Abstract
Cerebroma, a nervous system tumor located in the cranial cavity, seriously affects health and may even become life-threatening. The aim of the study was to investigate and discuss the expression levels of cluster of differentiation 44 (CD44) and Golgi protein 73 (GP73) in different types of cerebroma tissues and to study the correlations of CD44 and GP73. Reverse transcription-polymerase chain reaction (RT-PCR) was used to detect the mRNA expression levels of CD44 and GP73 in four kinds of cerebroma. The immunohistochemical streptavidin-biotin complex (SABC) method was applied to measure the expression levels of CD44 and GP73 in different types of cerebroma. Subsequently, immunofluorescence and western blot analysis were performed to detect the expression levels of CD44 and GP73 in four kinds of cerebroma tissues. Statistical Product and Service Solutions (SPSS) 17.0 software was used to analyze the differences in CD44 and GP73 expression levels of the four kinds of cerebroma tissues and normal brain tissues. Transcription of CD44 and GP73 mRNAs was detected in the four kinds of cerebroma tissues, and CD44 and GP73 proteins were expressed. The immunohistochemical results revealed that the expression levels of CD44 and GP73 in the four kinds of cerebroma were significantly increased compared with those in normal tissues. The immunofluorescence results indicated that both CD44 and GP73 were expressed in four kinds of cerebroma, and the expression of CD44 was higher than that of GP73. The results of analysis of variance showed that the differences in CD44 and GP73 expression levels in the four kinds of cerebroma tissues and normal brain tissues were statistically significant (P<0.01). The results show that the expression levels of CD44 and GP73 are obviously upregulated in four kinds of cerebroma tissues, suggesting that CD44 and GP73 have great value for cerebroma research and can provide a new direction for clinical study as well as the diagnosis and treatment of cerebroma.
Keywords: cerebroma, CD44, GP73, correlation research
Introduction
Cerebroma, when it appears as a nervous system tumor located in the cranial cavity, can have harmful effects (1). Cerebroma seriously affects health and may even become life-threatening (2). The onset of cerebroma is a complex process caused by multiple factors, including virus infections and radiation. At present the cause of the disease and its molecular mechanism has not been completely understood (3).
Cluster of differentiation 44 (CD44) is the forty-fourth cluster of differentiation molecule, which is widely distributed in T cells especially memory T cells (4). Previous studies showed that CD44 molecules are associated with tumor invasion and metastasis. The abnormal expression of CD44 may cause and accelerate the occurrence and development of tumors (5). Golgi protein (GP73) is a transmembrane protein located in Golgi. In a previous study, it was demonstrated that GP73 can be used as a serum marker of liver cancer, which plays an important role in HCC (6). As such, GP73 may be involved in other types of cancer.
In the present study, immunofluorescence and immunohistochemical methods were utilized to detect the expression intensities of CD44 and GP73 in cerebroma tissues. Reverse transcription-polymerase chain reaction (RT-PCR) and western blot analysis were performed to measure the expression levels of CD44 and GP73 mRNAs as well as proteins to understand the differences in the expression levels of CD44 and GP73 in varying tissues, discuss the changes of CD44 and GP73 expression levels in cerebroma tissues and analyze their correlation, thus providing new insights and directions for the genetic diagnosis and treatment of cerebroma.
Materials and methods
Collection of cerebroma tissues
The specimens were tissues resected during surgical procedures at Dezhou People's Hospital, fixed in 10% formaldehyde and then embedded in paraffin. Sixteen specimens of paraffin blocks were collected from patients with cerebroma, which was resected via surgery and confirmed by postoperative histopathological examination. Among the patients, 8 were men and 8 were women, aged 30–60 years. In addition, 4 specimens of normal brain tissues, provided by the Department of Neurosurgery of Dezhou People's Hospital (Shandong, China) were taken as the control group.
Main reagents
Reagents used in the study included: Bicinchoninic acid (BCA) Protein Assay kit (Beyotime, Shanghai, China); TRIzol® Total RNA extraction kit (Tiangen Biotech Co., Ltd., Beijing, China); RT-PCR reverse transcription kit (Tiangen Biotech Co., Ltd.); and the immunohistochemical staining kit was purchased from Zhongshan Goldenbridge Biotechnology Co., Ltd. (Beijing, China). 4,6-Diamino-2-phenyl indole (DAPI), anti-β-actin (β-actin) were used with the monoclonal antibodies for CD44 and GP73 and immunofluorescence was used for the secondary antibody (Cell Signaling Technology, Boston, MA, USA). For RT-PCR detection, the tissues in each group were added into the TRIzol reagent and ground to homogenate. The specimens were left to stand at room temperature for 5 min until they were completely lysed. This was followed by centrifugation at 12,000 × g for 5 min at 4°C, and the supernatant was extracted carefully. Chloroform was added to the supernatant and mixed thoroughly, after which the mixture was left to stand at room temperature for 5 min. This was followed by centrifugation at 12,000 × g for 15 min at 4°C, and the supernatant was extracted carefully. Next, isopropyl alcohol of the same volume was added and left to stand at room temperature for 10 min. The mixture was then centrifuged at 12,000 × g at 4°C for 10 min, and the precipitate was reserved. Then, 75% ethanol was added and mixed together, which was used to wash the RNA precipitate. Finally, RNase-free water was added to completely dissolve the precipitate. The value of optical density (OD) 260/280 and the concentration of RNA were measured. Amplification was performed step by step according to the instructions. The primer sequence template shown in Table I was used, and RT-PCR analysis was conducted to determine the reaction products.
Table I.
RT-PCR primer sequences for CD44 and GP73 mRNAs.
| Gene | Primer sequences |
|---|---|
| CD44 | U: CAGACCTGCCCAATGCCTTTGATGGACC |
| D: TCCACCTTCTTGACTCCCATGTGAGT | |
| GP73 | U: CGGGATCCATGATGGGCTTGGGAAACGGGCGTC |
| D: CGGAATTCTCAGAGTGTATGATTCCGCTTTTCAC | |
| β-actin | U: GAGCCGGGAAATCGTGCGT |
| D: GGAAGGAAGGCTGGAAGATG |
U, upstream; D, downstream.
Western blot analysis
An appropriate amount of the four kinds of cerebroma tissues to be tested was taken and washed with ice-cold normal saline. According to the instructions in the total protein extraction kit, lysis buffer for immunoprecipitation (IP) [containing phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors] was added and then placed on the ice to adequately grind the tissues. Next, the tissue homogenate was centrifuged at 12,000 × g for 10 min at 4°C, and the supernatant was taken to extract new supernatant by centrifugation at 12,000 × g at 4°C for another 20 min. After protein quantification was implemented to the supernatant according to the instructions in the protein kit, protein samples were loaded with an equal amount of total protein. The supernatant was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at a constant voltage of 220 V. Electrophoresis was not stopped until bromophenol blue reached the bottom of the gel. The gel was sliced in accordance with the molecular weight of the target protein and then transferred to a polyvinylidene fluoride (PVDF) membrane. Subsequently, it was placed in 5% skim milk and blocked at room temperature for 3 h on a shaking table, prior to being incubated overnight in mouse anti-human CD44, GP73 and β-actin primary monoclonal antibodies (dilution, 1:1,000; cat. nos. 5640, 97537 and 3700) at 4°C. The next day, after the membrane was thoroughly washed in Tween/Tris-buffered saline (TTBS) (3 times for 10 min), the horse anti-mouse secondary polyclonal antibody (dilution, 1:2,000; cat. no. 7076) was added and incubated at room temperature for 1 h. Enhanced chemiluminescence (ECL) was added for color development after the specimen was washed in TTBS (3 times for 10 min), and images were captured.
Immunohistochemistry
The section was placed in 0.01 mol/l sodium citrate buffer (pH 6.0) after dewaxing and hydration, and heated for antigen retrieval. The sections were then incubated in 0.3% hydrogen peroxide solution at 37°C for 30 min to inactivate endogenous peroxidase. Blocking buffer was added and blocked at room temperature for 30 min. Blocking buffer was then discarded, and primary antibody solution was added and incubated overnight in a wet box at 4°C. After the section was washed repeatedly, biotinylated secondary antibody working solution and horseradish peroxidase-conjugated streptavidin working solution were added, respectively, and the section was placed in diaminobenzidine (DAB) solution for color development. Finally, the section was mounted in buffered glycerol, observed and photographed under a microscope (Olympus Corporation, Tokyo, Japan).
Immunofluorescence
The paraffin section was dewaxed with xylene, hydrated with graded alcohol for antigen retrieval, and washed in 0.01 M phosphate-buffered saline (PBS) (pH 7.4) 3 times for 5 min. The section was then blocked in a wet box containing 10% bovine serum albumin (BSA) for 30 min at 37°C. The specimen, added with properly diluted fluorescence-labeled antibody (diluted at 1:70), was placed in the wet box and incubated overnight at 4°C. After the specimen was washed in PBS (pH 7.4) 3 times (5 min per time), the fluorescent secondary antibody (diluted at 1:100) was added in the dark and incubated in the wet box at 37°C for another 2 h. After being washed in PBS (pH 7.4) 3 times (5 min per time), the specimen was stained using DAPI in the dark for 15 min. Finally, the section was mounted in buffered glycerol, observed and photographed right under a fluorescence microscope.
Statistical analysis
Statistical analysis of the experimental data were presented as mean ± standard deviation (mean ± SD). SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis of the experimental results. A t-test was applied for mean comparisons between the two groups, and one-way analysis of variance (ANOVA) was used for mean comparisons among multiple groups. P-test was performed for pairwise comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
RT-PCR results
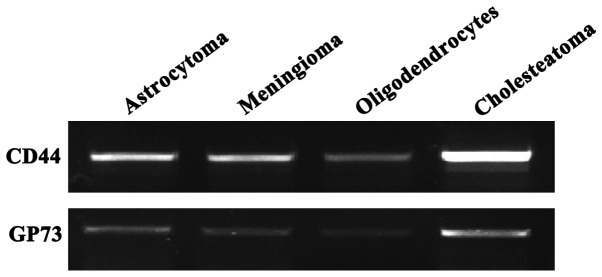
The total RNAs of astrocytoma, meningioma, oligodendrocyte and cholesteatoma were extracted, respectively. After the RT-PCR experiment, objective bands of CD44 and GP73 were detected in the four groups. In particular, the expression levels of CD44 and GP73 in cholesteatoma were very high, indicating that CD44 and GP73 mRNAs were transcribed in the four types of cerebroma tissues (Fig. 1).
Figure 1.
Expression levels of CD44 and GP73 genes in the four kinds of cerebroma.
Expression levels of CD44 and GP73 proteins
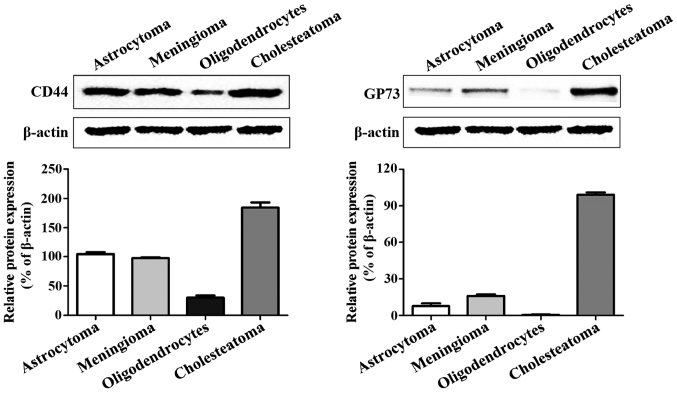
The western blot analysis results revealed that CD44 and GP73 proteins were expressed in four kinds of cerebroma tissues, of which CD44 was highly expressed in all four kinds of cerebroma, while the expression levels of GP73 in oligodendrocytes and cholesteatoma were higher than those in meningioma and astrocytoma (Fig. 2).
Figure 2.
Expression levels of CD44 and GP73 in the four kinds of cerebroma tissues.
Expression levels of CD44 and GP73 detected by streptavidin-biotin complex (SABC)
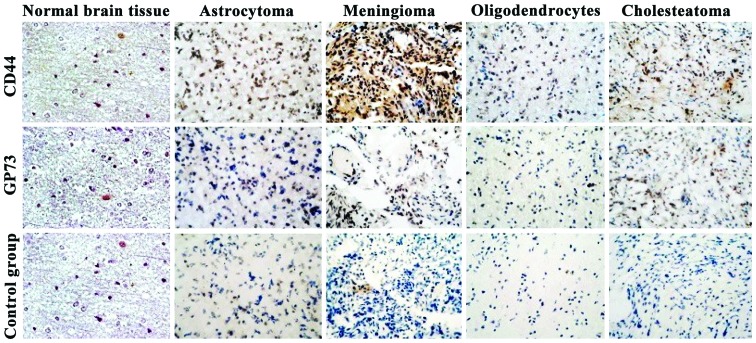
The expression levels of CD44 and GP73 proteins were mainly located in the cytoplasm (Fig. 3). By comparing the expression levels of CD44 and GP73 proteins in normal brain tissues with those in four kinds of cerebroma tissues, the differences were statistically significant (P<0.01) (Table II).
Figure 3.
Expression levels of CD44 and GP73 in tumor tissues and normal brain tissues [streptavidin-biotin complex (SABC); magnification, ×400].
Table II.
Expression levels of CD44 and GP73 in tumor tissues and normal brain tissues.
| Positive expression rate | |||||
|---|---|---|---|---|---|
| Proteins | Normal brain tissues | Astrocytoma | Meningioma | Oligodendrocytes | Cholesteatoma |
| CD44 | 4.90±1.20 | 68.40±4.10a | 62.12±2.13a | 60.12±6.83a | 40.12±2.21a |
| GP73 | 0.02±0.04 | 2.10±0.60a | 10.03±2.68a | 7.44±2.10a | 3.08±0.47a |
P<0.01 vs. normal brain tissues.
Expression levels of CD44 and GP73 in the four kinds of tumor tissues detected by immunofluorescence
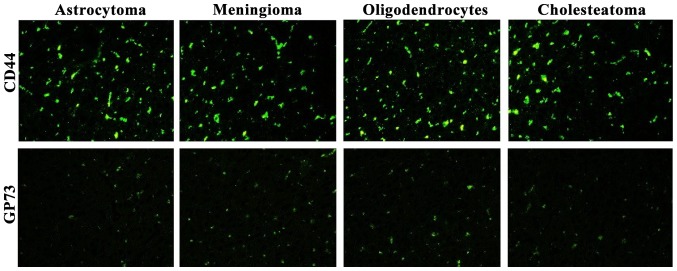
Fluorescence expression levels of CD44 and GP73 were detected in the four kinds of cerebroma tissues. CD44 was highly expressed in the four kinds of cerebroma tissues while the expression levels of GP73 were slightly decreased (Fig. 4).
Figure 4.
Expression levels of CD44 and GP73 in tumor tissues and normal brain tissues (magnification, ×400).
Discussion
Malignant cerebroma is a common tumor with extremely high malignancy in recent years. Characterized by high incidence rate and death rate, cerebroma is a malignant disease threatening people's health (7–8). The majority of patients are in the intermediate and advanced stage when they are diagnosed with cerebroma, which greatly limits the application of radical resection (9–10). Thus, it constitutes a global problem for the treatment of cerebroma. Therefore, it is imperative to identify a new, safe and effective method to succesfully treat cerebroma. Previous studies conducted on hepatocellular carcinoma have aimed to identify genes closely related to hepatocellular carcinoma. Appropriate intervention in the correspondence of these genes may become a novel direction for the treatment of hepatocellular carcinoma (11).
In recent years, with the rapid development of scientific research, an increasing number of genes have been found to be closely correlated with the occurrence of tumors. Previous findings have shown that CD44 plays an important role in tumors. CD44 protein has several different and crucial biological functions in organisms. The protein itself participates in organ development, multiple immune functions and hematopoietic functions, and also has important functions in tumors (12–14). CD44 not only has an important position in regulating growth of cerebroma tissues, but also has a crucial effect on survival, differentiation and migration of tumors, including promotion of adhesion, metastasis and dissemination, of tumor tissues (15–17). Previous findings have shown that GP73 also plays a key role in tumors. GP73 is a kind of transmembrane protein existing in Golgi apparatus, of which the content in different tissues (blood and body fluids) in vivo has an obviously rising trend when malignant cerebroma occurs (18–21). Thus, it can be seen that GP73 is closely related to the occurrence and development of cerebroma. Both CD44 and GP73 play an important role in tumors, and they can be used for the genetic diagnosis and treatment of tumors.
The results of the present study have shown that CD44 and GP73 were highly expressed in cerebroma tissues. RT-PCR and western blot results indicated that CD44 and GP73 were expressed in the four kinds of cerebroma tissues. By comparing the expression levels of CD44 and GP73 proteins in tissues of the cerebroma group with those of the blank control group, the differences were statistically significant (P<0.05). It is believed that this research on the expression levels of CD44 and GP73 proteins can provide a new theoretical basis for exploring the mechanisms of tumor infiltration and metastasis and offer a new direction for the diagnosis and treatment of cerebroma.
Helsinki Declaration of 1975, as revised in 2008. The study design was a multi institutional retrospective review of medical records. Research protocols were assessed and accepted by the institutional research boards of individual institutions.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 2.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Orian-Rousseau V, Ponta H. Perspectives of CD44 targeting therapies. Arch Toxicol. 2015;89:3–14. doi: 10.1007/s00204-014-1424-2. [DOI] [PubMed] [Google Scholar]
- 5.Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachert C, Fimmel C, Linstedt AD. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic. 2007;8:1415–1423. doi: 10.1111/j.1600-0854.2007.00621.x. [DOI] [PubMed] [Google Scholar]
- 7.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladson CL. The extracellular matrix of gliomas: Modulation of cell function. J Neuropathol Exp Neurol. 1999;58:1029–1040. doi: 10.1097/00005072-199910000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Diamandis P, Wildenhain J, Clarke ID, Sacher AG, Graham J, Bellows DS, Ling EK, Ward RJ, Jamieson LG, Tyers M, et al. Chemical genetics reveals a complex functional ground state of neural stem cells. Nat Chem Biol. 2007;3:268–273. doi: 10.1038/nchembio873. [DOI] [PubMed] [Google Scholar]
- 10.Sonoda Y, Ozawa T, Aldape KD, Deen DF, Berger MS, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674–6678. [PubMed] [Google Scholar]
- 11.Lagadec C, Vlashi E, Della Donna L, Meng Y, Dekmezian C, Kim K, Pajonk F. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res. 2010;12:R13. doi: 10.1186/bcr2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marhaba R, Zöller M. CD44 in cancer progression: Adhesion, migration and growth regulation. J Mol Histol. 2004;35:211–231. doi: 10.1023/B:HIJO.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, Prieto-Sánchez RM, Barba I, Martínez-Sáez E, Prudkin L, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Naor D, Sionov RV, Ish-Shalom D. CD44: Structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/S0065-230X(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 16.Iczkowski KA, Bai S, Pantazis CG. Prostate cancer overexpresses CD44 variants 7–9 at the messenger RNA and protein level. Anticancer Res. 2003;23:3129–3140. [PubMed] [Google Scholar]
- 17.Georgolios A, Batistatou A, Charalabopoulos A, Manolopoulos L, Charalabopoulos K. The role of CD44 adhesion molecule in oral cavity cancer. Exp Oncol. 2006;28:94–98. [PubMed] [Google Scholar]
- 18.Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431–1440. doi: 10.1053/jhep.2002.32525. [DOI] [PubMed] [Google Scholar]
- 19.Kladney RD, Tollefson AE, Wold WS, Fimmel CJ. Upregulation of the Golgi protein GP73 by adenovirus infection requires the E1A CtBP interaction domain. Virology. 2002;301:236–246. doi: 10.1006/viro.2002.1523. [DOI] [PubMed] [Google Scholar]
- 20.Hu JS, Wu DW, Liang S, Miao XY. GP73, a resident Golgi glycoprotein, is sensibility and specificity for hepatocellular carcinoma of diagnosis in a hepatitis B-endemic Asian population. Med Oncol. 2010;27:339–345. doi: 10.1007/s12032-009-9215-y. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Yang H, Mao Y, Xu H, Zhang J, Li G, Lu X, Sang X, Zhao H, Zhong S, et al. Increased Golgi protein 73 expression in hepatocellular carcinoma tissue correlates with tumor aggression but not survival. J Gastroenterol Hepatol. 2011;26:1207–1212. doi: 10.1111/j.1440-1746.2011.06733.x. [DOI] [PubMed] [Google Scholar]