grade, the Ki-67 labeling index (percentage of Ki-67 positive cells) and the Karnofsky performance scale (KPS) index] was evaluated using follow-up materials. The purpose of the present study was to investigate the potential of the MAGE-D4 gene as a cancer-associated tumor marker, which may represent a valuable prognostic biomarker and immunotherapeutic target for glioma.
Materials and methods
Patient characteristics and tissue specimens
The present study involved the analysis of 124 cases of glioma from patients aged 2–78 years (median age, 38 years) at the time of diagnosis. These patients consisted of 78 males and 46 females. Furthermore, 12 samples of normal brain biopsy tissue obtained from the Specimens Library of the Guangxi Medical University's Department of Histology and Embryology (Nanning, China) were included in the study as a control. None of the patients received preoperative radiotherapy or chemotherapy. Surgically resected tumor tissues were collected consecutively from patients at The First Affiliated Hospital and the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China) between September 2009 and June 2016, and were analyzed following study approval from the Ethics Committee of Guangxi Medical University and with written informed consent from the patients. Demographics, tumor size, WHO Classification of Tumors of the Central Nervous System (15), Ki-67 expression, p53 expression and KPS score were investigated from the retrospective database. Of the 124 patients, 84 were followed up via a telephone interview (Table I). Postoperative follow-up examinations, including a physical examination and enhanced computed tomography scan, were conducted every 3–6 months. A treatment strategy was selected from surgery, adjuvant radiotherapy and chemotherapy, according to the tumor status, tumor grade and patient condition. Collected tissue specimens were stored in liquid nitrogen at −80°C prior to analysis. Pathological evaluation with tumor staging was based on the 2007 WHO Classification of Tumors of the Central Nervous System (17).
Table I.
Clinical parameters of follow-up patients with gliomas (n=84) who were enrolled for survival analysis.
| Variable | Number of patients | Percentage of patients |
|---|---|---|
| Sex | ||
| Male | 56 | 66.67 |
| Female | 28 | 33.33 |
| Age, years | ||
| <38 | 41 | 48.81 |
| ≥38 | 43 | 51.19 |
| Tumor size, cm | ||
| <5 | 38 | 45.24 |
| ≥5 | 46 | 54.76 |
| WHO classificationa | ||
| I–II | 40 | 47.61 |
| III–IV | 44 | 52.38 |
| KPS | ||
| <70 | 35 | 41.67 |
| ≥70 | 49 | 58.33 |
According to the 2007 WHO Classification of Tumors of the Central Nervous System. WHO, World Health Organization; KPS, Karnofsky Performance Scale.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA extraction from frozen tissues and RT-qPCR were performed as previously described (18). Briefly, total RNA, which was derived from glioma tissues and normal brain specimens, was prepared using TRIzol® (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Then RNA was reverse transcribed into cDNA with RevertAid™ First Strand cDNA Synthesis kit (Fermentas, USA), according to the manufacturer's instructions. Primers of MAGE-D4 (19) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (20) were obtained from Applied Biosystems (Thermo Fisher Scientific, Inc.). The forward primer sequence for MAGE-D4 was 5′-CCAGCTTCTTCTCCTGGATC-3′ and the reverse primer sequence was 5′-GTAACACTGATACCCAAAACATG-3′. The forward primer sequence for GAPDH was 5′-GCACCGTCAAGGCTGAGAAC-3′ and the reverse primer sequence was 5′-TGGTGAAGACGCCAGTGGA-3′. qPCR was performed in a 20 µl total reaction volume using SYBR® Premix Ex Taq™ II kit (Takara Biotechnology Co., Ltd.) on the ABI Prism 7900 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cycling parameters were as follows: Initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec, and annealing and extension at 60°C for 1 min. Dissociation curves were performed to confirm specific product amplification. MAGE-D4 mRNA expression was standardized to that of GAPDH mRNA. All samples were tested in triplicate. Expression levels for samples were demonstrated as MAGE-D4 values standardized to the MAGE-D4/GAPDH ratio. The relative number of target transcripts was normalized to the number of human GAPDH transcripts in the same sample. The relative quantitation of target gene expression was performed using the standard curve or quantification cycle method (21).
Immunohistochemistry (IHC)
IHC was performed according to previous studies (15,16). Paraffin-embedded tissues were formalin fixed in 10% (v/v) formalin for 24 h at room temperature and cut into 4 µm-thick sections for IHC. Sections of formalin-fixed and paraffin-embedded tissues were dried in an oven at 60°C for 1 h, dewaxed with xylene and rehydrated through a descending series of alcohols (95, 80, 70 and 50%, v/v). The sections were then heated in EDTA (pH 8.0; 120°C for 3 min) in order to achieve antigen retrieval and washed by PBS prior to being treated with 3% H2O2 for 15 min at room temperature to block endogenous peroxidase activity. Non-specific binding was blocked by incubation with 5% (v/v) normal goat serum (OriGene Technologies, Inc., Beijing, China) for 10 min at 25°C. Subsequently, sections were incubated with the primary antibody (mouse anti-human monoclonal antibody IgG; cat. no. sc-393203 HRP; dilution, 1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Following washing three times with 0.01 M phosphate buffer (Beijing Solarbio Science & Technology Co., Ltd.; pH 7.2), Next, the sections were incubated at room temperature for 20 min with biotin conjugated goat anti-mouse IgG (cat. no. TA130009; dilution, 1:200; OriGene Technologies Inc.), which was used as the biotinylated secondary antibody. And then, horseradish peroxidase-labeled streptavidin (OriGene Technologies, Inc.) was performed for 20 min at room temperature. Finally, all antibody staining was developed using 3,3′-diaminobenzidine (DAB; Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China) as the chromogen for 5 min at room temperature and counterstained with hematoxylin. Omission of primary antibody was used as a negative control, and a known glioma tissue section with MAGE-D4-positive expression as a positive control.
According to the staining characteristics of MAGE-D4 protein, the majority was located within the cell cytoplasm and nucleus. The final score of protein expression was evaluated by the sum of the scores of staining intensity and the scores of the positive cell rate (22,23). The staining intensity was defined using the following scores: 0, negative; 1, very weak; 2, weak; 3, moderate; 4, intense; and 5, very intense. The rate of positive glioma cells were as follows: 0, absence of positive cells; 1, 1–5% positive cells; 2, 6–25% positive cells; 3, 26–50% positive cells; 4, 51–75% positive cells; and 5, 76–100% positive cells. Sum scores were as follows: 0–1, negative expression (−); 2–3, weak expression (+); 4–5, moderate expression (++); and 6–7, strong expression (+++). The assessment of positive immunoreactivity was achieved by two independent pathologists (Department of Pathology, First Affiliated Hospital of Guangxi Medical University, China) using a blind method. At least five independent foci of neoplastic infiltration in each tissue specimen were observed using an optical microscope (Nikon Corporation, Tokyo, Japan) at magnification, ×100 and ×200.
Bioinformatics and statistical analysis
ONCOMINE (https://www.oncomine.org) is currently the largest cancer microarray database and integrated data-mining platform globally, and was used to retrieve and analyze the gene expression profile microarray data for tumor samples (24). The expression levels of MAGE-D4 in a number of types of glioma were obtained from the Oncomine dataset (http://www.proteinatlas.org). A one-way analysis of variance (ANOVA) test was used to identify the significantly expressed differentially expressed genes (DEGs) in glioma samples, with a combination of P<0.05 and the |log2FC (fold change) |>2 used as the threshold. The immunohistochemical staining results of MAGE-D4 in glioma and normal tissues were selected from the Human Protein Atlas (www.proteinatlas.org) was applied to identify the total percentage of positive cells for MAGE-D4 protein. The MAGE-D4 protein expression associated with clinicopathological parameters was analyzed using the χ2 or Fisher's exact probability test. Data are expressed as the mean ± standard deviation. The data of normal brain tissues and different glioma groups were analyzed by an one-way ANOVA test followed by the post-hoc Student-Newman-Keuls test. The prognostic value of the MAGE-D4 protein expression was evaluated using regression analysis. The Kaplan-Meier method was used to calculate overall survival (OS) and recurrence-free survival (RFS) rates, whilst differences in survival curves were compared using the log-rank test. Cox proportional hazard models were used to detect prognostic factors during the multivariable regression analysis. Variables with P<0.05 were included in the final model. All the statistical analyses were conducted using the SPSS version 16 statistical software package for Windows (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
MAGE-D4 mRNA and protein expression analysis using bioinformatics
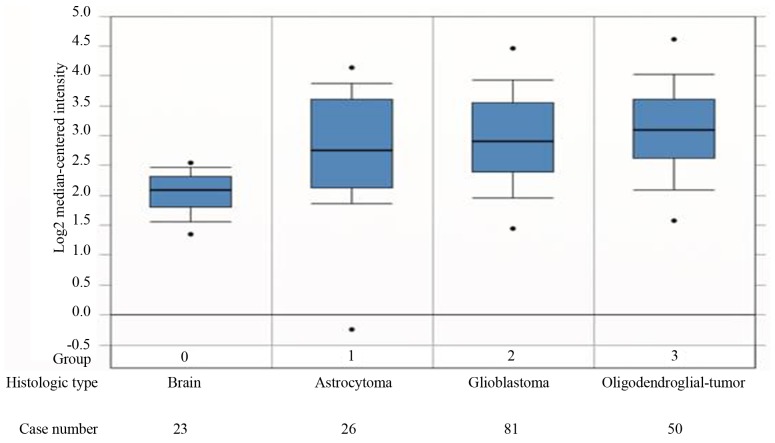
The differences in MAGE-D4 mRNA expression in various types of gliomas can be determined by searching the ONCOMINE microarray datasets. Query results from the Sun Brain 2 dataset (Fig. 1) indicated that MAGE-D4 mRNA expression in different types of glioma tissues (astrocytoma, glioblastoma and oligodendroglial) were all significantly increased compared with that in normal brain tissue (P<0.05; Student-Newman-Keuls post-hoc test). Furthermore, MAGE-D4 protein was detected in 10 cases of glioma, although not in normal brain tissue, using the Human Protein Atlas (HPA) Database (http://www.proteinatlas.org). The HPA database indicated that the total percentage of positive cells for MAGE-D4 protein was 80%. This consisted of 10% with strong expression, 30% with moderate expression and 40% with weak expression. The remaining 20% recorded negative expression. Staining for MAGE-D4 protein was mainly located within the cell nucleus.
Figure 1.
Melanoma-associated antigen-D4 mRNA upregulation in gliomas, including astrocytoma, glioblastoma and oligodendroglia, compared with normal brain tissues, according to the Sun Brain 2 dataset. The figure was generated using ONCOMINE (https://www.oncomine.org/). The 25th, 50th and 75th percentile values are depicted as horizontal lines; the 10th and 90th percentile values are depicted as whiskers; and the maximum and minimum values are depicted as dots.
MAGE-D4 mRNA expression analysis using RT-qPCR
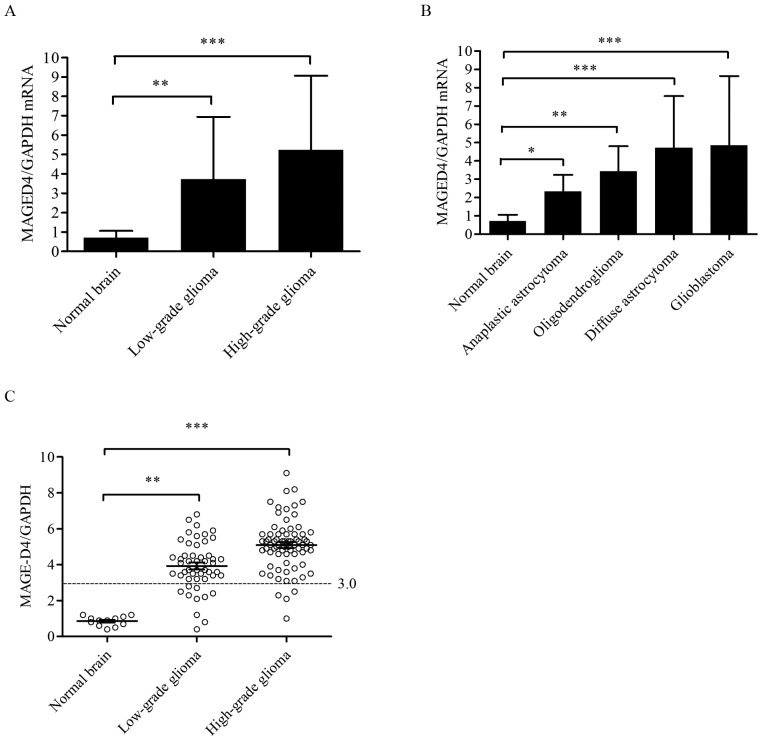
The expression levels of MAGE-D4 mRNA were analyzed in 124 glioma and 12 normal brain tissue specimens using RT-qPCR. The data from the normal brain tissues and the different glioma groups were analyzed using an one-way ANOVA test. There were significant differences between low-grade glioma and high-grade glioma compared with normal brain tissues (F=6.333; P<0.01). The median values of MAGE-D4 mRNA in low-grade glioma (3.733±0.10; P<0.01) and high-grade glioma (5.237±0.13; P<0.001) were significantly higher than that in normal brain tissues (0.710±0.07); however, there was no significant difference in MAGE-D4 mRNA between high-grade and low-grade glioma (P=0.486; Fig. 2A). For the expression levels of MAGE-D4 mRNA in each group, one-way ANOVA demonstrated significant differences between various types of glioma and normal brain tissues (F=5.922; P<0.001). The median value of MAGE-D4 mRNA was highest in glioblastoma (4.855±0.11; P<0.001), while diffuse astrocytoma had the second highest value (4.695±0.04; P<0.001), followed by oligodendroglioma (3.437±0.09; P<0.01) and anaplastic astrocytoma (2.333±0.13; P<0.05), all compared with normal brain tissue (0.710±0.07); however, there was no significant effect of multiple comparisons following a post-hoc test (Fig. 2B). As depicted in Fig. 2C, high expression of MAGE-D4 was defined as exceeding the median value of MAGE-D4/GAPDH in normal brain tissues by three-fold. The results demonstrated that the percentage of glioma tissues that exhibited high mRNA MAGE-D4 expression was 67.74%, whereas 0% of normal brain tissues had high expression levels (P<0.001). The percentage of high mRNA MAGE-D4 expression in low-grade (WHO, I–II; 24.19%) and high-grade gliomas (WHO, III–IV; 43.55%) was compared with that in normal brain tissue, respectively, with significant differences found (P<0.01 and P<0.001, respectively); however, no significant differences were found between high-grade and low-grade glioma (P=0.810). No apparent association was identified between the relative expression level and high expression ratio of MAGE-D4 with age, sex, tumor size and WHO grade (data not shown).
Figure 2.
MAGE-D4 mRNA expression using reverse transcription-quantitative polymerase chain reaction. (A) The median values of MAGE-D4 mRNA expression in low-grade glioma and high-grade glioma were significantly higher than that in normal brain tissue. Values are presented as the mean ± SD. The error bars represent the SD (**P<0.01 and ***P<0.001; Student-Newman-Keuls post-hoc test). (B) MAGE-D4 expression in different types of glioma demonstrated that the expression in glioblastoma was the highest; expression in diffuse astrocytoma was the second highest, followed by oligodendroglioma and anaplastic astrocytoma. Values are presented as the mean ± SD. The error bars represent the SD. (*P<0.05, **P<0.01 and ***P<0.001 compared with normal brain tissue; Student-Newman-Keuls post-hoc test). (C) High expression of MAGE-D4 was defined as exceeding the median value of MAGE-D4/GAPDH in normal brain tissues by three-fold. The cutoff value is 3.0 for MAGE-D4 high expression above the line. The percentage of high expression of MAGE-D4 mRNA in low-grade (WHO, I–II) and high-grade (WHO, III–IV) gliomas was compared with that in normal brain tissue (**P<0.01 and ***P<0.001; χ2 test with subsequent Bonferroni's correction). SD, standard deviation; MAGE, melanoma-associated antigen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
MAGE-D4 protein expression analysis using IHC
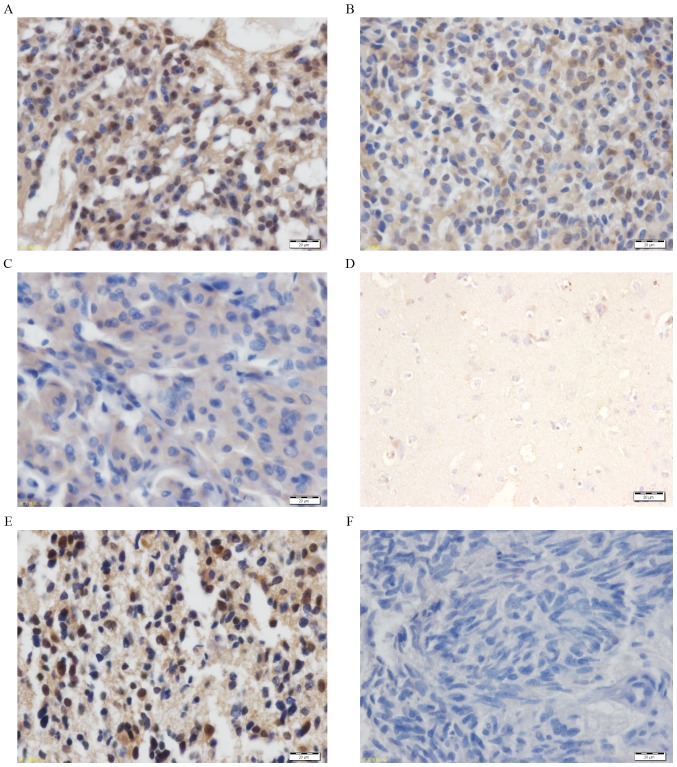
MAGE-D4 protein expression in 124 glioma tissues was analyzed using IHC. The results demonstrated that the total percentage of cells positive for MAGE-D4 protein was 78.23%. The majority of the staining for the MAGE-D4 protein was distributed in the cell cytoplasm and nucleus. The percentage of MAGE-D4-positive protein expression in cells in the low-grade glioma samples (WHO, I–II) was 28.23%, while in high-grade gliomas (WHO, III–IV) it was 50.00% (Table II). Based on age group, the percentage of positive cells in children (<16 years old) was 9.67% and in adults was 68.55%. According to the staining intensity, high expression of the MAGE-D4 protein (Fig. 3A and B) was demonstrated in 59.68% of patients, and low protein expression of MAGE-D4 (Fig. 3C) was demonstrated in the remaining 40.32%. Furthermore, 12 normal brain tissues were stained and the presence of MAGE-D4 was almost non-existent (Fig. 3D). A known glioma tissue section with MAGE-D4-positive expression was used as the positive control (Fig. 3E) and pre-immune rabbit serum was used as a negative control (Fig. 3F).
Table II.
Association between the MAGE-D4 protein and clinical charateristic of patients with glioma.
| Positive/total, n (%) | |||||
|---|---|---|---|---|---|
| Characteristics | Positive/total test, n (%) | Higha | Lowb | χ2 | P-value |
| Sex | |||||
| Male | 61/124 (49.19) | 45/124 (36.29) | 33/124 (26.61) | 0.749 | 0.862 |
| Female | 36/124 (29.03) | 29/124 (23.39) | 17/124 (13.71) | ||
| Age, years | |||||
| <38 | 49/124 (39.51) | 39/124 (31.45) | 23/124 (18.55) | 0.670 | 0.880 |
| ≥38 | 48/124 (38.71) | 35/124 (28.23) | 27/124 (21.77) | ||
| Age group | |||||
| Children | 12/124 (9.67) | 9/124 (7.26) | 11/124 (8.87) | 2.417 | 0.518 |
| Adults | 85/124 (68.55) | 55/124 (44.35) | 49/124 (39.52) | ||
| Tumor size, cm | |||||
| <5 | 47/124 (37.90) | 35/124 (28.23) | 24/124 (19.35) | 0.774 | 0.855 |
| ≥5 | 50/124 (40.32) | 43/124 (34.68) | 22/124 (17.74) | ||
| WHO classificationc | |||||
| I–II | 35/124 (28.23) | 23/124 (18.55) | 32/124 (25.80) | 16.116 | 0.001d |
| III–IV | 62/124 (50.00) | 51/124 (41.13) | 18/124 (14.52) | ||
| Ki-67 expression, % | |||||
| <10 | 51/124 (41.13) | 37/124 (29.84) | 33/124 (26.61) | 4.071 | 0.254 |
| ≥10 | 46/124 (37.10) | 37/124 (29.84) | 17/124 (13.71) | ||
| p53 expression, % | |||||
| ≤10 | 42/124 (33.87) | 27/124 (21.77) | 36/124 (29.03) | 19.969 | <0.001d |
| ≥10 | 55/124 (44.35) | 47/124 (37.90) | 14/124 (11.29) | ||
| KPS score | |||||
| <70 | 42/124 (33.87) | 33/124 (26.61) | 19/124 (15.32) | 0.615 | 0.893 |
| ≥70 | 55/124 (44.35) | 46/124 (37.10) | 26/124 (20.97) | ||
| Total | 97/124 (78.23) | 74/124 (59.68) | 50/124 (40.32) | ||
High MAGE-D4 protein expression (++/+++)
low MAGE-D4 protein expression (−/+)
classified by the 2007 WHO Classification of Tumors of the Nervous System
statistically significant (P<0.05). WHO, World Health Organization; KPS, Karnofsky Performance Scale.
Figure 3.
Immunohistochemical staining of MAGE-D4 protein in (A-C) glioma tissues and (D) normal brain tissue. Staining was primarily located in the cell cytoplasm and nucleus. Strong, moderate and weak immunoreactivities with the polyclonal MAGE-D4 antibody are depicted in images A-C, respectively. (D) MAGE-D4 protein staining was negative in normal brain tissue. (E) A known glioma tissue section that contained MAGE-D4-positive expression served as a positive control. (F) Pre-immune rabbit serum in place of the anti-MAGE-D4 antibody was used as a negative control. Scale bar, 20 µm. MAGE, melanoma-associated antigen.
Association between MAGE-D4 protein expression and clinicopathological parameters
The association of MAGE-D4 protein expression with clinical factors is demonstrated in Table II. There was a significant association between MAGE-D4 protein expression and WHO classification (P<0.001) and p53 expression (P<0.001). By contrast, MAGE-D4 protein expression was not associated with age (P=0.880), sex (P=0.862), age group (P=0.518), tumor size (P=0.855), Ki-67 expression (P=0.254) or KPS score (P=0.893).
Prognostic influence of overexpression of the MAGE-D4 protein
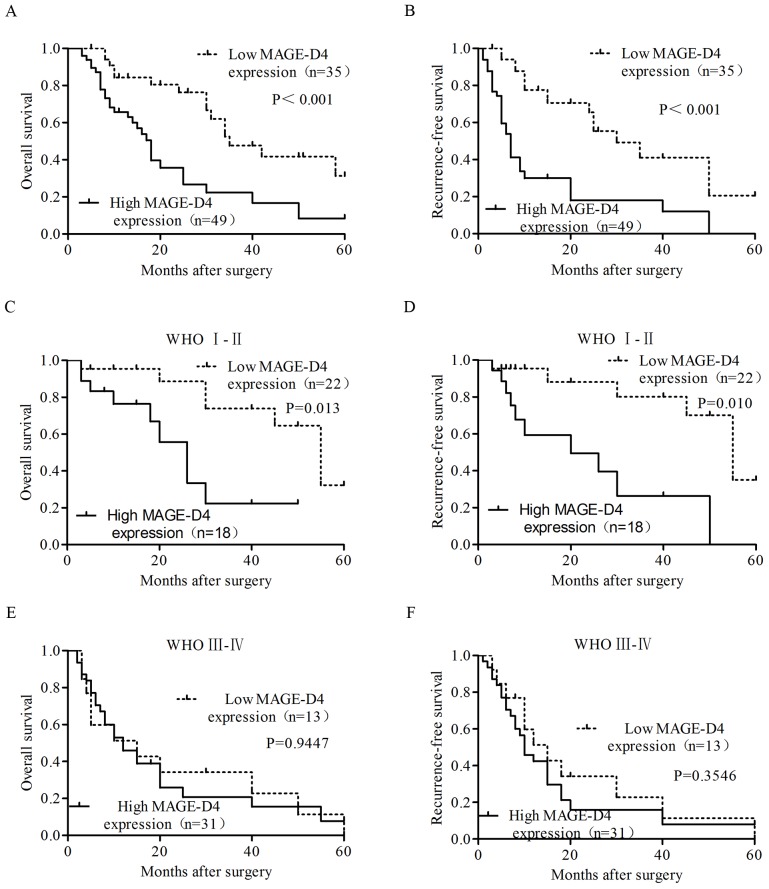
To determine the prognostic value for MAGE-D4, the impact of MAGE-D4 expression and tumor classification was assessed via Kaplan-Meier survival analysis. In total, 84/124 patients with glioma were followed up in the present study and had complete clinical records. The median duration of patient follow-up was 24.8 months (range, 1–69 months). Taking these clinical records into account, a significant positive association between MAGE-D4 protein expression and OS and RFS time was identified in patients with glioma. All patients with high MAGE-D4 expression in cancerous tissues had a significantly reduced median OS (18.00 vs. 33.29 months; P<0.001) and RFS (12.7 vs. 28.3 months; P<0.001) time compared with those with low MAGE-D4 expression (Fig. 4A and B). In the patients with low-grade (WHO, I–II) glioma, the median OS (26.11 vs. 35.85 months; P=0.013) and RFS (22.7 vs. 37.3 months; P=0.010) times were significantly reduced in patients with high MAGE-D4 protein expression (Fig. 4C and D); however, in high-grade gliomas (WHO, III–IV), there was no significant difference between high and low MAGE-D4 expression for OS and RFS (P>0.05; Fig. 4E and F) time. Patients were grouped by sex, age, tumor size (≥5.0 cm) (25), KPS score (<70), WHO classification (III and IV), Ki-67 expression (≥10%), p53 expression (≥10%) (26–28) and MAGE-D4 protein expression. Among these variables, an age of ≥38 years, WHO classification of III and IV, Ki-67 expression ≥10%, p53 expression ≥10% and high MAGE-D4protein expression were considered as significant prognostic parameters via univariate analysis. The survival of patients was impacted by a number of elements; therefore, the implementation of a multivariate analysis was necessary. The analysis indicated that the WHO classification and MAGE-D4 protein expression were significant impact factors in the prognosis of patients with glioma. There was a significant positive association between the high MAGE-D4 protein expression levels and the prognosis of patients with glioma; therefore, it was presumed that the MAGE-D4 protein expression could be identified as an independent prognostic factor for patients with glioma (Table III).
Figure 4.
Survival curves for 84 patients with glioma. Kaplan-Meier curves for OS and RFS time according to MAGE-D4 expression levels in (A and B) all follow-up patients, (C and D) patients with low-grade (WHO, I–II) and (E and F) high-grade (WHO, III–IV) glioma, respectively. (A and B) All patients with high MAGE-D4 expression levels in cancerous tissues experienced significantly shorter OS (P<0.001) and RFS (P<0.001) times than those with low MAGE-D4 expression levels. (C and D) Patients with glioma of WHO grade I–II with high MAGE-D4 expression levels in cancerous tissues experienced significantly shorter OS (P=0.013) and RFS (P=0.010) times than those with low MAGE-D4 expression levels. (E and F) There was no significant difference in OS and RFS (P>0.05) times between high and low MAGE-D4 expression levels in patients with gliomas of WHO grade III–IV. MAGE, melanoma-associated antigen; WHO, World Health Organization; RFS, recurrence-free survival; OS, overall survival.
Table III.
Univariate and multivariate analysis of different prognostic parameters.
| Univariatea | Multivariateb | ||||
|---|---|---|---|---|---|
| Variable | n | HR (95% CI)c | P-value | HR (95% CI)c | P-value |
| Sex, male | 56 | 1.164 (0.645–2.102) | 0.609 | ||
| Age, ≥38 years | 43 | 1.964 (1.102–3.501) | 0.018d | 1.353 (0.633–2.892) | 0.436 |
| Tumor size, ≥5 cm | 46 | 1.079 (0.536–2.172) | 0.829 | ||
| KPS score, <70 | 35 | 1.028 (0.580–1.821) | 0.924 | ||
| WHO classification, III + IV | 44 | 3.343 (1.794–5.861) | <0.001d | 3.698 (2.017–6.782) | <0.001d |
| Ki-67 expression, ≥10% | 36 | 1.885 (1.084–3.278) | 0.021d | 1.336 (0.668–2.671) | 0.413 |
| p53 expression, ≥10% | 44 | 2.079 (1.168–3.701) | 0.010d | 1.463 (0.805–2.656) | 0.212 |
| MAGE-D4 protein expression, high | 49 | 1.971 (1.102–3.527) | 0.019d | 2.384 (1.308–4.347) | 0.005d |
Univariate analysis was performed using the log-rank test
multivariate analysis was performed using the Cox proportional hazards model
HR, 95% CI for relative risk
statistically significant (P<0.05). WHO, World Health Organization; KPS, Karnofsky Performance Scale; HR, hazard ratio; CI, confidence interval; MAGE, melanoma-associated antigen.
Discussion
The diagnosis of glioma relies mainly on clinical manifestations and immunochemical indicators, including histopathological features, although few cancer-associated genes have been identified as valuable prognostic biomarkers (29). Recently, the molecular heterogeneity of cancer has been elucidated by microarray-based expression profiling studies. These microarray studies can be considered as high-throughput platforms for the analysis of gene expression with different tumor types (30,31). The identification of valuable prognostic biomarkers will benefit early diagnosis, tumor therapy response predictions, prognosis and eventually individualized treatment. The results of the microarray-based expression profiling studies demonstrated thatMAGE-D4 is a valuable prognostic biomarker and therapeutic target for glioma (30,31).
In the present study, it was first confirmed that the expression of MAGE-D4 mRNA and protein in different types of glioma specimens was significantly increased compared with that in normal brain tissue, as determined by bioinformatics analysis. Secondly, RT-qPCR and IHC analysis confirmed that MAGE-D4 mRNA and protein expression levels were significantly higher in glioma tissues than in normal brain tissues. The present results indicated that the percentage of glioma tissues with high expression of MAGE-D4 mRNA was 67.74% and that the percentage positive for MAGE-D4 protein expression was 78.23%. MAGE-D4 protein expression was also significantly associated with the age group (P<0.0001), WHO classification (P<0.001) and p53 expression (P<0.001). Finally, patients with high MAGE-D4 expression in cancerous tissues experienced significantly reduced median OS and RFS times compared with those with low MAGE-D4 expression.
MAGE-D4, which was initially defined as MAGE-E1, belongs to the MAGE family of genes and has been identified to exhibit significantly higher expression in tumor tissues than normal tissues (19). In exploring immunotherapeutic treatments, researchers initially identified that MAGE-D4 was overexpressed in a number of human malignant tumor types, including renal cell carcinoma, non-small cell lung cancer and oral squamous cell carcinoma (9,30,31), and the present study results were consistent with this aspect. Compared with our previous report (16), the present study demonstrated that the positive percentage of MAGE-D4 protein was greater than that of the mRNA expression, and there are several possibilities for explaining this inconsistency. According to the patterns of eukaryote gene expression, the level of mRNA does not always accurately reflect the protein level, which may be due to mRNA stability, translational regulation, post-transcriptional modifications, and proteasomal degradation (32,33). In this regard, it is more important to perform a protein analysis than an mRNA analysis; therefore, IHC was used to directly measure MAGE-D4 protein expression in glioma, and these protein levels were used in the subsequent statistical analysis. The present study demonstrated that MAGE-D4 protein expression was not associated with the age group (P>0.05). As there were fewer cases of gliomas in children, therefore, the number of patients included in such studies should be increased in the future.
In a type I MAGE family genetic study, Guo et al (5) reported that the protein expression levels of MAGE-A1, -A3 and -A11 in the glioma specimens were all significantly increased compared with those in the normal brain tissue. Patients with high MAGE-A1 and -A11 expression levels exhibited significantly reduced OS times compared with those with low expression levels (5). The results in terms of these aspects were consistent with the present study of a type II MAGE family. The results indicated that MAGE family types I and II may be used as ideal targets in immunotherapy for glioma, and may be potential markers for a poor prognosis of glioma. He et al (16) reported the differential expression of MAGE-D4 at the mRNA and protein level in glioma and normal brain tissues. There were 41 cases of glioma, with the number of cases being less than the present study with 124 cases of glioma and statistical efficiency being insufficient (16); however, the present study involved the analysis of 124 cases of glioma. In addition, the association of expression levels with clinicopathological parameters was analyzed and multivariate analysis and survival analysis, including OS and RFS time, of patients with glioma were performed. The present study is therefore superior to the study by He et al (16), and further verified that MAGE-D4 is a potential biomarker of glioma and an important factor in the prognosis of patients with glioma initiation and recurrence.
Additionally, with regard to the choice of a tumor size of ≥5 cm, Ki-67 expression of ≥10% and p53 expression of ≥10%, several factors were considered. Firstly, the average tumor size was 5 cm in the present study. Furthermore, according to the survival risk stratification standard for patients with low-grade glioma by the European Organization for Research and Treatment of Cancer (34). Karim et al (34) chose the ‘Greatest diameter=5 cm’ as the TNM staging classification standard of primary low-grade glioma, Daniels et al (35) considered that a tumor size of ≥6 cm was a be risk factor for OS time of low-grade glioma. Therefore, after comprehensive consideration, a tumor size of ≥5 cm was selected to be a prognostic parameter in the our present study. Secondly, according to the Human Protein Atlas (HPA) Database (http://www.proteinatlas.org), as IHC showed Ki-67 staining in nucleus, nucleoli and nuclear (https://www.proteinatlas.org/ENSG00000148773-MKI67/cell), and p53 staining in nucleoplasm (https://www.proteinatlas.org/ENSG00000141510-TP53/cell). Distribution of Ki-67 and p53 proteins was all located in the cell nucleus (36). Specimens with positive immunostaining of Ki-67 and p53 in glioma were quantified by image analysis. The ratio of immunostained surface was determined as previously described (26–28). Vascular endothelial cells, lymphocytic cells and necrotic areas were excluded from counts. From the five high magnification views of 100 cells, the percentage of positive cells was calculated and used in the following scoring method: 0–10%, 0 points; 11–24%, 1 points; 25–49%, 2 points; 50–74%, 3 points; ≥75%, 4 points. Thus, Ki-67<10%, low expression; Ki-67≥10%, high expression; p53<10%, low expression; and p53≥10%, high expression were used.
Additionally, there are 6 studies regarding survival analysis concerning MAGE-D4 in different tumor types (11–15,22), including non-small cell lung cancer, hepatocellular carcinoma, oral squamous cell carcinoma, breast cancer, squamous cell carcinoma of the esophagus and colorectal cancer. Of the 6 studies, 5 identified that MAGE-D4 was a major, independent prognostic factor for OS time (11–15) however, Ito et al (22) identified no significant difference between high and low MAGE-D4 expression for post-operative survival in non-small cell lung cancer. In the present study, there was no significantdifference between high and low MAGE-D4 expressionin high-grade gliomas (WHO, III–IV) for OS and RFS (P>0.05) time, possibly due to the relatively small patient number in the study; thus further studies should use an increased number of patients. To the best of our knowledge, the function of MAGE-D4 for survival analysis in patients with glioma has not previously been reported. The present study was the first to indicate MAGE-D4 as a major independent prognostic factor for OS and RFS time in glioma.
There were several limitations to the present study. Firstly, it is known that there is a significant association between the localization of protein and the biological functions of the cells (7). It has been indicated via immunofluorescence microscopy that MAGE-D4 is concentrated in the central-spindle and the mid-body regions from the telophase to the post-mitotic phase in lung cancer cells, which demonstrates that MAGE-D4 may serve a role in a particular cell cycle pattern and in cell division (22). Another study indicated that MAGE-D4 affected adhesion by causing changes in breast cancer cell morphology (11). In light of this other research, the present study is limited by its lack of functional analysis associated with the localization of the MAGE-D4 protein. Further study should focus on elucidating the cause of the morphological changes associated with glioma cells underlying the biological activities of MAGE-D4, including the mechanism underlying the action.
Secondly, in the present study, the focus was on understanding the differences in MAGE-D4 expression in glioma and normal brain tissue, including the influence of MAGE-D4 on the prognosis of patients with glioma. In order to identify the biological function of MAGE-D4, future in vitro and in vivo experiments should be designed using MAGE-D4-positive and -negative glioma cells. Finally, considering that the present study was conducted in a single institute, the number of patients included in the study should be increased and the classification of these patients should be balanced. In addition, it is equally important to detect the serum immunoreactivity of patients with glioma. The ultimate purpose of the experimental line of questioning is building up a foundation for immunotherapy based on MAGE-D4.
In conclusion, the present study demonstrated that MAGE-D4 protein was expressed at high levels in glioma, and its expression was significantly associated with the age group, WHO classification of glioma severity and p53 expression. Patients with high levels of MAGE-D4 expression generally had a poorer prognosis compared with those with a low level of expression. The multivariable analysis indicated that the high expression of MAGE-D4protein was an independent prognostic factor. These results indicated that MAGE-D4 may be a promising biomarker for glioma and an important factor for determining the prognosis of patients with new or recurring glioma.
Acknowledgements
The present study was supported by grants from the Guangxi Health and Family Planning Commission Research Award (no. Z2016037), the Guangxi Traditional Chinese Medicine Research Project (no. GZZC16-53) and the Youth Science Foundation of Guangxi Medical University (no. GXMUYSF201306).
Glossary
Abbreviations
- MAGE
melanoma-associated antigen
- IHC
immunohistochemistry
References
- 1.Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71:1437–1444. doi: 10.1001/jamaneurol.2014.1701. [DOI] [PubMed] [Google Scholar]
- 3.Lee KH, Ahn EJ, Oh SJ, Kim O, Joo YE, Bae JA, Yoon S, Ryu HH, Jung S, Kim KK, et al. KITENIN promotes glioma invasiveness and progression, associated with the induction of EMT and stemness markers. Oncotarget. 2015;6:3240–3253. doi: 10.18632/oncotarget.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diksin M, Smith SJ, Rahman R. The molecular and phenotypic basis of the glioma invasive perivascular niche. Int J Mol Sci. 2017;18:pii: E2342. doi: 10.3390/ijms18112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo L, Sang M, Liu Q, Fan X, Zhang X, Shan B. The expression and clinical significance of melanoma-associated antigen-A1, -A3 and -A11 in glioma. Oncol Lett. 2013;6:55–62. doi: 10.3892/ol.2013.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 7.Lee AK, Potts PR. A comprehensive guide to the MAGE family of ubiquitin ligases. J Mol Biol. 2017;429:1114–1142. doi: 10.1016/j.jmb.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 9.Krämer BF, Schoor O, Krüger T, Reichle C, Müller M, Weinschenk T, Hennenlotter J, Stenzl A, Rammensee HG, Stevanovic S. MAGED4-expression in renal cell carcinoma and identification of an HLA-A'25-restricted MHC class I ligand from solid tumor tissue. Cancer Biol Ther. 2005;4:943–948. doi: 10.4161/cbt.4.9.1907. [DOI] [PubMed] [Google Scholar]
- 10.Ma QY, Pang LW, Chen ZM, Zhu YJ, Chen G, Chen J. The significance of MAGED4 expression in non-small cell lung cancer as analyzed by real-time fluorescence quantitative PCR. Oncol Lett. 2012;4:733–738. doi: 10.3892/ol.2012.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germano S, Kennedy S, Rani S, Gleeson G, Clynes M, Doolan P, McDonnell S, Hughes L, Crown J, O'Driscoll L. MAGE-D4B is a novel marker of poor prognosis and potential therapeutic target involved in breast cancer tumorigenesis. Int J Cancer. 2012;130:1991–2002. doi: 10.1002/ijc.26200. [DOI] [PubMed] [Google Scholar]
- 12.Chong CE, Lim KP, Gan CP, Marsh CA, Zain RB, Abraham MT, Prime SS, Teo SH, Silvio Gutkind J, Patel V, Cheong SC. Over-expression of MAGED4B increases cell migration and growth in oral squamous cell carcinoma and is associated with poor disease outcome. Cancer Lett. 2012;321:18–26. doi: 10.1016/j.canlet.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 13.Takami H, Kanda M, Oya H, Hibino S, Sugimoto H, Suenaga M, Yamada S, Nishikawa Y, Asai M, Fujii T, et al. Evaluation of MAGE-D4 expression in hepatocellular carcinoma in Japanese patients. J Surg Oncol. 2013;108:557–562. doi: 10.1002/jso.23440. [DOI] [PubMed] [Google Scholar]
- 14.Oya H, Kanda M, Takami H, Hibino S, Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, et al. Overexpression of melanoma-associated antigen D4 is an independent prognostic factor in squamous cell carcinoma of the esophagus. Dis Esophagus. 2015;28:188–195. doi: 10.1111/dote.12156. [DOI] [PubMed] [Google Scholar]
- 15.Zhang QM, He SJ, Shen N, Luo B, Fan R, Fu J, Luo GR, Zhou SF, Xiao SW, Xie XX. Overexpression of MAGE-D4 in colorectal cancer is a potentially prognostic biomarker and immunotherapy target. Int J Clin Exp Pathol. 2014;7:3918–3927. [PMC free article] [PubMed] [Google Scholar]
- 16.He SJ, Gu YY, Yu L, Luo B, Fan R, Lin WZ, Lan XW, Lin YD, Zhang QM, Xiao SW, Xie XX. High expression and frequently humoral immune response of melanoma-associated antigen D4 in glioma. Int J Clin Exp Pathol. 2014;7:2350–2360. [PMC free article] [PubMed] [Google Scholar]
- 17.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang QM, Shen N, Xie S, Bi SQ, Luo B, Lin YD, Fu J, Zhou SF, Luo GR, Xie XX, Xiao SW. MAGED4 expression in glioma and upregulation in glioma cell lines with 5-aza-2′-deoxycytidine treatment. Asian Pac J Cancer Prev. 2014;15:3495–3501. doi: 10.7314/APJCP.2014.15.8.3495. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki M, Nakahira K, Kawano Y, Katakura H, Yoshimine T, Shimizu K, Kim SU, Ikenaka K. MAGE-E1, a new member of the melanoma-associated antigen gene family and its expression in human glioma. Cancer Res. 2001;61:4809–4814. [PubMed] [Google Scholar]
- 20.Parney IF, Hao C, Petruk KC. Glioma immunology and immunotherapy. Neurosurgery. 2000;46:778–792. doi: 10.1227/00006123-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 22.Ito S, Kawano Y, Katakura H, Takenaka K, Adachi M, Sasaki M, Shimizu K, Ikenaka K, Wada H, Tanaka F. Expression of MAGE-D4, a novel MAGE family antigen, is correlated with tumor-cell proliferation of non-small cell lung cancer. Lung Cancer. 2006;51:79–88. doi: 10.1016/j.lungcan.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Thaker PH, Deavers M, Celestino J, Thornton A, Fletcher MS, Landen CN, Kinch MS, Kiener PA, Sood AK. EphA2 expression is associated with aggressive features in ovarian carcinoma. Clin Cancer Res. 2004;10:5145–5150. doi: 10.1158/1078-0432.CCR-03-0589. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han ZG, Jiang WG, Hiscox S, Hallett MB, Isoai A, Mansel RE. Inhibition of motility and invasion of human lung cancer cells by invasion inhibiting factor 2. Surg Oncol. 1996;5:77–84. doi: 10.1016/S0960-7404(96)80004-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhao W, Zhou Y, Xu H, Cheng Y, Kong B. Snail family proteins in cervical squamous carcinoma: Expression and significance. Clin Invest Med. 2013;36:E223–E233. doi: 10.25011/cim.v36i4.19956. [DOI] [PubMed] [Google Scholar]
- 27.Song X, Zhang W, Zhang Y, Zhang H, Fu Z, Ye J, Liu L, Song X, Wu Y. Expression of semaphorin 3A and neuropilin 1 with clinicopathological features and survival in human tongue cancer. Med Oral Patol Oral Cir Bucal. 2012;17:e962–e968. doi: 10.4317/medoral.18168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu LK, Jiang XY, Zhou XX, Wang DM, Song XL, Jiang HB. Upregulation of vimentin and aberrant expression of E-cadherin/beta-catenin complex in oral squamous cell carcinomas: Correlation with the clinicopathological features and patient outcome. Mod Pathol. 2010;23:213–224. doi: 10.1038/modpathol.2009.160. [DOI] [PubMed] [Google Scholar]
- 29.Szopa W, Burley TA, Kramer-Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: Current status and future perspectives. Biomed Res Int. 2017;2017:8013575. doi: 10.1155/2017/8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheong SC, Chandramouli GV, Saleh A, Zain RB, Lau SH, Sivakumaren S, Pathmanathan R, Prime SS, Teo SH, Patel V, Gutkind JS. Gene expression in human oral squamous cell carcinoma is influenced by risk factor exposure. Oral Oncol. 2009;45:712–719. doi: 10.1016/j.oraloncology.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Tsai JR, Chong IW, Chen YH, Yang MJ, Sheu CC, Chang HC, Hwang JJ, Hung JY, Lin SR. Differential expression profile of MAGE family in non-small-cell lung cancer. Lung Cancer. 2007;56:185–192. doi: 10.1016/j.lungcan.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, McArdle SE. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010;10:2. [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.M200008-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Karim AB, Maat B, Hatlevoll R, Menten J, Rutten EH, Thomas DG, Mascarenhas F, Horiot JC, Parvinen LM, van Reijn M, et al. A randomized trial on dose-response in radiation therapy of low-grade cerebral glioma: European Organization for Research and Treatment of Cancer (EORTC) study 22844. Int J Radiat Oncol Biol Phys. 1996;36:549–556. doi: 10.1016/S0360-3016(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 35.Daniels TB, Brown PD, Felten SJ, Wu W, Buckner JC, Arusell RM, Curran WJ, Abrams RA, Schiff D, Shaw EG. Validation of EORTC prognostic factors for adults with low-grade glioma: A report using intergroup 86-72-51. Int J Radiat Oncol Biol Phys. 2011;81:218–224. doi: 10.1016/j.ijrobp.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouvier-Labit C, Chinot O, Ochi C, Gambarelli D, Dufour H, Figarella-Branger D. Prognostic significance of Ki67, p53 and epidermal growth factor receptor immunostaining in human glioblastomas. Neuropathol Appl Neurobiol. 1998;24:381–388. doi: 10.1046/j.1365-2990.1998.00137.x. [DOI] [PubMed] [Google Scholar]