Abstract
The aim of the current study was to investigate and discuss the function of T-box 3 (TBX3) gene expression in the pathogenesis of renal carcinoma. The carcinoma, adjacent and normal renal tissues of 210 patients with renal carcinoma who presented to The Central Hospital of Wuhan, Tongji Medical College from March, 2006 to March, 2012 were collected to extract total RNAs. The total RNAs were reverse-transcribed into complementary DNA (cDNA), and quantitative polymerase chain reaction (qPCR) was applied to detect the expression of TBX3 gene in these tissues, followed by its association with the prognosis of renal carcinoma as well as clinical features. A comparison of the renal carcinoma tissues with the adjacent tissues showed that TBX3 gene was obviously highly expressed in renal carcinoma tissues (P<0.05). In addition, compared with normal renal tissues, TBX3 gene was obviously highly expressed in renal carcinoma tissues (P<0.05). There was no significant difference in the expression levels of TBX3 gene in normal renal tissues and adjacent tissues (P=0.15). The expression of TBX3 gene in renal carcinoma tissues was not associated with patient age, sex and tumor size (P>0.05), but it was associated with tumor-node-metastasis (TNM) staging and lymph node metastasis (P<0.05). The Kaplan-Meier survival analysis revealed that the median survival time of patients in the positive TBX3 gene expression group (37.5 months) was shorter than that in the negative TBX3 gene expression group (66 months), and there was a statistical difference (P<0.05). The 3- and 5-year survival rates in the negative TBX3 gene expression group were 74 and 62%, respectively, and the 3- and 5-year survival rates in the positive TBX3 gene expression group were 52 and 32%, respectively, and the differences were significant (P<0.05). The results suggest that TBX3 gene is highly expressed in renal carcinoma tissues, and it is associated with TNM staging, lymph node metastasis and distant metastasis, which may be involved in the occurrence and metastasis of renal carcinoma.
Keywords: TBX3 gene, renal carcinoma, prognosis, qPCR, survival analysis
Introduction
Renal cell carcinoma (renal carcinoma for short) is a kind of tumor with a high malignancy in the urinary system, and it is one of the most common tumors. Also known as renal adenocarcinoma, it is a malignant tumor originated in the tubule epithelium in the renal parenchyma, accounting for 80–90% in renal malignant tumor (1). Its incidence and mortality rate are currently on the increase. Tumor metastasis has been identified in approximately half of the patients when they are examined, and a postoperative relapse rate as high as 90% is evident in more than half of the patients (2). Renal carcinoma exhibits special genetic and biological characteristics, heterogeneity and easy metastasis. Conventional chemoradiotherapy has no ideal effect on renal carcinoma with metastasis. Studies have found that satisfactory therapeutic effects have been obtained by applying molecular immunology, gene therapy and molecular-targeted therapy for tumor vascularization for advanced or metastatic renal carcinoma (3). The emergence of tumor-associated factors has a good application prospect for the diagnosis and treatment of renal carcinoma.
T-box (TBX) genes have the function of regulating transcription factors related to development, and TBX3 gene plays its role mainly by means of its expression product TBX3 protein, which is involved in each process of embryonic development in order to guarantee the normal differentiation and development of tissues and organs in the embryonic period (4). Previous findings showed that the expression of TBX3 mRNA in some tumor tissues is markedly higher than that in adjacent tissues (5). It has been concluded from statistical analyses that several clinical features, including lymph node metastasis and tumor-node-metastasis (TNM) staging, have a significant correlation with the abnormal expression of TBX3 gene (6). Some studies also indicate that TBX3 gene is an oncogene (7), and TBX3 protein is a major product of its function. Furthermore, TBX3 protein is a kind of inhibitory factor associated with transcription, and the normal differentiation and development of organs and tissues are dependent on its expression to some extent (8,9). Therefore, this study was conducted to investigate the correlation of TBX3 gene with the occurrence and metastasis of renal carcinoma by analyzing the expression level of TBX3 gene in renal carcinoma tissues of patients with the disease.
Materials and methods
Research objects and materials
In total, 210 patients with renal carcinoma who were admitted and treated in The Central Hospital of Wuhan, Tongji Medical College (Wuhan, China) from March, 2006 to March, 2012 were selected, and their carcinoma, adjacent and normal renal tissues (renal tissues more than 3 cm away from the tumor) were collected. Of the 210 patients, 86 patients were aged <60 years and 124 were aged ≥60 years, and 140 cases were males and 70 cases were females. Concerning TNM staging, 146 patients were in stage I–II and 64 were in stage III–IV. Regarding pathological typing, there were 165 cases of clear cell carcinoma and 45 cases of non-clear cell carcinoma, of which 95 cases were at low grade, and 115 were at middle and high grade.
Inclusion criteria for the study were: Patients who were histopathologically confirmed as renal carcinoma; patients who did not undergo chemoradiotherapy; and patients who voluntarily accepted this research and signed the informed consent. Exclusion criteria for the study were: Patients complicated with other malignant tumors; patients with congenital malformation or a long history of nephritis; patients with severe hepatic or renal dysfunction and coagulation disorders; and patients unable to cooperate with the research due to various reasons. All the patients received a long-term follow-up of >5 years. The study was approved by the Ethics Committee of The Central Hospital of Wuhan.
Major reagents
The quantitative polymerase chain reaction (qPCR) reagent used was, EvaGreen fluorescence quantitative PCR reagent (Biotium, Inc., Hayward, CA, USA). RNA extraction reagent TRIzol, and reverse transcription kit (Invitrogen, Carlsbad, CA, USA) as well as DNA Marker (100 bp ladder) (Beijing Dingguo Center for Biotechnology Development, Beijing, China) were also used.
Reverse transcription-polymerase chain reaction (RT-PCR)
Premier 6.0 software (Premier Biosoft International, Palo Alto, USA) was used as a reference for the primer sequences of TBX3 (primers were produced by Shanghai Yingjun Biological Technology Co., Ltd., Shanghai, China). The upstream and downstream sequences of TBX3 were: 5′-CCCGAAGAAGACGTAGAAGATGAC-3′, and 5′-CCCGAAGAAGAGGTGGAGGACGAC-3′. β-actin was used as an internal reference primer, and the upstream and downstream sequences were 5′-CCTCCATCGTCCACCGCAAATG-3′ and 5′-TGCTGTCACCTTCACCGTTCCA-3′.
Methods
SYBR-Green I Real-Time PCR kit was utilized to amplify target genes, of which the primers are listed above. The 2−∆∆Cq method was applied to detect the relative expression levels of relevant genes. The RNAs of samples were extracted in accordance with the methods and principles of qPCR. Ultraviolet adsorption was used to measure the concentration and purity of the RNA solution. Complementary DNA (cDNA) was produced according to the conventional methods. The cDNA of renal carcinoma tissue was marked as group A, adjacent tissues as group B, and normal renal tissues as group C.
Statistical analysis
In the present study, Statistical Product and Service Solutions (SPSS) 18.0 software (Chicago, IL, USA) was used for statistical analyses on the data of this research. The χ2 test was used for comparison between groups. Measurement data were presented as mean ± standard deviation. The t-test was used for comparison between groups. Cox regression model was used for multivariate analyses on factors influencing the prognosis of patients with renal carcinoma which were obtained from the univariate analyses. The Kaplan-Meier survival analysis was used to show the median survival time of patients in the two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of TBX3 gene in renal carcinoma tissues, adjacent tissues and normal renal tissues
The comparison of the renal carcinoma tissues with the adjacent tissues showed that TBX3 gene was obviously highly expressed in renal carcinoma tissues (P<0.05). Compared with that in normal renal tissues, TBX3 gene was obviously highly expressed in renal carcinoma tissues (P<0.05). There was no significant difference in the expression levels of TBX3 gene in normal renal tissues and adjacent tissues (P=0.15) (Table I).
Table I.
Expression levels of TBX3 gene in renal carcinoma tissues, adjacent tissues and normal renal tissues.
| Group | Renal carcinoma tissues | Adjacent tissues | Normal renal tissues |
|---|---|---|---|
| Expression level | 1.139±0.453 | 0.495±0.336 | 0.412±0.298 |
| P-value | P<0.05 | P=0.15 | |
The comparison of the renal carcinoma tissues with the adjacent tissues shows that TBX3 gene is obviously highly expressed in renal carcinoma tissues (P<0.05). Compared with that in normal renal tissues, TBX3 gene is obviously highly expressed in renal carcinoma tissues (P<0.05). There is no significant difference in the expression levels of TBX3 gene in normal renal tissues and adjacent tissues (P=0.15).
Relationship between the expression level of TBX3 gene in renal carcinoma tissues and clinical features
According to the median expression level (0.769) of TBX3 gene in renal carcinoma tissues, the patients were divided into the positive TBX3 gene expression group (105 patients in total, of which the TBX3 gene expression level was >0.769) and the negative TBX3 gene expression group (105 patients in total, of which the TBX3 gene expression level was <0.769). The TBX3 gene expression level in the carcinoma tissues with lymph node metastasis was obviously elevated compared with that in carcinoma tissues without lymph node metastasis (P<0.05). The expression of TBX3 gene in renal carcinoma tissues was not related to the patients' age, sex and depth of tumor invasion (P>0.05), but it was associated with TNM staging and lymph node metastasis (P<0.05) (Table II).
Table II.
Relationship between expression level of TBX3 gene in renal carcinoma tissues and clinicopathologic characteristics.
| TBX3 gene expression | |||
|---|---|---|---|
| Clinicopathologic characteristics | Positive (n) | Negative (n) | P-value |
| Age (years old) | 0.198 | ||
| <60 | 41 | 45 | |
| ≥60 | 64 | 60 | |
| Sex | 0.121 | ||
| Male | 69 | 71 | |
| Female | 36 | 34 | |
| TNM staging | 0.008a | ||
| I+II | 56 | 90 | |
| III+IV | 49 | 15 | |
| Lymph node metastasis | 0.012a | ||
| Yes | 74 | 11 | |
| No | 31 | 94 | |
| Pathological grading | 0.097 | ||
| Middle and high | 53 | 57 | |
| Low | 52 | 48 | |
| Tumor size | 0.061 | ||
| <7 | 45 | 66 | |
| ≥7 | 60 | 39 | |
| Pathological typing | 0.135 | ||
| Clear cell carcinoma | 79 | 86 | |
| Non-clear cell carcinoma | 26 | 19 | |
P<0.05.
Univariate analyses on factors influencing prognosis of renal carcinoma
The results showed that TNM staging, lymph node metastasis and 5-year survival rate of TBX3 gene had statistical significance (P<0.05), indicating that the factors affecting the prognosis of renal carcinoma. However, sex, age, degree of differentiation and tumor size had no significant differences on the 5-year survival rate of patients (P>0.05) (Table III).
Table III.
Clinicopathologic data of renal carcinoma and 5-year survival rate.
| Clinicopathologic characteristics | Case | 5-year survival rate (%) | χ2 test | P-value |
|---|---|---|---|---|
| Age (years old) | 3.68 | 0.126 | ||
| <60 | 86 | 50.1 | ||
| ≥60 | 124 | 48.3 | ||
| Sex | 4.14 | 0.121 | ||
| Male | 140 | 49.8 | ||
| Female | 70 | 51.2 | ||
| TNM staging | 12.35 | 0.000a | ||
| I+II | 146 | 80.5 | ||
| III+IV | 64 | 40.7 | ||
| Lymph node metastasis | 10.17 | 0.002a | ||
| Yes | 85 | 41.1 | ||
| No | 125 | 84.2 | ||
| Pathological grading | 7.56 | 0.189 | ||
| Middle and high | 110 | 52.9 | ||
| Low | 100 | 43.7 | ||
| Tumor size | 6.47 | 0.076 | ||
| <7 | 111 | 78.6 | ||
| ≥7 | 99 | 51.4 | ||
| Pathological typing | 7.13 | 0.147 | ||
| Clear cell carcinoma | 165 | 61.2 | ||
| Non-clear cell carcinoma | 45 | 49.8 | ||
| TBX3 gene expression | 10.21 | 0.006a | ||
| Positive | 105 | 62.0 | ||
| Negative | 105 | 32.0 |
P<0.05.
Multivariate analyses on factors influencing the prognosis of renal carcinoma
Cox regression model was used for multivariate analyses on factors influencing the prognosis of patients with renal carcinoma which were obtained from the univariate analyses (P<0.05). It was found that TNM staging, lymph node metastasis and TBX3 gene expression level were the independent risk factors for renal carcinoma (P<0.05) (Table IV).
Table IV.
Multivariate analyses on prognosis of renal carcinoma.
| Variables | Odds ratio (OR) | 95% confidence interval (95% CI) | P-value |
|---|---|---|---|
| TBX3 gene expression | 4.24 | 1.87–5.35 | 0.016a |
| Lymph node metastasis | 3.19 | 2.18–8.56 | 0.023a |
| TNM staging | 3.67 | 2.47–7.62 | 0.004a |
P<0.05.
Relationship between the expression of TBX3 gene in renal carcinoma and prognosis
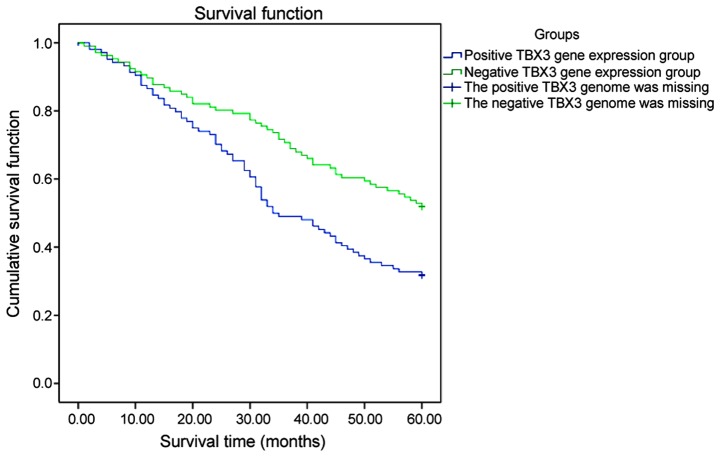
The Kaplan-Meier survival analysis revealed that the median survival time of patients in the positive TBX3 gene expression group (37.5 months) was lower than that in the negative TBX3 gene expression group (66 months), and there was a statistical difference (P<0.05). The 3- and 5-year survival rates in the negative TBX3 gene expression group were 74 and 62%, respectively. The 3- and 5-year survival rates in the positive TBX3 gene expression group were 52 and 32%, respectively, and the differences were significant (P<0.05) (Fig. 1).
Figure 1.
Survival curves of TBX3 gene. The 3- and 5-year survival rates in the negative TBX3 gene expression group are 74 and 62%, respectively. The 3- and 5-year survival rates in the positive TBX3 gene expression group are 52 and 32%, respectively.
Discussion
Studies in recent years have indicated that TBX3 gene is highly expressed in multiple tumors, and studies have been conducted with regard to breast cancer, pancreatic cancer, colorectal cancer, lung cancer and melanoma (10–14). However, to the best of our knowledge, there is no detailed research on TBX3 gene in renal carcinoma. Currently, the cause of renal carcinoma has yet to be elucidated, and most of the clinical manifestations of early renal carcinoma are not obvious. Most renal carcinomas are in advanced stage once they are identified, and the staging of the disease is closely associated with the 5-year survival rate (15,16). This experiment provided an important reference for early diagnosis, metastasis and other aspects of renal carcinoma by studying the expression level of TBX3 gene in renal carcinoma tissues. It also offered a better basis for assisting clinicians to treat the disease. In the age of precision medicine (17), TBX3 gene is to become a potentially effective therapeutic target and prognostic indicator in patients with renal carcinoma.
Current studies have revealed that TBX3 gene is closely associated with the occurrence and development of many kinds of malignant tumors. TBX3 gene has multiple oncogenic mechanisms, and studies have found that this gene can inhibit the expression of cancer suppressive factor p19ARF (human p19ARF) of inhibitor of CDK4/alternative reading frame (INK4α/ARF), in order to suppress cell growth (18). As a result, there is a small amount of TBX3 gene expression in normal cells, which is consistent with Smith et al (18). Some scholars have found that TBX3 gene can directly inhibit the cells by suppressing the promoters of cyclin p21 (Cip1/WAF1) (19). In addition, TBX3 gene can act on the transcriptional activation of p21 by attenuating p53 (20). In breast cancer, for example, it can also be observed that TBX3 gene is upregulated. Previous findings have revealed that TBX3 gene is associated with the Wnt/β-catenin signaling pathway for tumor formation, and the overexpression of TBX3 gene can inhibit the expression levels of suppressor genes in normal cells, thus accelerating the formation and development of malignant tumors (19,20). Other studies have found that TBX3 gene can strengthen the invasiveness of tumors by suppressing E-cadherin on the surface of cell membrane and upregulating β-catenin (21,22), thus leading to the invasion and metastasis of malignant tumors. In the present study, the TBX3 gene expression level in tumor tissues with lymph node metastasis of patients with renal carcinoma was obviously elevated, which may be associated with the fact that TBX3 gene can trigger tumor metastasis. Scholars have found that TBX3 gene can exert its carcinogenic effect by virtue of transforming growth factor-β1 (TGF-β1) (23–25). Therefore, TBX3 gene can promote migration and invasion, playing a vital role in the migration of malignant tumors (26).
In this study, the expression levels of TBX3 gene in renal carcinoma, adjacent and normal tissues were investigated using a large sample size. The specimens were collected in our hospital and the samples were strictly controlled in accordance with the inclusion and exclusion methods, to guarantee the reliability of the samples. The limitation of this study was that it was a single-center study, which had a certain sampling bias in terms of population and region. qPCR was applied in this experiment to reverse transcribe cDNA, in order to reversely infer the expression level of TBX3 gene. As a result, it had decided that this experiment has shortcomings such as money and time consumption. However, it was fully demonstrated in this research that compared with that in the adjacent tissues, TBX3 gene was obviously highly expressed in renal carcinoma tissues (P<0.05). There was no significant difference in the expression levels of TBX3 gene in normal renal and adjacent tissues (P=0.15). Findings of the present study were consistent with results of the studies conducted by Wang et al (27) and Shan et al (28), which revealed that TBX3 gene was highly expressed in tumors. It was shown in the present study that the expression of TBX3 gene in renal carcinoma tissues was not related to the patients' age, sex and tumor size (P>0.05), but it was associated with TNM staging and lymph node metastasis (P<0.05). The high expression of TBX3 gene may be associated with tumor metastasis, results obtained by Li and Varelas (20,21). The results of the presents study showed that the median survival time of patients in the positive TBX3 gene expression group (37.5 months) was lower than that in the negative TBX3 gene expression group (66 months), and there was a statistical difference (P<0.05). The 3- and 5-year survival rates in the negative TBX3 gene expression group were 74 and 62%, respectively. The 3- and 5-year survival rates in the positive TBX3 gene expression group were 52 and 32%, respectively, and the differences were significant (P<0.05).
In conclusion, TBX3 gene is highly expressed in renal carcinoma tissues, and is associated with TNM staging, lymph node metastasis and distant metastasis, and may be involved in the occurrence and metastasis of renal carcinoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors’ contributions
YW researched the literature, designed the study, analyzed and interpreted the patient data.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The Central Hospital of Wuhan. Patients voluntarily accepted this research and signed the informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fang Y, Bao W, Rao Q, Wang X, Xia Q, Shen Q, Zhou X, Yao B. TFE3 regulates renal adenocarcinoma cell proliferation via activation of the mTOR pathway. Mol Med Rep. 2017;16:2721–2725. doi: 10.3892/mmr.2017.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM, Li Y, Nelson RA, Mu B, Onami SH, et al. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PLoS One. 2012;7:e35661. doi: 10.1371/journal.pone.0035661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo P. Renal cell carcinoma: Presentation, staging, and surgical treatment. Semin Oncol. 2000;27:160–176. [PubMed] [Google Scholar]
- 4.Papaioannou VE. The T-box gene family: Emerging roles in development, stem cells and cancer. Development. 2014;141:3819–3833. doi: 10.1242/dev.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219:105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 6.Ansari D, Rosendahl A, Elebro J, Andersson R. Systematic review of immunohistochemical biomarkers to identify prognostic subgroups of patients with pancreatic cancer. Br J Surg. 2011;98:1041–1055. doi: 10.1002/bjs.7574. [DOI] [PubMed] [Google Scholar]
- 7.Boyd SC, Mijatov B, Pupo GM, Tran SL, Gowrishankar K, Shaw HM, Goding CR, Scolyer RA, Mann GJ, Kefford RF, et al. Oncogenic B-RAF(V600E) signaling induces the T-Box3 transcriptional repressor to repress E-cadherin and enhance melanoma cell invasion. J Invest Dermatol. 2013;133:1269–1277. doi: 10.1038/jid.2012.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Esmailpour T, Shang X, Gulsen G, Liu A, Huang T. TBX3 over-expression causes mammary gland hyperplasia and increases mammary stem-like cells in an inducible transgenic mouse model. BMC Dev Biol. 2011;11:65. doi: 10.1186/1471-213X-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R, Yang A, Jin Y. Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extra embryonic endoderm differentiation in mouse embryonic stem cells. J Biol Chem. 2011;286:8425–8436. doi: 10.1074/jbc.M110.202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhawan P, Singh AB, Ellis DL, Richmond A. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 2002;62:7335–7342. [PubMed] [Google Scholar]
- 11.Renard CA, Labalette C, Armengol C, Cougot D, Wei Y, Cairo S, Pineau P, Neuveut C, de Reynies A, Dejean A, et al. Tbx3 is a downstream target of the Wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer. Cancer Res. 2007;67:901–910. doi: 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- 12.Yarosh W, Barrientos T, Esmailpour T, Lin L, Carpenter PM, Osann K, Anton-Culver H, Huang T. TBX3 is overexpressed in breast cancer and represses p14 ARF by interacting with histone deacetylases. Cancer Res. 2008;68:693–699. doi: 10.1158/0008-5472.CAN-07-5012. [DOI] [PubMed] [Google Scholar]
- 13.Wang HC, Meng QC, Shan ZZ, Yuan Z, Huang XY. Overexpression of Tbx3 predicts poor prognosis of patients with resectable pancreatic carcinoma. Asian Pac J Cancer Prev. 2015;16:1397–1401. doi: 10.7314/APJCP.2015.16.4.1397. [DOI] [PubMed] [Google Scholar]
- 14.Shan ZZ, Yan XB, Yan LL, Tian Y, Meng QC, Qiu WW, Zhang Z, Jin ZM. Overexpression of Tbx3 is correlated with epithelial-mesenchymal transition phenotype and predicts poor prognosis of colorectal cancer. Am J Cancer Res. 2015;5:344–353. [PMC free article] [PubMed] [Google Scholar]
- 15.Redmond KL, Crawford NT, Farmer H, D'Costa ZC, O'Brien GJ, Buckley NE, Kennedy RD, Johnston PG, Harkin DP, Mullan PB. T-box 2 represses NDRG1 through an EGR1-dependent mechanism to drive the proliferation of breast cancer cells. Oncogene. 2010;29:3252–3262. doi: 10.1038/onc.2010.84. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Lu J, Sui L, Wang D, Yin M, Hoffmann I, Legler A, Pflugfelder GO. The orthologous Tbx transcription factors Omb and TBX2 induce epithelial cell migration and extrusion in vivo without involvement of matrix metalloproteinases. Oncotarget. 2014;5:11998–12015. doi: 10.18632/oncotarget.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer K, Pflugfelder GO. Putative breast cancer driver mutations in TBX3 cause impaired transcriptional repression. Front Oncol. 2015;5:244. doi: 10.3389/fonc.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, Mowla S, Prince S. Basal transcription of the human TBX3 gene, a key developmental regulator which is overexpressed in several cancers, requires functional NF-Y and Sp1 sites. Gene. 2011;486:41–46. doi: 10.1016/j.gene.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Capparelli C, Chiavarina B, Whitaker-Menezes D, Pestell TG, Pestell RG, Hulit J, Andò S, Howell A, Martinez-Outschoorn UE, Sotgia F, et al. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, ‘fueling’ tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle. 2012;11:3599–3610. doi: 10.4161/cc.21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Li B, Xu B, Han B, Xia H, Chen QM, Li LJ. Expression of p53, p21(CIP1/WAF1) and eIF4E in the adjacent tissues of oral squamous cell carcinoma: Establishing the molecular boundary and a cancer progression model. Int J Oral Sci. 2015;7:161–168. doi: 10.1038/ijos.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varelas X, Bouchie MP, Kukuruzinska MA. Protein N-glycosylation in oral cancer: Dysregulated cellular networks among DPAGT1, E-cadherin adhesion and canonical Wnt signaling. Glycobiology. 2014;24:579–591. doi: 10.1093/glycob/cwu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Weinberg MS, Zerbini L, Prince S. The oncogenic TBX3 is a downstream target and mediator of the TGF-β1 signaling pathway. Mol Biol Cell. 2013;24:3569–3576. doi: 10.1091/mbc.E13-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, Berndt SI, Bézieau S, Brenner H, Butterbach K, et al. Colon Cancer Family Registry and the Genetics and Epidemiology of Colorectal Cancer Consortium: Identification of genetic susceptibility loci for colorectal tumors in a Genome-Wide meta-analysis. Gastroenterology. 2013;144:799–807.e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S. The highly homologous T-Box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer. 2010;1:272–282. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, Prince S. The anti-proliferative function of the TGF-β1 signaling pathway involves the repression of the oncogenic TBX2 by its homologue TBX3. J Biol Chem. 2014;289:35633–35643. doi: 10.1074/jbc.M114.596411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker AS, Johnson EK, Maykel JA, Stojadinovic A, Nissan A, Brucher B, Champagne BJ, Steele SR. Future directions for the early detection of colorectal cancer recurrence. J Cancer. 2014;5:272–280. doi: 10.7150/jca.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang HC, Meng QC, Shan ZZ, Yuan Z, Huang XY. Overexpression of Tbx3 predicts poor prognosis of patients with resectable pancreatic carcinoma. Asian Pac J Cancer Prev. 2015;16:1397–1401. doi: 10.7314/APJCP.2015.16.4.1397. [DOI] [PubMed] [Google Scholar]
- 28.Shan ZZ, Yan XB, Yan LL, Tian Y, Meng QC, Qiu WW, Zhang Z, Jin ZM. Overexpression of Tbx3 is correlated with Epithelial-Mesenchymal Transition phenotype and predicts poor prognosis of colorectal cancer. Am J Cancer Res. 2014;5:344–353. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.