Abstract
Hepatocellular carcinoma is the sixth most prevalent malignant tumor and the third most common cause of cancer-associated mortality. Statins have been investigated for carcinoma prevention and treatment. In addition, receptor-interacting protein 140 (RIP140) has been observed to inhibit the Wnt/β-catenin signaling pathway and cell growth. The present study aimed to investigate whether simvastatin (SV) is able to induce SMCC-7721 cell apoptosis through the Wnt/β-catenin signaling pathway. Initially, a cell model of RIP140 overexpression was established, and then cells were treated with SV. The cell growth, viability and apoptosis were measured by cell counting kit-8 and flow cytometry. Furthermore, the expression levels of RIP140, β-catenin, c-myc and cyclin D1 were detected by reverse transcription-quantitative polymerase chain, western blot analysis and immunofluorescence. The results demonstrated that SV significantly increased the expression of RIP140 in SMCC-7721 cells; however, β-catenin, c-myc and cyclin D1 levels were significantly decreased. Furthermore, the immunofluorescence assay of β-catenin confirmed that SV decreased the content of this protein in SMCC-7721 cells. Notably, RIP140 exerted a synergistic effect on the apoptosis rate induced by SV (RIP140 + SV group), while the alteration in RIP140, β-catenin, c-myc and cyclin D1 levels was more evident in the combination group as compared with the RIP140 or SV alone groups. In conclusion, these results suggested that SV is able to induce the apoptosis of SMCC-7721 cells through the Wnt/β-catenin signaling pathway, as well as that RIP140 and SV exert a synergistic effect on the inhibition of cell proliferation and survival.
Keywords: simvastatin, receptor-interacting protein 140, β-catenin, SMCC-7721 cells
Introduction
Statins, which function as 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) inhibitors, are well-recognized for their efficacy in the prevention and treatment of cardiovascular disease (1). However, a growing body of studies and in vitro experiments suggested that statins may have an unprecedented beneficial effect on cancer cell inhibition and thus serve as an efficient treatment of various types of cancer, including hepatocellular carcinoma (HCC), as well as prostate, breast, lung and colorectal carcinomas (2–6). HCC is the sixth most prevalent malignant tumor and the third most common cause of cancer-associated mortality worldwide, with a poor 5-year survival rate (7). However, there are currently no effective chemotherapy treatments available for this tumor (3). Therefore, it is necessary to further investigate the pathogenesis of HCC and identify efficient therapeutic protocols.
Receptor-interacting protein 140 (RIP140), also known as nuclear receptor interacting protein 1, is a coregulator of numerous transcription factors and a signal transduction regulator (8,9). This molecule is mainly found in metabolic tissues, including liver, muscle and adipose tissues. RIP140 is able to negatively regulate the energy homeostasis by affecting the storage of lipids and inhibiting the expression of genes involved in fatty acid oxidation and glucose metabolism (10). However, numerous studies had identified that RIP140 served an important role in the development of cancer through inhibiting the Wnt/β-catenin signaling pathway (11,12). Wnt/β-catenin signaling inactivates glycogen synthase kinase 3β (GSK3β) for the co-receptor Frizzled/low-density lipoprotein receptor-related protein 1 stimulated by Wnt ligands. Inactivation of GSK3β results to inability of β-catenin phosphorylation, which would decrease the ubiquitination and proteolysis of β-catenin. Therefore, β-catenin is accumulated by translocation from the cytoplasm into the nucleus, where it forms a complex with T-cell factor 4 (TCF4), and activates the transcription of the target genes, including c-myc and cyclin D1. Consequently, this results in cell proliferation and cancer development (11,12). Whereas, RIP140 can negative regulate these genes expression by interact with the β-catenin, and inhibit the activity of β-catenin (13).
As statins are able to induce cell apoptosis, RIP140 simultaneously inhibits cell proliferation through the Wnt/β-catenin signaling pathway. However, whether simvastatin (SV) affects the Wnt/β-catenin signaling and RIP140 expression in HCC remains unclear and requires further investigation. Therefore, in the present study, a RIP140 overexpression cell model was established in the HCC SMCC-7721 cell line. These cells were then treated with the SV, and the results revealed that SV was able to inhibit cell proliferation by increasing the expression of RIP140 and inhibiting the Wnt/β-catenin signaling.
Materials and methods
Determination the IC50 of SV concentration to SMCC-7721 cells by cell counting kit-8
SMCC-7721 cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Tianjin Haoyang Biological Products Technology Co., Ltd., Tianjin, China) and penicillin and streptomycin (100 U/ml and 0.1 mg/ml, respectively; P1400, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), and incubated at 37°C in a humidified atmosphere containing 5% CO2. For cell growth and viability assays, SMCC-7721 cells, at the same confluence (30–40%) for every well, were plated onto 96-well flat-bottomed plates (Beaver Nano-Technologies Co., Ltd., Suzhou, China). Next, different concentrations of SV, including 0, 4, 8, 12, 16 and 20 µmol/l, were added into each well and cultured to measure the cell growth and viability. Following incubation for 48 h, 20 µl cell counting kit-8 (CCK-8; Beijing Zoman Biotechnology Co., Ltd., Beijing, China) solution was added into each well and incubated at 37°C in the dark for 2 h. The absorbance of each well was measured using a microplate reader (Multiskan FC; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 450 nm. Furthermore, the half maximal inhibitory concentration (IC50) of SV was calculated. Each assay was repeated at least three times.
Transfection and grouping
In order to construct a RIP140 overexpression cell model, RIP140-overexpressing plasmids [pcDNA3.1(+)], which were constructed by Magus Technology (Shanghai, China), were transfected into SMCC-7721 cells. Transfection was performed according to the manufacturer's protocol. At 12 h before transfection, cells were seeded onto 6-well plates containing antibiotic-free medium and were allowed to reach 60–70% confluence prior to transfection. The RIP140-overexpressing (4 µg) or negative (4 µg; control group) plasmids and 5 µl Entranster™-D-4000 (Engreen Biosystem New Zealand Ltd., Auckland, New Zealand) were diluted with 50 µl DMEM (FBS- and antibiotic-free). After 5 min, the diluted plasmids (4 µg) and Entranster™-D-4000 were mixed together, and were incubated at 37°C for 20 min, prior to being added to the wells. The DMEM medium was replaced after 6 h with DMEM supplemented with 10% FBS, and cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 for 48 h. The Control and RIP140 groups were then further divided into groups treated with or without 8 µmol/l SV, and the following groups were obtained: Control, RIP140, Control + SV (referred to as the SV group) and RIP140 + SV groups. After 48 h of incubation, all cells were harvested and used in subsequent experiments.
CCK-8 and flow cytometry analysis for cell growth and viability
In order to identify whether RIP140 and SV are able to alter the HCC cell proliferation, the growth and viability of SMCC-7721 cells was determined by CCK-8, while apoptosis was examined by flow cytometry (14). For cell growth and viability determination, the CCK-8 assay was conducted as described earlier, with the exception of the groups investigated, which included the Control, RIP140, SV and RIP140 + SV groups, and the SV concentration (8 µmol/l) used. For the determination of cell apoptosis by flow cytometry analysis, cells (1×106/ml) were seeded into 6-well plates and cultured for 12 h, followed by treatment with SV at 8 µmol/l for 48 h. Following trypsin digestion, cells were collected, washed twice by PBS and analyzed using a BD FACSCalibur™ flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's protocol of the FITC Annexin V Apoptosis Detection kit (cat. no. 556547; Promega Corporation, Madison, WI, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the cells using the RNAiso Plus reagent (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. Next, total RNA (2 µg) was reverse transcribed into cDNA in a total reaction volume of 20 µl using a RevertAid First Strand cDNA Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.) following qualification by NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Primers for β-actin, RIP140, cyclin D1 and c-myc were synthesized by Shenggong Biology Engineering Technology Service, Ltd. (Shanghai, China), and their sequences are displayed in Table I (15,16). In order to determine the expression of these genes, SYBR® Premix Ex Taq™ was used as a fluorescent dye (cat. no. RR42LR; Takara Biotechnology Co., Ltd.). qPCR was conducted with an ABI 7500 sequence detection system (Thermo Fisher Scientific, Inc.) as follows: 95°C for 3 min, and 40 cycles of 95°C for 10 sec and 60°C for 1 min. Fluorescent information and melting curves were obtained. All samples were analyzed in triplicate, and the quantification cycle (Cq) value was defined as the number of cycles required for the fluorescent signal to reach the threshold. Using the comparative Cq method, the relative expression levels were calculated using the formula for 2−ΔΔCt (17). Experimental data represent the mean ± standard deviation of three biological replicates. In the current study, the gene expression was normalized to the expression of β-actin.
Table I.
Primers sequences used in polymerase chain reaction.
| Gene | Primer sequence |
|---|---|
| β-actin | 5′-GATCATTGCTCCTCCTGAGC-3′ (forward) |
| 5′-ACTCCTGCTTGCTGATCCAC-3′ (reverse) | |
| RIP140 | 5′-ATAGCCCTCAGTCATGATT-3′ (forward) |
| 5′-CAGCACATGACAACGGTTCA-3′ (reverse) | |
| CyclinD1 | 5′-GGCGGAGGAGAACAAACAGA-3′ (forward) |
| 5′-TGTGAGGCGGTAGTAGGACA-3′ (reverse) | |
| c-myc | 5′-CCCTCCACTCGGAAGGACTA-3′ (forward) |
| 5′-GCTGGTGCATTTTCGGTTGT-3′ (reverse) |
RIP140, receptor-interacting protein 140.
Western blot analysis
Total proteins were extracted from the cells using a radioimmunoprecipitation assay buffer, and the protein concentration was measured using an bicinchoninic acid assay (cat. no. CW0014; Kangwei Century Biotechnology Co., Ltd., Beijing, China). Equal amounts of clear lysates (~50 µg protein) were separated by SDS-PAGE on 10% gel, and then transferred onto polyvinylidene fluoride membranes (PVDF; Immobilon-P; EMD Millipore, Billerica, MA, USA). Equal transfer was validated by staining with Ponceau red staining (cat. no. CW0057S; Kangwei Century Biotechnology Co., Ltd.). The membranes were blocked at room temperature for 1 h with 10% skimmed milk in Tris-buffered saline (TBS), prior to being incubated with primary antibodies in TBS containing 0.05% Tween 20, 2% bovine serum albumin (cat. no. A8010; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) and 0.05% sodium azide overnight at 4°C. The following primary antibodies were used: RIP140 (cat. no. sc-8997; dilution, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); apoptosis-associated proteins B-cell lymphoma 2 (Bcl-2; cat. no. 12789-1-AP; dilution, 1:2,000; ProteinTech Group, Inc., Chicago, IL, USA) and Bcl-2-associated X protein (Bax; cat. no. 50599-2-Ig; dilution, 1:2,000; ProteinTech Group, Inc.); and the Wnt/β-catenin signaling pathway-associated proteins, β-catenin (cat. no. 51067-2-AP; dilution, 1:2,000; ProteinTech Group, Inc.), c-myc (cat. no. 10828-1-AP; dilution, 1:2,000; ProteinTech Group, Inc.) and cyclin D1 (cat. no. 60186-1-Ig; dilution, 1:5,000; ProteinTech Group, Inc.). A β-actin antibody (cat. no. 60008-1-Ig; dilution, 1:10,000; ProteinTech Group, Inc.) served as the internal control. Subsequently, the PVDF membranes were incubated at room temperature for 1 h with secondary horseradish peroxidase-coupled rabbit antibodies (dilution, 1:10,000; ProteinTech Group, Inc.) in TBS containing 0.05% Tween 20. Signals were revealed using an enhanced chemiluminescence reagent (cat. no. CW0049M; Century Biotech Co., Ltd.) and an autoradiography system (Chemiluminescence Imaging System; Bio-Rad Laboratories, Inc., Hercules, CA, USA) (18). The Image Lab software (version 5.1; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to analyze the protein bands and the results were normalized by the b-actin. Each assay was repeated at least three times.
Immunofluorescence analysis of β-catenin expression
SMCC-7721 cells cultured in a 24-well plate were transfected with RIP140 plasmids and were then treated by SV, followed by fixation in 4% (w/v) paraformaldehyde for 20 min at room temperature. Next, a β-catenin antibody (cat. no. 51067-2-AP; dilution, 1:100; ProteinTech Group, Inc.) was added to the cells and incubated overnight at 4°C. Subsequent to washing with PBS, cells were stained with FITC-conjugated secondary antibodies (cat. no. BA1105; dilution, 1:50; Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 20 min. DAPI staining (cat. no. AR1177; Wuhan Boster Biological Technology, Ltd.) was then performed at a concentration of 300 nM for 5 min (19). Finally, an inverted fluorescence microscope (Olympus IX71; Olympus Corp., Tokyo, Japan) was used to analyze the results.
Statistical analysis
Data are presented as the mean ± standard deviation. All statistical analyses were performed using SPSS version 17.0 software (SPSS, Inc., Chicago, IL, USA). The Student's t-test was used for single comparisons. For multiple comparisons, one-way analysis of variance with Tukey's or Games-Howell post-hoc analysis was used. Statistically significant differences were considered to be indicated by P<0.05.
Results
Appropriate concentration of SV
The results displayed in Fig. 1A indicated that SV treatment significantly inhibited the SMCC-7721 cell growth, with the proliferation rate decreasing from 91.58 to 21.56% upon increase of the SV dose between 4 and 20 µmol/l, respectively. Therefore, the inhibition efficiency of SV was dose dependent. Furthermore, the IC50 of SV in SMCC-7721 cells was calculated to be 12.57 µmol/l. However, to significantly demonstrate whether RIP140 is able to improve the sensibility of SV to SMCC-7721 cells, the treatment concentration of SV selected for further experiments in the present study was 8.0 µmol/l.
Figure 1.
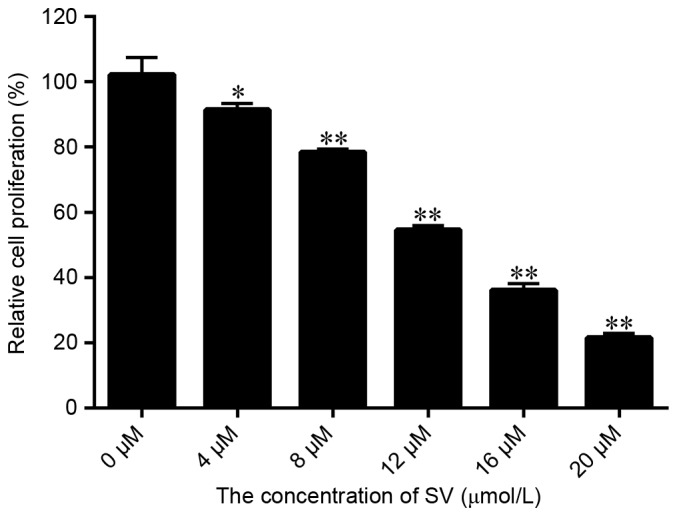
Selection of the appropriate concentration of SV. The growth and viability of SMCC-7721 cells following treatment by different doses of SV are reported. Values are represented as the mean ± standard deviation (n=6). *P<0.05 and **P<0.001, vs. control group. SV, simvastatin.
RIP140 expression in plasmid-transfected and SV-treated cells
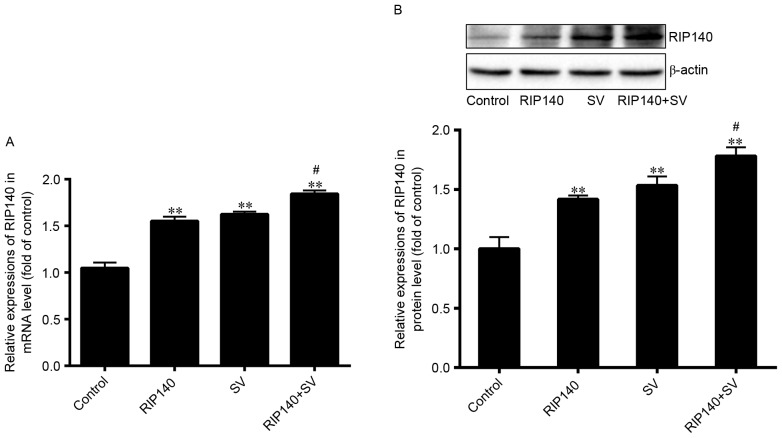
In the current study, RIP140 overexpression plasmids were transfected into SMCC-7721 cells. Additionally, the present study determined the expression of RIP140 following treatment with SV, using RT-qPCR and western blot analysis. As demonstrated in Fig. 2, the mRNA and protein expression levels of RIP140 were significantly upregulated in the RIP140, SV and RIP140 + SV groups as compared with those in the control group (P<0.001; n=4). The mRNA level of RIP140 increased by ~55.32, 62.48 and 84.36% in the RIP140, SV and RIP140 + SV groups, respectively, whereas the protein levels increased by ~41.71, 53.39 and 78.24%, respectively. In addition, the RIP140 expression in the RIP140 + SV group was further increased in comparison with that in the RIP140 or SV groups (P<0.05; n=4). These results suggested that both the RIP140 plasmid transfection and SV treatment were able to increase the expression of RIP140 in SMCC-7721 cells (P<0.001; n=4).
Figure 2.
(A) mRNA and (B) protein expression levels of RIP140 were analyzed by reverse transcription-quantitative polymerase chain reaction and western blot analysis, respectively. Values are expressed as the mean ± standard deviation (n=4). **P<0.001 vs. control group; #P<0.05 vs. SV group. SV, simvastatin; RIP140, receptor-interacting protein 140.
HCC cell proliferation and apoptosis are affected by RIP140 overexpression or and SV treatment
SV induces cell apoptosis, while RIP140 is also able to inhibit the cell proliferation; thus, the present study investigated the combined effect of RIP140 and SV on the growth and viability of the HCC SMCC-7721 cells by CCK-8 and flow cytometry assays, respectively. To examine the apoptosis of SMCC-7721 cells, flow cytometry assays were conducted in each group (Fig. 3A-D), and the apoptosis rate was calculated (Fig. 3E). The apoptosis rates in the RIP140, SV and RIP140 + SV groups were 13.44, 22.08 and 41.70%, respectively. The data shown in Fig. 3E indicated that RIP140 overexpression or SV treatment alone led to evidently enhanced cell apoptosis compared with the control group cells (P<0.05 and P<0.001, respectively; n=3). It was also observed that the apoptosis rate in the RIP140 + SV group was significantly higher when compared with that in the RIP140 or SV group alone (P<0.001; n=3). Notably, the rate increase in the RIP140 + SV group was higher than the combined increase observed in the RIP140 and SV groups by 6.18% (Fig. 3E). As observed in Fig. 3F, RIP140 overexpression or SV treatment alone were able to decrease the proliferation of SMCC-7721 cells (P<0.05 and P<0.001, respectively; n=6). However, in the RIP140 + SV group, overexpression of RIP140 significantly promoted the proliferation inhibition induced by SV (Fig. 3F; P<0.001; n=6). The inhibition rates in the RIP140, SV and RIP140 + SV groups were 17.54, 25.99 and 50.01, respectively. Furthermore, the rate increase in the RIP140 + SV group was higher by 6.48% than the combined total increase of the RIP140 and SV groups (Fig. 3F). These findings confirmed that both RIP140 overexpression and SV treatment were able to induce apoptosis and decrease the proliferation of SMCC-7721 cells, while an enhanced effect was observed in the RIP140 + SV group.
Figure 3.
Cell apoptosis in the (A) control, (B) RIP140, (C) SV and (D) RIP140 + SV groups was analyzed by flow cytometry. (E) Quantified apoptosis rate and (F) cell proliferation results are presented. (G) Protein expression levels of Bcl-2 and Bax were analyzed by western blot analysis. Values are expressed as the mean ± standard deviation (n=4 or 6). *P<0.05 and **P<0.001, vs. control group; ##P<0.001 vs. SV group. SV, simvastatin; RIP140, receptor-interacting protein 140; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
These results were further verified by examining the expression levels of two apoptosis-associated proteins, Bcl-2 and Bax. As displayed in Fig. 3G, RIP140 overexpression or SV treatment alone were able to increase the expression of Bax protein, as well as decrease the expression of Bcl-2 (P<0.05 and P<0.001). Similarly, the effect in the RIP140 + SV group was markedly higher compared with that in the SV or RIP140 group alone (P<0.001; n=4; Fig. 3G).
β-catenin content is decreased by RIP140 and SV treatment
β-catenin is able to regulate the transcription of several genes associated with cell proliferation (11,12); therefore, the present study analyzed the expression of β-catenin in SMCC-7721 cells. The relative content of β-catenin was initially analyzed by immunofluorescence. As demonstrated in Fig. 4A, the relative content of β-catenin in the RIP140, SV and RIP140 + SV group was reduced when compared with the control group. It was also observed that the RIP140 + SV group exhibited lower fluorescence in comparison with the groups treated with RIP140 overexpression or SV alone. Furthermore, western blot analysis revealed that the expression of β-catenin was significantly decreased in the RIP140, SV and RIP140 + SV groups as compared with the control group (P<0.05 and P<0.001; n=4), while expression in the RIP140 + SV group was also markedly lower in comparison with the RIP140 or SV group alone (P<0.05; n=4; Fig. 4B). Thus, the western blot analysis results were in agreement with the immunofluorescence results.
Figure 4.
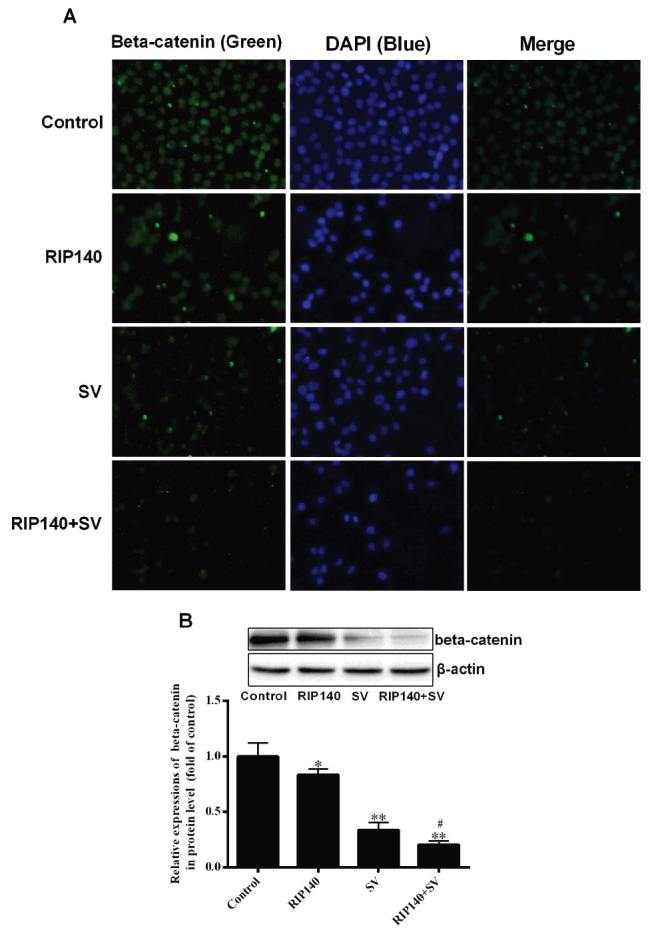
Content of β-catenin in the SMCC-7721 cells. (A) Immunofluorescence analysis of β-catenin. (B) Protein expression levels of β-catenin detected by western blot analysis. Values are presented as the mean ± standard deviation (n=4). *P<0.05 and **P<0.001, vs. control group; #P<0.05 vs. SV group. SV, simvastatin; RIP140, receptor-interacting protein 140.
Expression of c-myc and cyclin D1
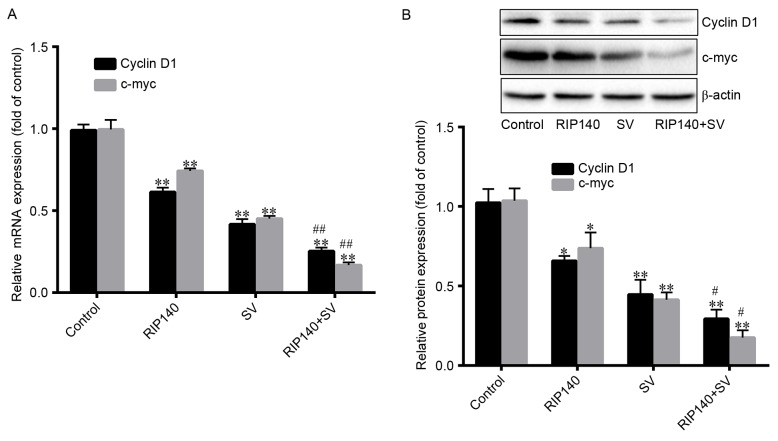
β-catenin is known to activate the transcription of the target genes, including cyclin D1 and c-myc. Thus, the current study investigated the expression levels of cyclin D1 and c-myc. As demonstrated in Fig. 5A, cyclin D1 mRNA expression levels were significantly decreased by ~38.60, 58.29 and 74.72% in the RIP140, SV and RIP140 + SV groups, respectively (P<0.001; n=4), compared with the control groups. However, the protein levels of cyclin D were significantly decreased by ~34.21, 55.37 and 71.73%, respectively (P<0.001; n=4; Fig. 5B). Furthermore, Fig. 5A also demonstrates that the mRNA levels of c-myc were significantly decreased by ~25.32, 54.46 and 82.83% (P<0.001; n=4), in the RIP140, SV and RIP140 + SV groups, respectively, compared with the control groups. Additionally, c-myc protein levels were significantly decreased by ~29.88, 62.37, 86.19%, respectively (P<0.001; n=4; Fig. 5B). Finally, the protein and mRNA expression levels in the RIP140 + SV group were significantly lower in comparison with those in the RIP140 or SV groups alone (P<0.001; n=4; Fig. 5A and B).
Figure 5.
(A) mRNA and (B) protein expression levels of c-myc and cyclin D1 were analyzed by reverse transcription-quantitative polymerase chain reaction and western blot analysis, respectively. Values are expressed as the mean ± standard deviation (n=4). *P<0.05 and **P<0.001, vs. control group; #P<0.05 and ##P<0.001, vs. SV group. SV, simvastatin; RIP140, receptor-interacting protein 140.
Discussion
In the present study, a RIP140 overexpression cell model was initially constructed and then cells were treated by the SV. The results of growth and viability assays displayed that RIP140 overexpression and SV treatment alone were able to inhibit SMCC-7721 cell proliferation. However, RIP140 overexpression enhanced the effect of SV treatment on these cells when these two were applied in combination. In addition, RIP140 overexpression and SV treatment applied together or separately on the cells resulted in decreased β-catenin, c-myc and cyclin D1 levels as compared with the control cells. These results suggested that β-catenin participated in the growth and viability regulation of the SV on the SMCC-7721 cells.
Statins are widely used to treat cardiovascular diseases since they decrease the biosynthesis of cholesterol through inhibition the enzyme HMGCR (1). In addition, statins have been investigated for carcinoma prevention or as cancer treatments (2–6). However, certain studies have suggested that statin-induced inhibition of cancer growth is due to the decrease of the cholesterol content in the plasma or cells (20,21). Cholesterol is vital for cell membrane integrity, cellular metabolism and cell signaling in cellular proliferation (21). Therefore, inhibition of certain signal transmissions is the primary mechanism underpinning the anticancer activity of statins. For instance, Huang et al (22) indicated that SV significantly promoted apoptosis in HCC cells through a mechanism that may involve the upregulation of the Notch1 gene in the Akt-dependent signaling pathway. Lee et al (23) also revealed that NS398 and SV co-administration produced greater anti-proliferative and pro-apoptotic effects against Hep3B and Huh-7 cells via inhibition of the NF-κB and Akt pathways, and activation of the caspase cascade. Besides these signaling pathway, statins are also able to regulate other signaling pathways, including the Hippo and PKC pathways (24,25).
In the present study, it was demonstrated that RIP140 and SV reduced the content of β-catenin (Fig. 4), suggesting that this protein may be involved in the apoptosis of SMCC-7721 cells. β-catenin is a subunit of the cadherin protein complex and functions as an intracellular signal transducer in the Wnt signaling pathway, which is involved in the expression of certain genes associated with cell proliferation (11,12). The findings of the present study also indicated that SV treatment in the SMCC-7721 cells increased the expression of RIP140 (Fig. 2). RIP140 is a transcriptional co-regulator that is involved in the negative regulation of energy homeostasis by affecting the storage of lipids and inhibiting the expression of genes involved in fatty acid oxidation and glucose metabolism (9–11), thus negatively regulating carcinomas, obesity, diabetes, atherosclerosis and other metabolic diseases (12,13,26). The possible mechanism underlying the negative regulation of carcinomas is the suppressive role of RIP140 on the pathogenesis of carcinomas by interacting with β-catenin and negatively regulating Wnt/β-catenin/TCF signaling (15,26). In the present study, the overexpression of RIP140 would strengthen the suppression of β-catenin expression and, eventually, of the β-catenin/TCF signaling. Furthermore, when SMCC-7721 cells were treated with SV, the content of β-catenin was also significantly decreased (Fig. 4). The results further revealed that the apoptosis rate of the RIP140 + SV group was higher when compared with RIP140 or SV group alone (Figs. 3 and 4). Therefore, it is suggested that RIP140 and SV exerted a synergistic effect on the apoptosis of SMCC-7721 cells, and that RIP140- and SV-induced apoptosis was associated with the Wnt/β-catenin signaling pathway due to the fact that the content of β-catenin was decreased in cells following combined treatment, compared with monotherapy (Figs. 2 and 4).
In conclusion, the present study results revealed that SV induced SMCC-7721 cell apoptosis through increasing the expression of RIP140 and decreasing the expression of β-catenin, and this effect may also be associated with the Wnt/β-catenin signaling pathway. Furthermore, RIP140 exerted a synergistic effect along with SV on the inhibition of the HCC cell proliferation and survival.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81560151) and the Jiangxi Provincial Department of Science and Technology (grant no. 20142BAB205014).
References
- 1.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Lee KH, Lee GK, Lee SH, Lim KY, Joo J, Go YJ, Lee JS, Han JY. Randomized phase II study of afatinib plus simvastatin versus afatinib alone in previously treated patients with advanced nonadenocarcinomatous non-small cell lung cancer. Cancer Res Treat. 2017;49:1001–1011. doi: 10.4143/crt.2016.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou YY, Zhu GQ, Wang Y, Zheng JN, Ruan LY, Cheng Z, Hu B, Fu SW, Zheng MH. Systematic review with network meta-analysis: Statins and risk of hepatocellular carcinoma. Oncotarget. 2016;7:21753–21762. doi: 10.18632/oncotarget.7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demierre MF, Higgins PD, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–942. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 5.Blanc-Brude OP, Mesri M, Wall NR, Plescia J, Dohi T, Altieri DC. Therapeutic targeting of the survivin pathway in cancer: Initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin Cancer Res. 2003;9:2683–2692. [PubMed] [Google Scholar]
- 6.Li G, Zheng J, Xu B, Ling J, Qiu W, Wang Y. Simvastatin inhibits tumor angiogenesis in HER2-overexpressing human colorectal cancer. Biomed Pharmacother. 2017;85:418–424. doi: 10.1016/j.biopha.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Zhang Y, Wang W, Hua Y, Liu L, Shen S, Peng B. Thrombocytopenia and the outcomes of hepatectomy for hepatocellular carcinoma: A meta-analysis. J Surg Res. 2017;210:99–107. doi: 10.1016/j.jss.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira-Silva GÁ, Lages CC, Sartorelli P, Hasegawa FR, Soares MG, Ionta M. Casearin D inhibits ERK phosphorylation and induces downregulation of cyclin D1 in HepG2 cells. Toxicol In Vitro. 2017;38:27–32. doi: 10.1016/j.tiv.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Liu WH, Lee YM, Lam KK, Chen YF, Wang JJ, Yen MH, Cheng PY. The role of receptor-interacting protein 140 in the accumulation of fat in ovariectomised rats. Obes Surg. 2011;21:935–940. doi: 10.1007/s11695-010-0241-9. [DOI] [PubMed] [Google Scholar]
- 10.Mejhert N, Laurencikiene J, Pettersson AT, Kaaman M, Stenson BM, Rydén M, Dahlman I. Role of receptor-interacting protein 140 in human fat cells. BMC Endocr Disord. 2010;10:1. doi: 10.1186/1472-6823-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalán V, Gómez-Ambrosi J, Lizanzu A, Rodríguez A, Silva C, Rotellar F, Gil MJ, Cienfuegos JA, Salvador J, Frühbeck G. RIP140 gene and protein expression levels are downregulated in visceral adipose tissue in human morbid obesity. Obes Surg. 2009;19:771–776. doi: 10.1007/s11695-009-9834-6. [DOI] [PubMed] [Google Scholar]
- 12.Lapierre M, Docquier A, Castet-Nicolas A, Gitenay D, Jalaguier S, Teyssier C, Cavaillès V. The emerging role of the transcriptional coregulator RIP140 in solid tumors. Biochim Biophys Acta. 2015;1856:144–150. doi: 10.1016/j.bbcan.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Aziz MH, Chen X, Zhang Q, DeFrain C, Osland J, Luo Y, Shi X, Yuan R. Suppressing NRIP1 inhibits growth of breast cancer cells in vitro and in vivo. Oncotarget. 2015;6:39714–39724. doi: 10.18632/oncotarget.5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang CR, Jin ZX, Dong L, Tong XP, Yue S, Kawanami T, Sawaki T, Sakai T, Miki M, Iwao H, et al. Cisplatin augments FAS-mediated apoptosis through lipid rafts. Anticancer Res. 2010;30:2065–2071. [PubMed] [Google Scholar]
- 15.Zhang D, Wang Y, Dai Y, Wang J, Suo T, Pan H, Liu H, Shen S, Liu H. Downregulation of RIP140 in hepatocellular carcinoma promoted the growth and migration of the cancer cells. Tumour Biol. 2015;36:2077–2085. doi: 10.1007/s13277-014-2815-y. [DOI] [PubMed] [Google Scholar]
- 16.Jafari N, Zargar SJ, Yassa N, Delnavazi MR. Induction of apoptosis and cell cycle arrest by dorema glabrum root extracts in a gastric adenocarcinoma (AGS) cell line. Asian Pac J Cancer Prev. 2016;17:5189–5193. doi: 10.22034/APJCP.2016.17.12.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, Ding Y, Gong J, Yan N. Sphingomyelin synthase 2 affects CD14-associated induction of NF-κB by lipopolysaccharides in acute lung injury in mice. Mol Med Rep. 2016;14:3301–3306. doi: 10.3892/mmr.2016.5611. [DOI] [PubMed] [Google Scholar]
- 19.Berwick DC, Javaheri B, Wetzel A, Hopkinson M, Nixon-Abell J, Grannò S, Pitsillides AA, Harvey K. Pathogenic LRRK2 variants are gain-of-function mutations that enhance LRRK2-mediated repression of β-catenin signaling. Mol Neurodegener. 2017;12:9. doi: 10.1186/s13024-017-0153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohda T, Iwata K, Hirano G, Sakurai K, Yokoyama K, Morihara D, Takeyama Y, Irie M, Shakado S, Sakisaka S. 3-Hydroxyl-3-methylglutaryl-coenzyme A reductase is up regulated in hepatocellular carcinoma associated with paraneoplastic hypercholesterolemia. Med Mol Morphol. 2013;46:239–242. doi: 10.1007/s00795-013-0042-z. [DOI] [PubMed] [Google Scholar]
- 21.Furuya Y, Sekine Y, Kato H, Miyazawa Y, Koike H, Suzuki K. Low-density lipoprotein receptors play an important role in the inhibition of prostate cancer cell proliferation by statins. Prostate Int. 2016;4:56–60. doi: 10.1016/j.prnil.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Ma J, Xu J, Su Q, Zhao J. Simvastatin induces growth inhibition and apoptosis in HepG2 and Huh7 hepatocellular carcinoma cells via upregulation of Notch1 expression. Mol Med Rep. 2015;11:2334–2340. doi: 10.3892/mmr.2014.2976. [DOI] [PubMed] [Google Scholar]
- 23.Lee SJ, Hwang JW, Yim H, Yim HJ, Woo SU, Suh SJ, Hyun JJ, Jung SW, Koo JS, Kim JH, et al. Synergistic effect of simvastatin plus NS398 on inhibition of proliferation and survival in hepatocellular carcinoma cell line. J Gastroenterol Hepatol. 2014;29:1299–1307. doi: 10.1111/jgh.12503. [DOI] [PubMed] [Google Scholar]
- 24.Higashi T, Hayashi H, Kitano Y, Yamamura K, Kaida T, Arima K, Taki K, Nakagawa S, Okabe H, Nitta H, et al. Statin attenuates cell proliferative ability via TAZ (WWTR1) in hepatocellular carcinoma. Med Oncol. 2016;33:123. doi: 10.1007/s12032-016-0845-6. [DOI] [PubMed] [Google Scholar]
- 25.Kim W, Yoon JH, Kim JR, Jang IJ, Bang YJ, Kim YJ, Lee HS. Synergistic anti-tumor efficacy of lovastatin and protein kinase C-beta inhibitor in hepatocellular carcinoma. Cancer Chemother Pharmacol. 2009;64:497–507. doi: 10.1007/s00280-008-0897-1. [DOI] [PubMed] [Google Scholar]
- 26.Lei JJ, Peng RJ, Kuang BH, Yuan ZY, Qin T, Liu WS, Guo YM, Han HQ, Lian YF, Deng CC, et al. NOP14 suppresses breast cancer progression by inhibiting NRIP1/Wnt/β-catenin pathway. Oncotarget. 2015;6:25701–25714. doi: 10.18632/oncotarget.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]