Abstract
In order to examine the prognostic significance of miR-33a in patients with hepatocellular carcinoma (HCC), total RNA was extracted from 149 HCC biopsies, 36 of which were paired with para-carcinoma tissues, and miR-33a expression was measured by reverse transcription-quantitative polymerase chain reaction. The results demonstrated that miR-33a expression was decreased in HCC biopsies compared with normal liver tissue samples. It was also demonstrated that miR-33a expression was significantly associated with tumor foci number. Furthermore, overall and progression-free survival time was decreased in patients expressing low miR-33a with multiple tumor foci. Taken together, the low expression of miR-33a may be a potential risk factor for HCC.
Keywords: microR-33a expression, hepatocellular carcinoma, overall survival, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer worldwide, and is associated with an extremely poor prognosis (1–3). A principal reason for the high mortality of this disease is the failure of early diagnosis for patients with HCC and the lack of effective therapies for patients with HCC in advanced stages. Despite a number of advances made in therapeutic strategies and surgery, the overall prognosis for patients with HCC remains poor due to the high rate of metastasis and recurrence (4–6). Hence, the characterization of the molecular mechanisms in HCC is urgently required to allow the development of novel treatment methods for patients with HCC.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs (21–23 nucleotides) that are transcribed as precursors in the nucleus and are subsequently processed into mature miRNAs in the cytoplasm. Mature miRNAs primarily function by sequence specific interactions with the 3′-untranslated regions of mRNAs, leading to the translational suppression or degradation of the mRNAs (7). An increasing number of studies have suggested that miRNAs serve an essential role as prognostic and predictive biomarkers in various types of cancer. For example, miR-1290 and miR-196b have been demonstrated to predict the chemotherapeutic response of patients with lung adenocarcinoma (8). miR-149, an anti-tumor miRNA, has been identified as dysregulated in a variety of types of cancer, including gastric (9), breast (10) and colorectal cancer (11,12).
It is also reported that a number of are associated with HCC carcinogenesis, progression and metastasis, including miR-15b (13), miR-122 (14) and miR-29 (15). Furthermore, the low expression of miR-124 is significantly associated with a more aggressive phenotype and a poorer prognosis (16), whereas a high expression of miR-182 has been associated with intrahepatic metastasis and poor prognosis in HCC (17). However, the mechanism of these miRNAs and their regulatory networks in HCC remain elusive, and a number of miRNAs that are associated with HCC have yet to be considered.
miR-33a was originally demonstrated to regulate lipid and cholesterol metabolism (18), and is an intronic miRNA located within the sequence of the sterol regulatory element binding protein 2 (SREBP-2) gene. miR-33a has also been implicated as a tumor suppressor miRNA; Kuo et al (19) identified that miR-33a is downregulated in lung cancer cells and inhibits osteolytic bone metastasis by targeting parathyroid hormone. miR-33a has also been demonstrated to act as a tumor suppressor miRNA, and may downregulate the expression of the oncogenic kinase Pim-1 in K562 lymphoma cells and colon carcinoma (20,21). However, the role of miR-33a in HCC is not yet fully characterized. In the present study, the miR-33a expression was quantified, and its significance in the prediction of a prognosis was assessed in patients with HCC.
Materials and methods
Patients and tissue samples
A total of 149 HCC biopsies with a median age of 65.4 years (range 45 to 81 years), 36 of which were pairs of tissue with para-carcinoma tissues, were extracted between July 2004 and October 2013 from Tissue Bank, China-Japan Union Hospital, Jilin University (Jilin, China). All tissues were snap-frozen immediately in liquid nitrogen and stored at −80°C until the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay. None of the participants had undergone chemotherapy or radiotherapy prior to surgery. Clinical features were recorded, including each participant's characteristics (age, sex,), tumor characteristics (foci number, diameter, tumor differentiation, and distant metastasis) and miR-33a expression status, as included in Table I. The HCC tissues were staged based on 7th edition of the American Joint Committee on Cancer Tumor-Node-Metastasis staging system for HCC (22). All patients were grouped as ≥60 or <60 years of age, and tumors were grouped as ≥5 or <5 cm according to the tumor diameter.
Table I.
The association between miR-33a expression and clinicopathological characteristics in patients with hepatocellular carcinoma.
| miR-33a expression, n (%) | ||||
|---|---|---|---|---|
| Parameters | Cases, n (%) | Low | High | P-value |
| Age, years | 0.211 | |||
| <60 | 66 (44.9) | 29 (19.7) | 37 (25.2) | |
| ≥60 | 81 (55.1) | 44 (29.9) | 37 (25.2) | |
| Unknown | 2 | |||
| Sex | 0.045 | |||
| Male | 117 (79.6) | 63 (42.9) | 54 (36.7) | |
| Female | 30 (20.4) | 10 (6.8) | 20 (13.6) | |
| Unknown | 2 | |||
| Number of foci | 0.007 | |||
| Single | 97 (68.3) | 39 (27.5) | 58 (40.8) | |
| Multiple | 45 (31.7) | 29 (20.4) | 16 (11.3) | |
| Unknown | 7 | |||
| Diameter | 0.078 | |||
| <5 cm | 91 (68.3) | 40 (27.2) | 51 (34.7) | |
| ≥5 cm | 56 (31.7) | 33 (22.4) | 23 (15.6) | |
| Unknown | 2 | |||
| Tumor differentiation | 0.105 | |||
| Poor | 10 (7.7) | 7 (5.4) | 3 (2.3) | |
| Moderate | 96 (73.8) | 51 (39.2) | 45 (34.6) | |
| Well | 24 (18.5) | 8 (6.2) | 16 (12.3) | |
| Unknown | 19 | |||
| Distant metastasis | 0.223 | |||
| Absence | 89 (91.8) | 26 (26.8) | 63 (64.9) | |
| Presence | 8 (8.2) | 4 (4.1) | 4 (4.1) | |
| Unknown | 52 | |||
miR-33a, microRNA-33a.
A 150-month follow-up was conducted. The information required in order to complete the follow-up was received via outpatient visits or telephone calls, and was updated at three-month intervals. OS (overall survival) was defined as the time period from diagnosis to the time of mortality, irrespective of the cause. PFS (progression free survival) was defined from the initial date of diagnosis to the time of tumor progression, as assessed by a computed tomography (CT) scan, or to the time of mortality due to HCC.
Ethics statement
The present study was approved by the Ethics Committee of Shanghai Tenth People's Hospital, Tongji University School of Medicine (Shanghai, China; approval no., SHSY-IEC-pap-15-18). Each participant provided written informed consent prior to participating in the study. All samples were handled anonymously, according to applicable ethical and legal standards.
RNA extraction
Total RNA was isolated from HCC and para-carcinoma tissue specimens using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. The RNA concentration was determined using a Nanodrop 1000 spectrophotometer (Nanodrop; Thermo Fisher Scientific, Inc.; Wilmington, DE, USA) and the purity was identified with 1.5% denaturing agarose gel.
RT-qPCR
RT reactions were performed using AMV Reverse Transcriptase (Takara Biotechnology Co., Ltd., Dalian, China) and qPCR was performed on the Applied Biosystems 7900HT Sequence Detection System (Thermo Fisher Scientific, Inc.). TaqMan probe-based qPCR was carried out using TaqMan MicroRNA Reverse Transcription kit (cat. no. 4366597) and Universal Master Mix II (cat. no. 4440048; Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the protocol of the manufacturer (23). Specific RT primers and TaqMan probes for hsa-miR-33a (cat. no. 000424) and U6 (cat. no. 4427975; (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used to quantify the expression of hsa-miR-33a. The specific primers are as follows: hsa-miR-33a, forward, 5′-GCACTTTCATGATACAAGCCG-3′ and reverse, 5′-GACCACTCAGTTTAGAGCCA-3′; U6, forward, 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′. Thermocycling conditions were as follows: Initial denaturation at 94°C for 10 min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, with a final extension at 72°C for 10 min. Each reaction was independently tested a minimum of three times. U6 was used as the internal control and miR-33a levels were quantified using the 2−ΔΔCq method (24). miR-33a level was summarized and recorded as high vs. low levels of expression based on the median value.
Statistical analysis
Statistical analysis was conducted using SPSS 20.0 (IBM Corp., Armonk, NY, USA) and the production of figures was performed with GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). An independent t-test was used to evaluate differences between two groups, and a χ2 test was used to examine differences between multiple groups. OS and DFS curves were generated by GraphPad 5.0 software. Univariate and multivariate survival analyses were performed with Cox proportional hazard regression. P<0.05 was considered to indicate a statistically significant difference.
Results
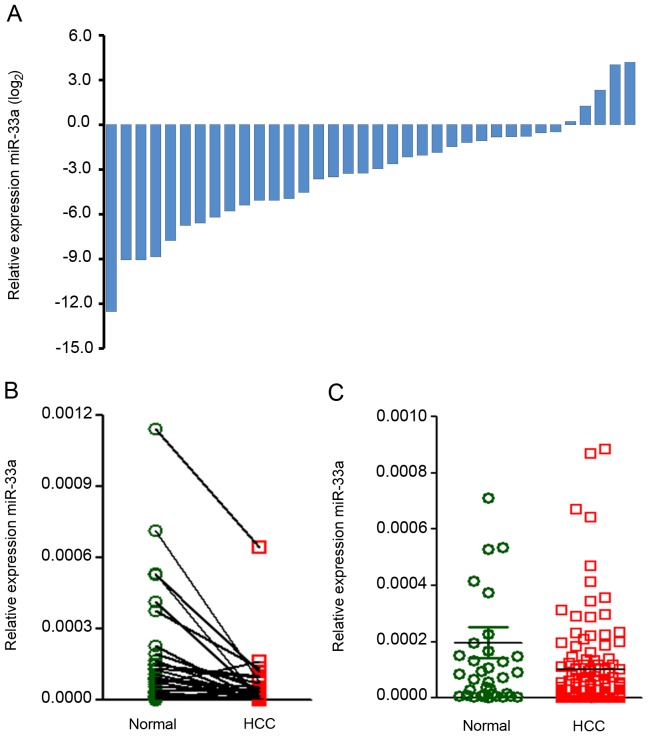
miR-33a expression in HCC and para-carcinoma tissue samples. In order to investigate the expression and prognostic significance of miR-33a in patients with HCC, the present study evaluated miR-33a expression levels via RT-qPCR in 36 biopsy pairs and 113 unpaired HCC biopsies. The miR-33a expression levels were classified as high or low compared with the median value. miR-33a expression levels were significantly lower in the 36 tumor biopsies than in the paired adjacent non-tumor tissues [fold change (FC), 0.11; P=0.004; Fig. 1A and B]. It was also identified that the miR-33a expression was significantly downregulated in 149 HCC tissue samples relative to 36 non-tumor tissue samples (FC, 0.51, P=0.043; Fig. 1C).
Figure 1.
miR-33a expression in normal liver and HCC tissues. (A) miR-33a expression levels in cancer vs. adjacent non-tumor tissue samples (n=36). (B) miR-33a expression levels in paired HCC and adjacent non-tumor tissue samples (n=36). (C) miR-33a expression in tumor samples (n=149) and paired adjacent non-tumor tissue (n=36). miR-33a, microRNA-33a; HCC, hepatocellular carcinoma.
Association of miR-33a expression with the clinical characteristics of HCC
The associations between miR-33a expression and individual clinical characteristics were investigated, and are listed in Table I. The results revealed that miR-33a expression levels were significantly correlated with sex (P=0.045) and tumor foci number (P=0.007) in HCC samples. However, there was no association of miR-33a expression with patient age, tumor diameter, tumor differentiation or distant metastasis (P>0.05).
Association between clinical characteristics and prognosis in HCC. The association between the clinical characteristics and HCC prognosis were further investigated by univariate survival analyses based on a Cox proportional hazard regression model. In accord with our prior hypothesis, multiple tumor foci [hazard ratio (HR), 1.995; 95% confidence interval (CI), 1.208–3.293; P=0.007] increased tumor size (HR, 1.945; 95% CI, 1.170–3.246; P=0.011), poorly differentiated tumors (HR, 0.471; 95% CI, 0.262–0.842; P=0.011) and distant metastasis (HR, 3.468; 95% CI, 1.130–10.644; P=0.032) were positively associated with a poor prognosis (Table II).
Table II.
Cox regression model analysis for OS and PFS based on various clinical characteristics in patients with hepatocellular carcinoma.
| Univariate analysis for OS | Multivariate analysis for OS | Univariate analysis for PFS | Multivariate analysis for PFS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age | 1.193 | 0.709–2.009 | 0.507 | 1.062 | 0.692–1.655 | 0.979 | ||||||
| Sex | 0.493 | 0.225–1.084 | 0.079 | 0.609 | 0.330–1.124 | 0.113 | ||||||
| Number of foci | 1.995 | 1.208–3.293 | 0.007 | 16.665 | 6.330–43.873 | <0.001 | 1.634 | 1.040–2.569 | 0.033 | 5.589 | 2.975–10.503 | <0.001 |
| Diameter | 1.945 | 1.170–3.246 | 0.011 | 1.549 | 0.991–2.419 | 0.055 | ||||||
| Tumor differentiation | 0.471 | 0.262–0.842 | 0.011 | 0.512 | 0.308–0.851 | 0.011 | ||||||
| Distant metastasis | 3.468 | 1.130–10.644 | 0.032 | 4.879 | 2.199–10.825 | <0.001 | ||||||
| miR-33a expression | 0.072 | 0.033–0.159 | <0.001 | 0.194 | 0.118–0.317 | <0.001 | ||||||
OS, overall survival; PFS, progression free survival; HR, hazard regression; CI, confidence interval.
Low expression of miR-33a is a prognostic marker for patients with HCC
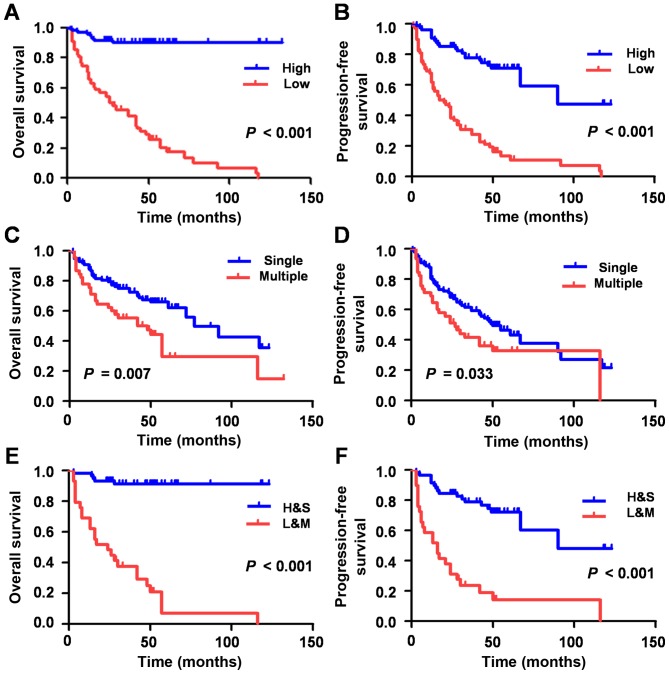
As univariate survival (Table II) and Kaplan-Meier survival analyses (Fig. 2A-D) demonstrated, the lower expression of miR-33a group exhibited a shorter OS (HR, 0.072; 95% CI, 0.033–0.159; P<0.001) and PFS (HR, 0.194, 95% CI, 0.118–0.317; P<0.001), which indicated that the low expression of miR-33a may be a negative factor for HCC prognosis.
Figure 2.
Analysis of the effect of miR-33a expression and clinical parameters on the survival of patients with HCC. Univariate survival analysis of (A) OS and (B) PFS in HCC, as determined by Kaplan-Meier plots stratified by miRNA-33a expression. Univariate survival analysis of (C) OS and (D) PFS in HCC, as determined by Kaplan-Meier plots stratified by the number of foci. Multivariate analysis of (E) OS and (F) PFS by Kaplan-Meier survival analysis stratified by the number of foci and miRNA-33a expression. miR-33a, microRNA-33a; HCC, hepatocellular carcinoma; OS, overall survival; PFS, progression free survival.
In order to explore whether the expression of miR-33a may unite with other clinical factors to influence HCC survival, a multivariate Cox proportional hazards regression analysis was performed. Associations between miR-33a expression and parameters (including age, sex, tumor foci number, tumor diameter, tumor differentiation and distant metastasis) that are predictive of HCC prognosis were initially analyzed by the model. As presented in Table I, the results demonstrated that the patient sex (P=0.045) and tumor foci number (P=0.007) were positively correlated with the miR-33a expression levels in the HCC tissue samples. A forward stepwise univariate survival analysis revealed that tumor foci number, tumor diameter, differentiation, distant metastasis and miR-33a expression were positively associated with HCC prognosis. The above results demonstrated that the number of tumor foci and the miR-33a expression level may jointly influence HCC prognosis.
The 149 HCC patients were subsequently divided into two groups. One group was comprised of patients with low miR-33 expression levels and multiple tumor foci (L+M), whereas patients with high miR-33 expression levels and single tumor foci, comprised the other group (H+S). The L+M group experienced a significantly shorter OS (HR, 16.665; 95% CI, 6.330–43.873; P<0.001) and PFS (HR, 5589; 95% CI, 2.975–10.503; P<0.001) time compared with H+S, as determined via multivariate Cox regression model and Kaplan-Meier survival analyses (Table II; Fig. 2).
In summary, the data of the present study demonstrated that the expression of miR-33a may serve a significant role in HCC progression, and that miR-33a may have exhibited potential as a tumor biomarker in the determination of the prognosis of HCC.
Discussion
Molecular biomarkers have started to serve an important role in the selection of patient therapeutics; they may serve as indicators of a patient's individual likelihood of a chemotherapeutic response. The HCC mortality rate is the fastest growing among all types of cancer and it is the third most common cause of tumor-associated mortality (25–27). Although there have been improvements in surgery and other therapeutic methods, the 5-year survival rate has remained <15% for a number of years (28–31). In order to seek a more efficient and individualized treatment, it is essential for researchers to comprehensively understand the molecular mechanisms of HCC progression.
Accumulating studies have demonstrated that the expression of miRNA is dysregulated in various types of human cancer, and may be associated with oncogenesis. It has been reported that miRNAs can indirectly repress the expression of a number of cancer-associated genes, and directly work as tumor suppressors or oncogenes (32). A number of studies have described the role of miRNAs in cancer treatment and diagnosis as a prognostic indicator. For example, a seven-miRNA signature of could be a robust predictor for OS and relapse-free survival in gastric cancer (33), or the low expression of miR-26 in the diagnosis of HCC (34).
A number of miRNAs have a confirmed effect on the initiation and progression of HCC. For example, the overexpression of miR-149 suppressed the migration and invasion of HCC by targeting protein phosphatase, Mg2+/Mn2+ dependent 1F directly (35). miR-148a induces hepatocytic differentiation by inhibiting the inhibitor of nuclear factor κα/NUMB/NOTCH pathway (36).
miR-33a serves a significant role in fatty acid metabolism and cholesterol synthesis (37,38), and inhibiting miR-33a has been considered as a method to reduce the risk of cardiovascular disease (39). It is suggested that miR-33 may function as a tumor-associated molecule, as miR-33b can inhibit cell growth and induce apoptosis through suppressing the activity of WNT/β-catenin signaling in lung adenocarcinoma cells (40), and also inhibit the proliferation and migration of osteosarcoma cells by targeting hypoxia-inducible factor-1α (41). With further in-depth study of miR-33a, it was identified that mir-33a can also affect cell proliferation and cell cycle progression in tumors by regulating cyclin-dependent kinase 5, cyclin D1 and Pim-1 (42,43), inhibit bone metastasis by targeting parathyroid hormone-related protein (19), and inhibit cancer cell growth, invasion and metastasis by regulating the expression of high mobility group AT-hook 2 (44) and β-catenin (45). However, to the best of our knowledge, no report previously existed regarding the role of miR-33a in HCC.
In the present study, HCC and para-carcinoma tissues were examined to determine the prognostic significance of miR-33a expression. Kaplan-Meier survival curve analysis indicated that patients with lower miR-33a expression exhibited significantly poorer survival. miR-33a expression was significantly associated with the number of tumor foci, which is an important clinical determinant of the prognosis of patients with HCC. In a univariate Cox model, it was identified that low miR-33a expression was an independent predictive factor for the OS time of HCC patients. In a multivariate Cox model, it was identified that the presence of multiple foci was associated with the low expression of miR-33a, and the decreased OS and PFS time of patients with HCC.
In summary, the data of the present study revealed that the miR-33a expression level was significantly associated with the number of tumor foci, and the combination of low miR-33a expression with multiple foci number was associated with significantly decreased OS and PFS time. miR-33a may, therefore, promote regional metastasis and serve as a potential prognostic biomarker for HCC in clinical practice. However, further study with a larger cohort is required in order to validate this view.
Acknowledgements
The present study was supported in part by grants from the National Natural Science Foundation of China (81201535, 81302065, 81472202, 81772932, 81472209 and 81702243), Jilin Provincial Science and Technology Department (20140414061GH), Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900) and Shanghai Municipal Commission of Health and Family Planning (201440398 and 201540228).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Hessheimer AJ, Isabel Real M, Bruix J. Treatment of hepatocellular carcinoma. Crit Rev Oncol Hematol. 2006;60:89–98. doi: 10.1016/j.critrevonc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Guglielmi A, Ruzzenente A, Conci S, Valdegamberi A, Vitali M, Bertuzzo F, De Angelis M, Mantovani G, Iacono C. Hepatocellular carcinoma: Surgical perspectives beyond the barcelona clinic liver cancer recommendations. World J Gastroenterol. 2014;20:7525–7533. doi: 10.3748/wjg.v20.i24.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishizawa T, Hasegawa K, Aoki T, Takahashi M, Inoue Y, Sano K, Imamura H, Sugawara Y, Kokudo N, Makuuchi M. Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology. 2008;134:1908–1916. doi: 10.1053/j.gastro.2008.02.091. [DOI] [PubMed] [Google Scholar]
- 7.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 8.Saito M, Shiraishi K, Matsumoto K, Schetter AJ, Ogata-Kawata H, Tsuchiya N, Kunitoh H, Nokihara H, Watanabe S, Tsuta K, et al. A three-microRNA signature predicts responses to platinum-based doublet chemotherapy in patients with lung adenocarcinoma. Clin Cancer Res. 2014;20:4784–4793. doi: 10.1158/1078-0432.CCR-14-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J, Xia L, Wang R, Cai X, Hu H, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693. doi: 10.1371/journal.pone.0041693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–4507. doi: 10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y, Qin HL. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229:12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 12.Øster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET, Sandoval J, Moran S, Esteller M, Hansen TF, et al. Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer. 2013;132:2303–2315. doi: 10.1002/ijc.27921. [DOI] [PubMed] [Google Scholar]
- 13.Chung GE, Yoon JH, Myung SJ, Lee JH, Lee SH, Lee SM, Kim SJ, Hwang SY, Lee HS, Kim CY. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23:113–119. [PubMed] [Google Scholar]
- 14.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 16.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo PL, Liao SH, Hung JY, Huang MS, Hsu YL. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochim Biophys Acta. 2013;1830:3756–3766. doi: 10.1016/j.bbagen.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M, Lange-Grünweller K, Weirauch U, Gutsch D, Aigner A, Grünweller A, Hartmann RK. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2012;31:918–928. doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 22.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 23.Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, Gilissen C, Jansen BJ, Adema GJ, Kögler G, Le Sage C, Agami R, van der Reijden BA, Jansen JH. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19:877–885. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L, et al. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896–11905. doi: 10.18632/oncotarget.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Y, Fu D, Tang WQ, Cai Y, Ma D, Wang H, Xue R, Liu T, Huang X, Dong L, et al. Ubiquitin C-terminal hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559–572. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 27.Wu SD, Ma YS, Fang Y, Liu LL, Fu D, Shen XZ. Role of the microenvironment in hepatocellular carcinoma development and progression. Cancer Treat Rev. 2012;38:218–225. doi: 10.1016/j.ctrv.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Ma Y, Fang Y, Wu S, Liu L, Fu D, Shen X. Regulatory T cell: A protection for tumor cells. J Cell Mol Med. 2012;16:425–436. doi: 10.1111/j.1582-4934.2011.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y, Mu J, Ma Y, Ma D, Fu D, Shen X. The interaction between ubiquitin C-terminal hydrolase 37 and glucose-regulated protein 78 in hepatocellular carcinoma. Mol Cell Biochem. 2012;359:59–66. doi: 10.1007/s11010-011-0999-7. [DOI] [PubMed] [Google Scholar]
- 30.Liu LL, Fu D, Ma YS, Shen XZ. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011;20:2023–2030. doi: 10.1089/scd.2011.0012. [DOI] [PubMed] [Google Scholar]
- 31.Fang Y, Fu D, Shen XZ. The potential role of ubiquitin C-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, −143 and −145 expression in colorectal cancer. Dis Markers. 2009;26:27–34. doi: 10.1155/2009/921907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 34.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo G, Chao YL, Tang B, Li BS, Xiao YF, Xie R, Wang SM, Wu YY, Dong H, Liu XD, Yang SM. miR-149 represses metastasis of hepatocellular carcinoma by targeting actin-regulatory proteins PPM1F. Oncotarget. 2015;6:37808–37823. doi: 10.18632/oncotarget.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung KH, Zhang J, Zhou C, Shen H, Gagea M, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Beretta L. Differentiation therapy for hepatocellular Carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology. 2016;63:864–879. doi: 10.1002/hep.28367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling; Proc Natl Acad Sci USA; 2011; pp. 9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suárez Y, de Cabo R, Gorospe M, Fernández-Hernando C. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891–2902. doi: 10.1128/MCB.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu J, Li M, An J, Zhao B, Zhong W, Gu Q, Cao L, Yang H, Hu C. MicroRNA-33b inhibits lung adenocarcinoma cell growth, invasion, and epithelial-mesenchymal transition by suppressing Wnt/β-catenin/ZEB1 signaling. Int J Oncol. 2015;47:2141–2152. doi: 10.3892/ijo.2015.3187. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol Res. 2017;25:397–405. doi: 10.3727/096504016X14743337535446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramírez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11:922–933. doi: 10.4161/cc.11.5.19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Zhou X, Shan B, Han J, Wang F, Fan X, Lv Y, Chang L, Liu W. Downregulation of microRNA-33a promotes cyclin-dependent kinase 6, cyclin D1 and PIM1 expression and gastric cancer cell proliferation. Mol Med Rep. 2015;12:6491–6500. doi: 10.3892/mmr.2015.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice SJ, Lai SC, Wood LW, Helsley KR, Runkle EA, Winslow MM, Mu D. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1) J Biol Chem. 2013;288:16348–16360. doi: 10.1074/jbc.M113.474643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu C, Zhao Y, Zhang Z, Ni Y, Li X, Yong H. MicroRNA-33a inhibits lung cancer cell proliferation and invasion by regulating the expression of β-catenin. Mol Med Rep. 2015;11:3647–3651. doi: 10.3892/mmr.2014.3134. [DOI] [PubMed] [Google Scholar]