Abstract
Postoperative monitoring for patients with colorectal cancer (CRC) requires sensitive biomarkers that are associated with medical response and adjuvant therapy following surgery. Conventional tumor biomarkers [including carcinoembryonic antigen (CEA), CA19-9 and CA125] are widely used, but none of the markers provide high sensitivity or specificity. Previous studies indicated that circulating tumor DNA (ctDNA) is useful for postoperative monitoring of patients with cancer. However, the majority of previous studies involved patients with lung cancer, and therefore further studies are required which investigate patients with CRC. The present study enrolled 11 patients with CRC. All patients underwent surgery, and a number of patients were treated with postoperative chemotherapy. Tumor tissues and serial blood samples were collected from each patient, and somatic mutations of each sample were obtained using next-generation sequencing. The mutation landscape and dynamic changes in mutations for each patient were analyzed, and these results were compared with the changes of CEA levels. A number of driver genes were selected, including tumor protein P53 (TP53), APC and KRAS, to monitor the postoperative outcome of the 11 patients with CRC. Driver mutations were detected in preoperative plasma in 7 patients, with markedly decreased mutation rates detected in postoperative plasma compared with preoperative plasma. Driver mutations were not detected in 4 patients in the preoperative or postoperative plasma. In 1 patient with metastatic rectal cancer, the rate of TP53 mutation increased from 8.95 (preoperative) to 71.4% (postoperative), and a new phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α mutation emerged. This patient succumbed to mortality six months following surgery, however there were no marked changes in CEA levels during periodic detection of CEA levels. In summary, ctDNA has a high sensitivity and specificity in prediction of the prognosis of patients with CRC.
Keywords: colorectal cancer, circulating tumor DNA, postoperative monitoring, next generation sequencing, liquid biopsy
Introduction
Colorectal cancer (CRC) is one of the most common types of gastrointestinal cancer in China (1). CRC increased from being the fifth most common cause of mortality in 2000 to the third most common cause in 2015 (2). As colonoscopy is not a popular screening method in China, a majority of patients are diagnosed at advanced stages of caner (3). In addition, effective treatment protocols for patients following surgery or chemotherapy have not developed successfully (4,5). The patients that are diagnosed at an early-stage CRC have a better prognosis, and the five-year survival rate is ~90% following surgery (6). Radical resection has been the primary treatment for early-stage CRC (including stage I–II) (7). However, adjuvant chemotherapy has additionally become an important treatment (8–10). Regardless of whether patients receive adjuvant therapy, dynamic monitoring following surgery is associated with the improved survival rates of patients with early-stage CRC (11–13). The sensitivity of existing tumor biomarkers [including carcinoembryonic antigen (CEA), cancer antigen 125 (CA125) and carbohydrate antigen 19-9] and other tests [including computed tomography (CT)] used for monitoring CRC is low (14–16). Therefore, identifying a highly sensitive clinical biomarker for the detection of CRC is of utmost importance.
Circulating tumor DNA (ctDNA) is fragmented DNA that is released from exfoliated tumor cells into the blood (17). ctDNA carries genetic information from the primary tumor and displays the genetic mutations of the tumor, which overcomes the problem of tumor heterogeneity (18). CtDNA is additionally used to monitor responses to treatment and to make decisions on further therapy during follow-up visits (19–23). Previous studies have revealed that mutations in plasma ctDNA and tumor biopsies were similar for patients with early breast cancer and CRC, and the majority of patients (89%) with driver mutations detected in ctDNA would recur 2–4 weeks postoperatively (24,25).
As a noninvasive detection method, ctDNA as a marker reveals distinct advantages in patients with advanced cancer over other detection methods. However, using ctDNA analysis to monitor therapy is a less sensitive method for patients in the early stages of cancer, which is likely as early-stage tumors release only a few DNA fragments into the blood (26). However, other sequencing methods additionally have problems that limit early detection (17,27). In the present study, next generation sequencing (NGS) technology was used to reveal significant associations between changes in ctDNA and different stages of CRC progression in order to examine the value of ctDNA in the clinical diagnosis and treatment of patients with CRC.
Materials and methods
Ethics statement
The present study was approved by the Shenzhen People's Hospital Ethics Committee, and the protocols were performed in accordance with the approved guidelines (28). Written informed consent was obtained from each patient for the use of blood and tumor tissues with the approval of the Shenzhen People's Hospital Ethics Committee (Shenzen, China). All samples and clinical data used in the present study were double-blind.
Patients and collection of clinical samples
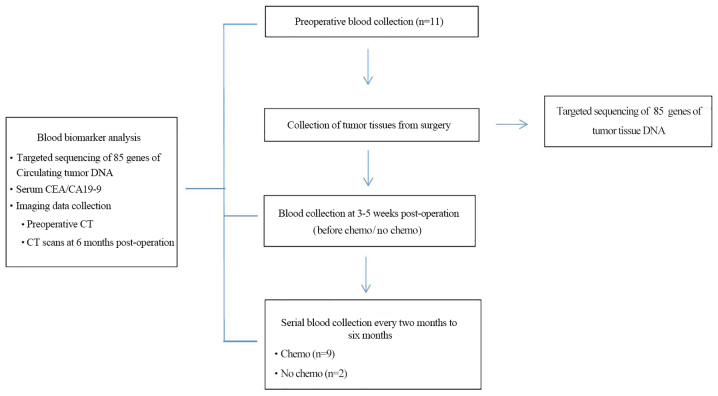
The present study recruited 11 patients with primary CRC who underwent radical resection in the Department of Gastrointestinal Surgery at the Shenzhen People's Hospital from September 2016 to April 2017. Exclusion criteria: i) Pregnant or lactating women; ii) patients who had received prior treatment with a MEK inhibitor, and iii) a history of clinically significant interstitial lung disease or pneumonitis. Tumor tissues were collected during surgery, quickly frozen in liquid nitrogen and stored at −80°C. Blood samples (20 ml) for ctDNA and CEA analysis were collected from 4–10 weeks postoperatively, with serial twice-monthly blood sample collection, which lasted for up to six months (Fig. 1). Adjuvant chemotherapy was used at the discretion of the treating clinician, who was blinded to the ctDNA results. As per protocol, follow-up included a twice-monthly clinical review, detection of CEA levels and CT imaging for six months. Subsequently, follow-up visits were performed according to the standard of care at participating institutions, which included three-month and six-month CT visits for two years.
Figure 1.
Patient enrolment and sample collection. Flow chart of patient enrollment and sample collection. CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9; CT, computed tomography; chemo, chemotherapy.
DNA extraction and sequencing
Tumor DNA was isolated from surgically resected CRC specimens. Blood samples were collected in tubes containing EDTA and centrifuged at 1,600 × g for 10 min at 4°C within 2 h of collection. Peripheral blood lymphocyte (PBL) debris was stored at −20°C until further use. The supernatants were further centrifuged at 10,000 × g for 10 min at 4°C, and plasma was harvested and stored at −80°C until further use. PBL DNA was extracted using the RelaxGene Blood DNA system (Tiangen Biotech Co., Ltd., Beijing, China), and ctDNA was extracted from ≥2 ml of plasma using the QIAamp Circulating Nucleic Acid kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. DNA was quantified using the Qubit 2.0 fluorometer and the Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol.
DNA was collected from PBLs and was sheared using the enzyme dsDNA Fragmentase (New England BioLabs, Inc., Ipswich, MA, USA). Size selection of the DNA fragments (150–250 bp) was then performed using Ampure XP beads (Beckman Coulter, Inc., Brea, CA, USA), which has the additional benefits of higher recovery and greater speed. DNA fragments and ctDNA were used for library construction using the KAPA Library Preparation kit (Kapa Biosystems, Inc., Wilmington, MA, USA) according to the manufacturer's protocol. Agencourt AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA) were used for all the cleanup steps. End repair and 3′-end A-tailing were performed following DNA fragmentation. Notably, T-tailed adapters were used and a 3′dA overhang was added enzymatically onto the fragmented DNA sample. Ligation was performed for 15 min at 20°C. Single-step size selection was performed by adding 50 µl (×1) of PEG/NaCl SPRI Solution buffer (Beckman Coulter, Inc.) to enrich ligated DNA fragments. The ligated fragments were then amplified using 1× KAPA HiFi Hot Start Ready mix (Kapa Biosystems, Inc.) and Pre-LM-PCR Oligos (Kapa Biosystems, Inc.) in 50 µl reactions, and 7–8 PCR cycles were performed depending on the amount of DNA input. The thermocycling conditions were as follows: Initial Denaturation, 98°C for 30 sec; 7–8 cycles at 98°C for 10 sec; 60°C for 10 sec; 68°C for 30 sec, and the final extension at 68°C for 60 sec. The purity and concentration of the DNA fragments were assessed using the Qubit 2.0 fluorometer and the Qubit dsDNA HS Assay kit. Fragment length was determined on a 4200 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) using the DNA 1000 kit (Agilent Technologies, Inc., Santa Clara, CA, USA). DNA sequencing was then performed on the Illumina NextSeq 500 system according to the manufacturer's recommendations at a depth of 7,000x.
Variant calling and analysis
Sequenced genes and their genebank accession numbers are provided in Table I (Colorectal cancer gene panel, 85 genes). Sequencing data were de-multiplexed and aligned to the hg19 genome (GRch37) using Burrows-Wheeler Aligner (29) version 0.7.15-r1140 using default settings. Pileup files for properly paired reads with mapping quality ≥60 were generated using Samtools (http://www.htslib.org/) (30). Somatic variants call were created using VarScan2 (http://varscan.sourceforge.net/) (31) [the minimum read depth at a position=100; the variant allele frequency (VAF) is the proportion of reads at a site which contain the variant allele. VAF threshold ≥0.05; somatic-P-value ≤0.01; strand-filter ≥1; otherwise, default parameters]. Allele frequencies were calculated for all Q30 bases. Using a custom Python script, previously identified tumor DNA mutations were intersected with a Samtools mpileup file generated for each plasma DNA sample; the number and frequency were then calculated for each mutation. For detection of mutations from plasma DNA without tumor DNA present, a mutation was identified if ≥4 mutant reads were identified in plasma and ≥1 mutant read was identified on each strand. No mutant reads were observed in matched genomic DNA from white blood cells.
Table I.
Colorectal cancer gene panel (85 genes).
Statistical analysis
A likelihood ratio test was used for comparing the difference of the number of nonsynonymous mutations between cancer stages. P<0.05 was considered to indicate a statistically significant difference. Analyses were performed using the R (version 3.1.2, https://www.R-project.org) (32).
Results
Patient characteristics
A total of 11 patients with CRC were enrolled, including 9 patients with colon cancer and 2 patients with rectal cancer. The cohort included 4 patients with stage I, 3 patients with stage II, 3 patients with stage III and 1 patient with stage IV using a pathological staging system (33). CT imaging revealed that 1 patient with CRC had liver metastasis at surgery. A total of 9 patients received postoperative chemotherapy following surgery. In total, 11 primary tumor tissues and 42 plasma samples were collected. Complete information is presented in Tables II and III.
Table II.
Clinical characteristics of 11 patients with CRC.
| Patient no. | Sex | Age at surgery (years) | Histology | TNM | Tumor stage | Size of tumor tissue (cm) | Maximum diameter (cm) | Adjuvant chemotherapy at surgery | Family history of cancer |
|---|---|---|---|---|---|---|---|---|---|
| H1001 | Male | 69 | Colon | T4bN0M0 | IIC | 4.0×3.0×2.0 | 4.0 | N | N |
| H1002 | Female | 50 | Colon | T2N1aM0 | IIIA | 2.1×1.5×0.6 | 2.1 | Oxaliplatin+5-FU (20 weeks) | N |
| H1003 | Male | 73 | Rectum | T3N2aM1b | IVB | 5.5×4.0×0.8 | 5.5 | Capecitabine (10 weeks) | Y |
| H1004 | Female | 49 | Rectum | T2N0M0 | I | 2.0×2.0×0.5 | 2.0 | Oxaliplatin+5-FU (2 weeks) | N |
| Capecitabine (12 weeks) | |||||||||
| H1005 | Female | 60 | Colon | T3N1cM0 | IIIB | 2.2×1.8×1.0 | 2.2 | Oxaliplatin (16 weeks) | NA |
| H1006 | Female | 69 | Colon | T1N0M0 | I | 1.5×1.5×1.0 | 1.5 | N | Y |
| H1007 | Male | 53 | Colon | T2N0M0 | I | 4.3×2.5×1.2 | 4.3 | Oxaliplatin+5-FU (4 weeks) | N |
| Capecitabine (18 weeks) | |||||||||
| H1008 | Female | 61 | Colon | T1N0M0 | I | 7.5×4.0×3.5 | 7.5 | Capecitabine (18 weeks) | N |
| H1009 | Male | 67 | Colon | T3N0M0 | IIA | 11.0×4.0×1.2 | 11 | Capecitabine (18 weeks) | N |
| H010 | Female | 35 | Colon | T3N1aM0 | IIIB | 3.0×2.5×0.7 | 3 | Oxaliplatin+5-FU (20 weeks) | N |
| H1011 | Female | 48 | Colon | T3N0M0 | IIA | 2.5×4.5×1.5 | 4.5 | Capecitabine (24 weeks) | N |
N, no; Y, yes; TNM, tumor-node metastasis; 5-FU, fluorouracil; NA, unknown.
Table III.
Patient and tumor characteristics.
| Characteristics | Patients (n=11) |
|---|---|
| Age at surgery | |
| Years, median (range) | 57.6 (35–73) |
| Sex, n (%) | |
| Male | 4 (36.4) |
| Female | 7 (63.6) |
| Tumor stage, n (%) | |
| I | 4 (36.3) |
| II | 3 (27.3) |
| III | 3 (27.3) |
| IV | 1 (9.1) |
| Localization, n (%) | |
| Colon | 9 (81.8) |
| Rectum | 2 (18.2) |
| Adjuvant chemotherapy, n (%) | |
| Yes | 9 (81.8) |
| No | 2 (18.2) |
| Family history, n (%) | |
| Yes | 2 (18.2) |
| No | 8 (72.7) |
| NA | 1 (9.1) |
NA, unknown.
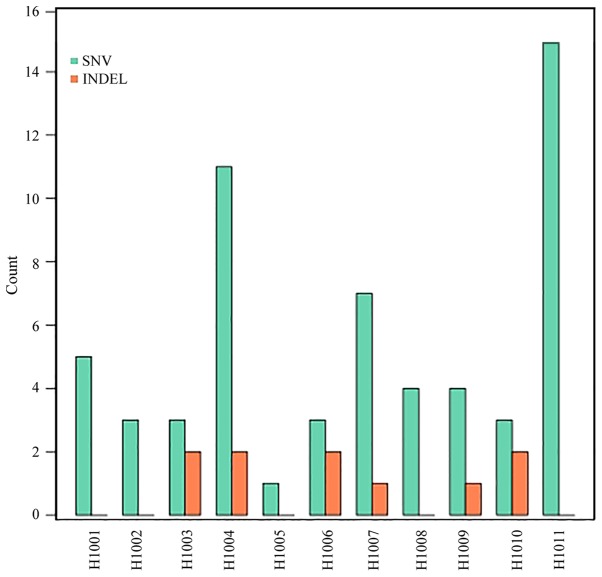
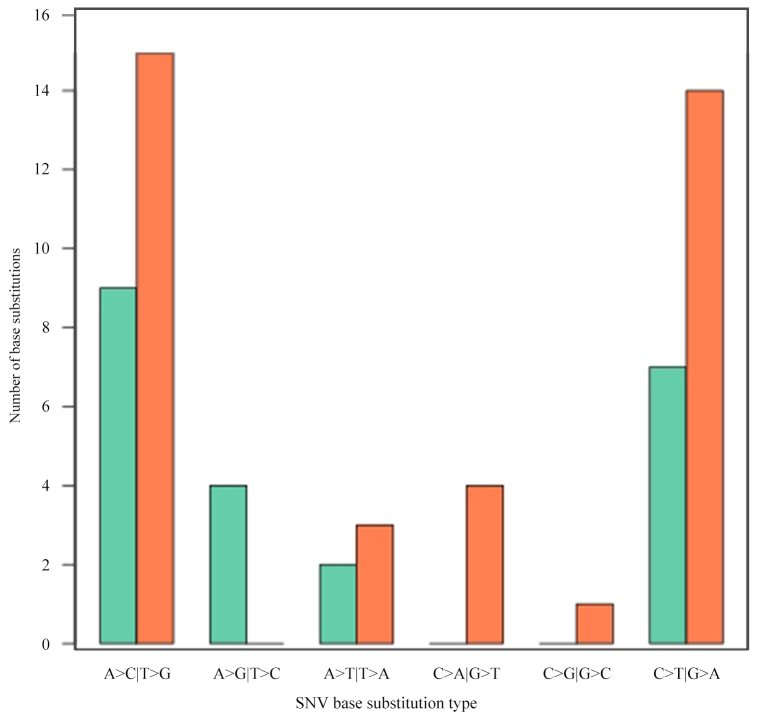
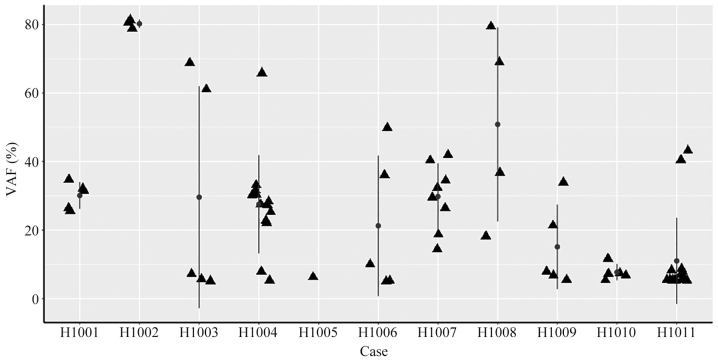
Somatic mutations in tumor tissue samples. Varscan2 (somatic P-value=0.01, minimum variant frequency=5%, strand filter=true, and otherwise default parameters) was used to detect tumor somatic single nucleotide variants (SNVs) and short insertion/deletion events captured by a panel of 85 genes (0.03% of the human genome; Table I). Within this small target, the number of nonsynonymous mutations detected in tumors ranged from 1 to 15, with a median of 5, including SNVs (59–86%) and small deletions (10–14%; Fig. 2; Table IV). The number of nonsynonymous mutations was not associated with the CRC stage (P=0.451). Additionally, it was identified that A>C|T>G transversion occurred most frequently (24 and 41%, respectively), followed by C>T|G>A transition (Fig. 3).
Figure 2.
Mutation statistics of tumor samples from 11 patients with colorectal cancer. The y-axis indicates the number of mutations and the x-axis indicates the patient ID. The type of mutations (SNV and INDEL) is indicated in green and orange, respectively. SNV, single nucleotide variants; INDEL, small insertions and deletions.
Table IV.
Dynamic detection of tumor tissue DNA and serial plasma circulating tumor DNA driver mutations in 11 patients with colorectal cancer.
| Case | Gene | Chromosome | Transcript | Exon | Nucleotide variation | Amino acid variation | Mutation frequency of tumor tissue (%) | Cosmid | Mutation frequency of preoperative plasma (%) | ALT-Depth | Depth | Mutation frequency of 1 month postoperative plasma (%) | ALT-Depth | Depth | Mutation frequency of 3 month postoperative plasma(%) | ALT-Depth | Depth | Mutation frequency of 5 mo postoperative plasma(%) | ALT-Depth | Depth |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1001 | ERBB2 | chr17 | NM_004448 | exon21 | c.G2524A | p.V842I | 34.81 | COSM14065 | 1.26 | 35 | 2,787 | 0 | 0 | 0 | – | – | – | 0 | 0 | 0 |
| H1001 | APC | chr5 | NM_000038 | exon6 | c.G568T | p.E190X | 32.16 | COSM258874 | 0.87 | 23 | 2,653 | 0 | 0 | 0 | – | – | – | 0 | 0 | 0 |
| H1001 | TP53 | chr17 | NM_000546 | exon5 | c.G524A | p.R175H | 31.4 | COSM10648 | 1.02 | 16 | 1,575 | 0.46 | 5 | 1,081 | – | – | – | 0 | 0 | 0 |
| H1001 | ERBB4 | chr2 | NM_005235 | exon28 | c.T3492A | p.N1164K | 26.48 | 1.03 | 33 | 3,215 | 0 | 0 | 0 | – | – | – | 0 | 0 | 0 | |
| H1001 | KRAS | chr12 | NM_004985 | exon2 | c.G35A | p.G12D | 25.66 | COSM521 | 0.44 | 10 | 2,254 | 0 | 0 | 0 | – | – | – | 0 | 0 | 0 |
| H1002 | TP53 | chr17 | NM_000546 | exon8 | c.G818A | p.R273H | 81.35 | COSM10660 | 0 | 0 | 0 | 0 | 0 | 0 | 0.13 | 3 | 2,352 | 0 | 0 | 0 |
| H1002 | APC | chr5 | NM_000038 | exon16 | c.G4222T | p.E1408X | 80.49 | COSM18822 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1002 | APC | chr5 | NM_000038 | exon16 | c.T2735A | p.L912X | 78.89 | COSM4167198 | 0 | 0 | 0 | 0.26 | 2 | 768 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1003 | TP53 | chr17 | NM_000546 | exon8 | c.A857G | p.E286G | 61.08 | COSM43565 | 8.95 | 233 | 2,602 | 11.32 | 124 | 1,100 | 42.39 | 993 | 2,342 | 71.4 | 1,990 | 2,787 |
| H1003 | MLH3 | chr14 | NM_014381 | exon2 | c.G1163A | p.S388N | 7.23 | 0.11 | 3 | 2,790 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1003 | KMT2D | chr12 | NM_003482 | exon31 | c.G7489A | p.A2497T | 5.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1003 | PIK3CA | chr3 | NM_006218 | exon9 | c.G1624A | p.E542K | 0 | COSM760 | 0 | 0 | 0 | 0 | 0 | 0 | 12.66 | 195 | 1,542 | 19.96 | 505 | 2,532 |
| H1003 | APC | chr5 | NM_000038 | exon6 | c.617delC | p.T206fs | 68.83 | 8.58 | 159 | 1,854 | 15.69 | 96 | 612 | 0 | 0 | 0 | 76.53 | 2,045 | 2,672 | |
| H1003 | PTEN | chr10 | NM_001304717 | exon1 | c.19_21del | p.7_7del | 5.76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | CTNNB1 | chr3 | NM_001904 | exon3 | c.A121G | p.T41A | 65.85 | COSM5664 | 0.27 | 9 | 3,292 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1004 | BRCA2 | chr13 | NM_000059 | exon10 | c.A832G | p.S278G | 33.18 | 0.22 | 6 | 2,735 | 0.53 | 6 | 1,125 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | PIK3CA | chr3 | NM_006218 | exon2 | c.A331G | p.K111E | 31.66 | COSM13570 | 0.81 | 25 | 3,076 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1004 | KRAS | chr12 | NM_004985 | exon2 | c.G38A | p.G13D | 30.32 | COSM532 | 1 | 19 | 1,897 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1004 | BRCA2 | chr13 | NM_000059 | exon13 | c.C7006T | p.R2336C | 30.17 | COSM432312 | 0.56 | 15 | 2,702 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1004 | FZR1 | chr19 | NM_016263 | exon7 | c.C542T | p.A181V | 28.37 | 0.33 | 9 | 2,733 | 0.21 | 2 | 937 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | SLIT1 | chr10 | NM_003061 | exon11 | c.G1054A | p.A352T | 27.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0.27 | 2 | 740 | 0 | 0 | 0 | |
| H1004 | CYP19A1 | chr15 | NM_000103 | exon10 | c.C1285T | p.P429S | 27.34 | 0.41 | 10 | 2,458 | 0.34 | 4 | 1,178 | 0.52 | 6 | 1,163 | 0 | 0 | 0 | |
| H1004 | KMT2D | chr12 | NM_003482 | exon11 | c.C3868T | p.R1290W | 22.82 | COSM4042398 | 0.59 | 12 | 2,031 | 0 | 0 | 0 | 0.22 | 2 | 913 | 0 | 0 | 0 |
| H1004 | KMT2B | chr19 | NM_014727 | exon28 | c.T6737G | p.V2246G | 7.95 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | KMT2B | chr19 | NM_014727 | exon28 | c.T6746G | p.V2249G | 5.39 | 0.51 | 3 | 587 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | ARID1A | chr1 | NM_006015 | exon1 | c.433delC | p.P145fs | 25.28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1004 | AKAP9 | chr7 | NM_005751 | exon19 | c.5010_5013del | p.G1670fs | 22.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1005 | EPCAM | chr2 | NM_002354 | exon1 | c.T25G | p.F9V | 6.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | – | – | – | |
| H1006 | RB1 | chr13 | NM_000321 | exon1 | c.A13C | p.T5P | 10.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1006 | KMT2B | chr19 | NM_014727 | exon28 | c.T6746G | p.V2249G | 5.22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1006 | ARID1A | chr1 | NM_006015 | exon1 | c.A653C | p.N218T | 5.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1006 | APC | chr5 | NM_000038 | exon16 | c.4217_4218del | p.Q1406fs | 49.86 | COSM5007948 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1006 | TP53 | chr17 | NM_000546 | exon7 | c.749_756del | p.P250fs | 36.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1007 | SMAD4 | chr18 | NM_005359 | exon9 | c.G988C | p.E330Q | 41.96 | COSM14240 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1007 | TP53 | chr17 | NM_000546 | exon5 | c.G473A | p.R158H | 40.38 | COSM10690 | 0.21 | 2 | 972 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1007 | PIK3CA | chr3 | NM_006218 | exon5 | c.A1034T | p.N345I | 34.47 | COSM94978 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.48 | 5 | 1,058 |
| H1007 | FBXW7 | chr4 | NM_018315 | exon8 | c.C1018T | p.H340Y | 32.41 | COSM28798 | 0.28 | 3 | 1,086 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1007 | KRAS | chr12 | NM_004985 | exon2 | c.G35T | p.G12V | 26.5 | COSM520 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1007 | CDH1 | chr16 | NM_004360 | exon9 | c.G1259A | p.G420D | 18.71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1007 | ATM | chr11 | NM_000051 | exon17 | c.T2570G | p.L857R | 14.45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1007 | SMAD2 | chr18 | NM_005901 | exon9 | c.1009_1024del | p.R337fs | 29.56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1008 | KRAS | chr12 | NM_004985 | exon3 | c.A183T | p.Q61H | 79.42 | COSM555 | 3.26 | 52 | 1,593 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1008 | SMAD4 | chr18 | NM_005359 | exon12 | c.T1619G | p.L540R | 68.95 | COSM14165 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1008 | UGT1A1 | chr2 | NM_000463 | exon1 | c.G323A | p.R108H | 36.8 | COSM1018132 | 0.46 | 7 | 1,508 | 0.2 | 4 | 1,992 | 0.17 | 3 | 1,756 | 0 | 0 | 0 |
| H1008 | ATIC | chr2 | NM_004044 | exon6 | c.G515A | p.R172H | 18.27 | COSM1183927 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1009 | KRAS | chr12 | NM_004985 | exon2 | c.G35A | p.G12D | 33.98 | COSM521 | 0.54 | 3 | 553 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1009 | RB1 | chr13 | NM_000321 | exon1 | c.A13C | p.T5P | 7.94 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1009 | ARID1A | chr1 | NM_006015 | exon1 | c.A317C | p.N106T | 6.77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1009 | MSH6 | chr2 | NM_000179 | exon1 | c.A256C | p.T86P | 5.56 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1009 | ARID1A | chr1 | NM_006015 | exon20 | c.5125_5131del | p.L1709fs | 21.35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1010 | KMT2B | chr19 | NM_014727 | exon1 | c.T266G | p.V89G | 7.46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1010 | KMT2B | chr19 | NM_014727 | exon28 | c.T6746G | p.V2249G | 6.78 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1010 | KMT2D | chr12 | NM_003482 | exon11 | c.T2987G | p.V996G | 5.41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1010 | TP53 | chr17 | NM_000546 | exon7 | c.715_720del | p.239_240del | 11.72 | COSM45055 | 0 | 0 | 0 | 0 | 0 | 0 | 0.19 | 3 | 1,624 | 0 | 0 | 0 |
| H1010 | APC | chr5 | NM_000038 | exon16 | c.2025delA | p.T675fs | 7.35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | TP53 | chr17 | NM_000546 | exon8 | c.C844T | p.R282W | 43.17 | COSM10704 | 0.39 | 6 | 1,533 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1011 | PIK3CA | chr3 | NM_006218 | exon5 | c.T1035A | p.N345K | 40.51 | COSM754 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1011 | RB1 | chr13 | NM_000321 | exon1 | c.A13C | p.T5P | 8.73 | 9.42 | 13 | 138 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | TP53 | chr17 | NM_000546 | exon5 | c.G404A | p.C135Y | 8.28 | COSM10801 | 0.19 | 3 | 1,548 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1011 | KMT2B | chr19 | NM_014727 | exon1 | c.T248G | p.L83R | 7.96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | KMT2B | chr19 | NM_014727 | exon28 | c.T6746G | p.V2249G | 7.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | ERBB2 | chr17 | NM_004448 | exon1 | c.T25G | p.W9G | 6.37 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | KMT2B | chr19 | NM_014727 | exon28 | c.T6737G | p.V2246G | 5.67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | CREBBP | chr16 | NM_004380 | exon30 | c.A4931C | p.N1644T | 5.61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | APC | chr5 | NM_000038 | exon16 | c.G4057T | p.E1353X | 5.57 | COSM19048 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H1011 | PTCH1 | chr9 | NM_000264 | exon1 | c.A26C | p.E9A | 5.41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | PTPRK | chr6 | NM_002844 | exon15 | c.C2432T | p.T811I | 5.39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | TYMP | chr22 | NM_001953 | exon8 | c.T941G | p.L314R | 5.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | RB1 | chr13 | NM_000321 | exon1 | c.A24C | p.K8N | 5.29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| H1011 | TYMP | chr22 | NM_001953 | exon9 | c.T1175G | p.V392G | 5.22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
ALT, alteration.
Figure 3.
SNV base substitution statistics. Counts are indicated in columns. The base substitution types are indicated in rows, and different subtypes of base substitutions are represented in aquamarine and coral. SNV, single nucleotide variant.
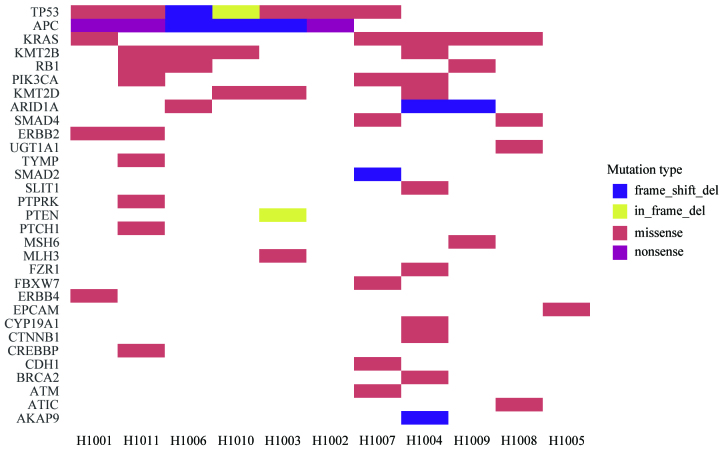
Different driver gene mutations revealed inter-individual tumor genetic heterogeneity. For example, KRAS mutations in codons 12 and 13 were observed in 3 patients and 1 patient, respectively. Tumor protein p53 (TP53) mutations in exons 5, 7 and 8 were observed in 3, 2 and 2 patients, respectively. A heatmap was constructed in order to demonstrate the somatic mutations occurring in the tumor tissues (Fig. 4). Different mutation loci and their corresponding mutation rates indicated intratumor genetic heterogeneity, implying tumor evolution (Fig. 5).
Figure 4.
Mutation heatmap. Mutation spectrum of tumor tissues from 11 patients with colorectal cancer. The mutation frequencies of genes with high mutation rates among these 11 patients are indicated. The names of the mutated genes are indicated in the y-axis, and the patient no. is shown in the x-axis. The type of mutations, including frame-shift deletion, in-frame deletion, missense, nonsense and frame-shift insertions are represented by the colors blue, yellow, red, orange and pink, respectively.
Figure 5.
Summary of VAF in 11 tumor cases. Scatter plots of the VAF in tumor tissues from 11 patients with colorectal cancer. The VAF values are indicated in the y-axis, and the patient ID is shown in the x-axis. The vertical line with dots represent the mean ± standard deviation of VAF. VAF, variant allele fraction.
Somatic mutation detection in preoperative plasma
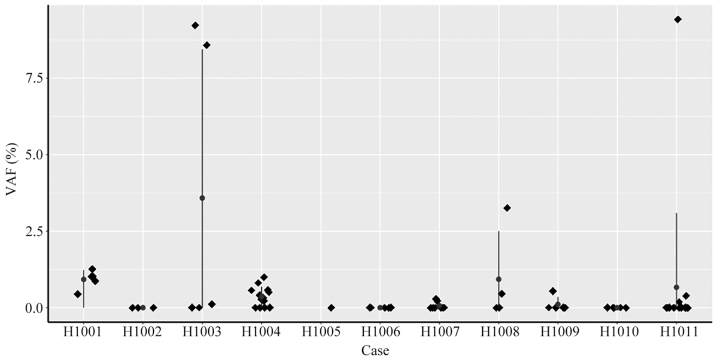
Next, ctDNA levels in preoperative plasma were examined. At a mean sequencing depth of x~7,000 (pre-duplication), a custom Python script was used to identify whether somatic tumor mutations were present in preoperative plasma. For a total of 7 patients, at least one mutation in tumor tissue was detected in preoperative plasma, and VAF in plasma ranged from 0.11 to 9.42% (Fig. 6). For the other 4 patients (H1002, H1005, H1006 and H1010), who were diagnosed with stage II or stage III cancer, no mutations that matched the tumor mutations were detected in the plasma.
Figure 6.
VAF in pre-operative plasma from 11 patients with CRC. Scatter plots of VAF in ctDNA of pre-operative plasma from 11 patients with CRC. The patient ID is presented on the x-axis, and the VAF is indicated on the y-axis. The vertical line with dots represent the mean ± standard deviation of the VAF. CRC, colorectal cancer; VAF, variant allele fraction.
Dynamic monitoring of postoperative patients with CRC using ctDNA
In order to determine prognosis for patients with CRC and decide whether or not to administer adjuvant chemotherapy, ctDNA levels were monitored periodically for six months. Driver gene mutations were detected at low mutation rates in preoperative plasma ctDNA from 7 patients (7/11; 63.6%), whilst no driver mutations were detected in the remaining 4 patients (4/11; 36.4%; patient nos. ID H1002, H1005, H1006 and H1010; see Table IV).
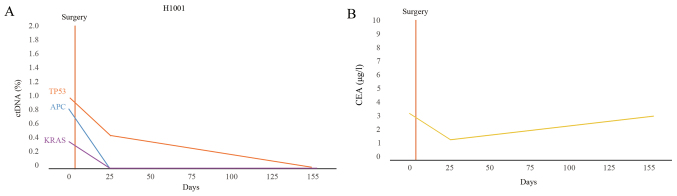
Following surgery, the number of mutations and the corresponding mutation rate decreased compared with preoperative plasma in postoperative plasma in the majority of patients. Driver mutations were not detected in 6 patients (6/11; 54.5%). To illustrate this, the changes in the percentage of ctDNA and CEA levels in patient H1001 were plotted (Fig. 7A and B). In patient H1001, the CEA levels initially declined and then increased slowly, but the levels remained in the normal range (<5 µg/l; Fig. 7B).
Figure 7.
Dynamic ctDNA monitoring of patient H1001. (A) ctDNA and (B) CEA levels were assessed preoperatively and postoperatively for patient no. H1001. Patient no. H1001 exhibited no recurrence. The number of days following surgery is indicated on the x-axis. (B) The concentration of CEA is indicated on the y-axis. ctDNA, circulating tumor DNA; CEA, carcinoembryonic antigen.
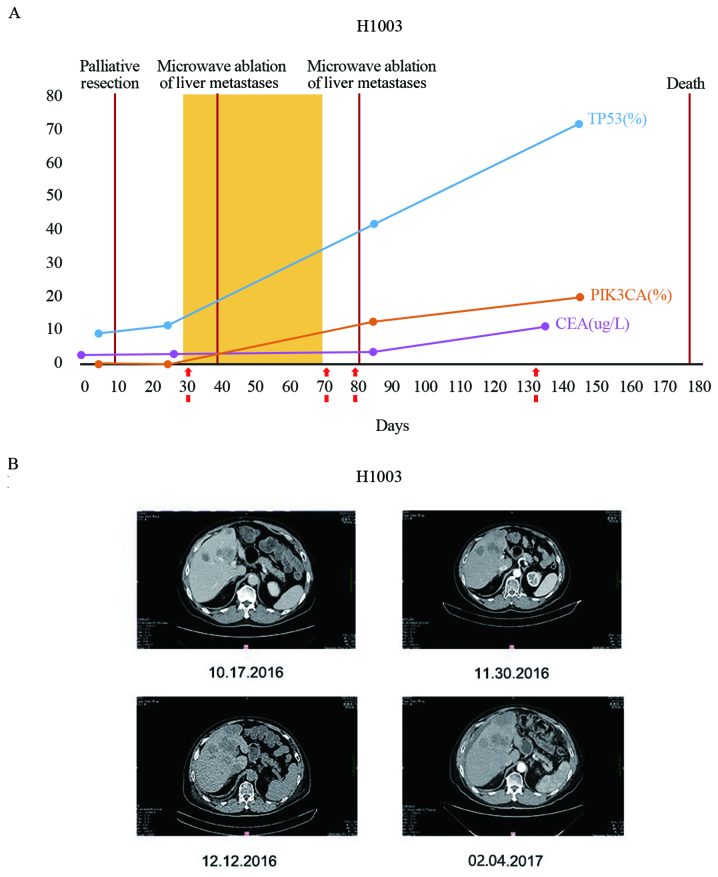
The VAF values for patient H1003 are presented in Fig. 8A. For patient H1003, the mutation rate of NM_0,011,26113.2 (TP53):c.857A>G (p.Glu286Gly) in ctDNA increased from 8.95 (preoperation) to 11.32% (post-operation) (Fig. 8A). Liver metastases were detected in patient H1003 at the time of surgery (Fig. 8B). Following surgery, patient H1003 received one round of chemotherapy and two rounds of microwave ablation for liver metastases, but treatments were subsequently terminated. The periodic monitoring of plasma ctDNA in patient H1003 revealed an increase in the mutation rate of TP53, and the development of a new mutation, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA). NM_006218.3 (PIK3CA):c.1624G>A (p.Glu542Lys) (Fig. 8A). In the final ctDNA sample from this patient, the mutation rate of TP53 and PIK3CA reached 71.4 and 19.96%, respectively (Fig. 8A). Patient H1003 succumbed to mortality six months following surgery. Furthermore, CEA levels were lower compared to the threshold (cut-off, 5 µg/l) in the three tests prior to mortality (2.63, 2.02 and 3.45 µg/l), which was not revealed to be associated with the patient's condition. However, the CEA level increased at the time of last sampling (11.12 µg/l; Fig. 8B). This finding suggests that ctDNA is more sensitive compared with CEA as a biomarker for predication of disease progression.
Figure 8.
Dynamic ctDNA monitoring of patient H1003. (A) ctDNA and CEA levels were assessed pre-operatively and postoperatively for patient H1003. H1003 presented with recurrence during colonoscopy and in contrast-enhanced CT. For patient H1003, the TP53 and PIK3CA mutation rate changes in ctDNA are presented at the indicated times, and compared with CEA. The number of days following surgery is indicated on the x-axis is indicated on the x-axis. The VAF values are indicated on the y-axis. (B) CT images of liver metastases in patient H1003, in whom the level of ctDNA was four times compared with other patients in the study without metastases. Images were captured on October 17th, 2016 (30 days after surgery), November 30th, 2016 (43 days after surgery), December 12th, (55 days after surgery) and February 4th, 2017 (98 days after surgery). CT, computed tomography; ctDNA, circulating tumor DNA; CEA, carcinoembryonic antigen; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; TP53, tumor protein P53; VAF, variant allele fraction.
Discussion
In the present study, somatic mutations in tumor tissues and preoperative plasma were highly associated as represented by the status of KRAS gene. In previous studies, ctDNA detection rates for patients in the early stages of cancer were low (16,27,34). Detection was improved in the present study by increasing the depth of ctDNA sequencing (~7,000X), which allowed for the monitoring of ctDNA dynamically to follow disease progression. ctDNA analysis using NGS was demonstrated to be sensitive for monitoring early therapeutic responses of CRC and metastatic breast cancer (35). Driver mutations in genes such as TP53, KRAS and adenomatosis polyposis coli tumor suppressor as observed in the 11 patients in the present study, were previously reported in patients with CRC (36). These mutations, thus, appear to be useful biomarkers for postoperative monitoring of CRC. As the sample size is relatively small, statistical bias is difficult to avoid.
For patients with early-stage CRC, administration of adjuvant chemotherapy following surgical resection remains controversial. Highly sensitive and specific biomarkers to predict the response to surgery are urgently required. Biomarkers for the early detection of recurrences would also be clinically valuable. In addition to previous research (16), the present study indicates that plasma ctDNA analysis has the potential for the detection of the early stages of cancer. Tie et al (37) demonstrated that ctDNA detection subsequent to stage II (33) colon cancer resection provided evidence of residual disease and identified patients at a substantially high risk of recurrence. Similar results were observed in the present study. However, the previous study by Tie et al (37) made predictions by targeting 15 genes and one single mutation in the ctDNA was selected for further analysis. By contrast, the present study examined preoperative and postoperative ctDNA for a whole spectrum of mutations. By comparison, the present study not only predicted the risk of recurrence, but was indicative of the tumor evolution for patients with cancer recurrence, which increases the current understanding of mechanism underlying recurrence in cancer.
As tumor-specific DNA is only a small fraction of cell-free DNA, ctDNA mutations are difficult to detect in patients with early stage cancer (37). For the patients in the present study with no preoperative driver gene mutations, we hypothesized that tumor cells may have invaded the serous layer of the intestinal wall. During the six months of monitoring, no changes were observed in the blood of patients with early-stage cancer. These findings indicate a good surgical outcome consistent with a greater survival of patients with early-stage CRC following surgery (37,38).
With the rapid progression of patients with late-stage cancer, driver mutations may quickly and substantially change in the blood. A high frequency of TP53 somatic mutation was detected in preoperative blood and tumor tissue of patient H1003. One month after surgery, the rate of TP53 mutations increased, which was associated with the presence of liver metastases observed on CT imaging subsequent to surgery. Conversely, decreased VAF of TP53 mutation were observed in all the other patients that did not have metastases following surgery. These results suggest that ctDNA ought to be analyzed in plasma at least every six months following surgery in order to monitor for disease progression in patients with early-stage cancer. Plasma CEA levels of patients with early or even advanced CRC were often below the normal threshold, implying that ctDNA has a higher sensitivity and specificity for detecting CRC compared with conventional CEA analysis.
Sequencing error rates are reported to be >0.1% in data analysis (39). In the present study, the correct mutations were selected by increasing the sequencing depth and a VAF threshold of <5% was used to filter the mutations detected. According to the tissue mutation results, ctDNA mutations were filtered using a VAF threshold of <0.1%, which may limit the sensitivity of ctDNA and omit a number of low-frequency mutations. Although the number of patients in the present study is small, it demonstrates the potential of using ctDNA to monitor the medical response and disease progression following surgery. ctDNA analysis will be applied to a greater number of clinical applications in the future.
Acknowledgements
The authors would like to thank Mr. Liwei Deng and Mr. Cheng Jin for enhancing the diagram and charts. The present study was supported by the grants from the Gastrointestinal Department of Shenzhen People's Hospital (grant no. 201601016), the Creative Design Program of Nanshan Shenzhen (grant no. KC2015JSJS0028A) for data collection, and the Special Funds for Future Industries of Shenzhen (grant no. JSGG20160229123927512) for sequencers and reagents. Clinicians of the Shenzhen People's Hospital were involved in study design, sample collection, and clinical information collection, while researchers and engineers from HaploX were involved in the study design and sequencing data analysis.
References
- 1.Chen W, Zheng R, Zuo T, Zeng H, Zhang S, He J. National cancer incidence and mortality in China, 2012. Chin J Cancer Res. 2016;28:1–11. doi: 10.1007/978-3-319-42740-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Wong SH. Colorectal cancer screening in Asia. Br Med Bull. 2013;105:29–42. doi: 10.1093/bmb/lds040. [DOI] [PubMed] [Google Scholar]
- 4.Garborg K. Colorectal cancer screening. Surg Clin North Am. 2015;95:979–989. doi: 10.1016/j.suc.2015.05.007. [DOI] [PubMed] [Google Scholar]; Leung WK, Lau JY, Suen BY, Wong GL, Chow DK, Lai LH, To KF, Yim CK, Lee ES, Tsoi KK, et al. Repeat-screening colonoscopy 5 years after normal baseline-screening colonoscopy in average-risk Chinese: A prospective study. Am J Gastroenterol. 2009;104:2028–2034. doi: 10.1038/ajg.2009.202. [DOI] [PubMed] [Google Scholar]
- 5.Soon MS, Kozarek RA, Ayub K, Soon A, Lin TY, Lin OS. Screening colonoscopy in Chinese and Western patients: A comparative study. Am J Gastroenterol. 2005;100:2749–2755. doi: 10.1111/j.1572-0241.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 6.National Cancer Institute's SEER database.[EB/OL] http://seer.cancer.gov/ [Aug 26;2016 ]. http://seer.cancer.gov/
- 7.Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandalà M, Cervantes A, Arnold D, ESMO Guidelines Working Group Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi64–vi72. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- 8.Twelves C, Wong A, Nowacki MP, Abt M, Burris H, III, Carrato A, Cassidy J, Cervantes A, Fagerberg J, Georgoulias V, et al. Capecitabine as adjuvant treatment for stage III colon cancer. N Engl J Med. 2005;352:2696–2704. doi: 10.1056/NEJMoa043116. [DOI] [PubMed] [Google Scholar]
- 9.Murphy CC, Harlan LC, Lund JL, Lynch CF, Geiger AM. Patterns of colorectal cancer care in the united states: 1990–2010. J Natl Cancer Inst. 2015;107:pii: djv198. doi: 10.1093/jnci/djv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffery M, Hickey BE, Hider PN, See AM. Follow-up strategies for patients treated for non-metastatic colorectal cancer. Cochrane Database Syst Rev. 2016;11:CD002200. doi: 10.1002/14651858.CD002200.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 12.Pita-Fernández S, Alhayek-Aí M, González-Martín C, López-Calviño B, Seoane-Pillado T, Pértega-Díaz S. Intensive follow-up strategies improve outcomes in non-metastatic colorectal cancer patients after curative surgery: A systematic review and meta-analysis. Ann Oncol. 2015;26:644–656. doi: 10.1093/annonc/mdu543. [DOI] [PubMed] [Google Scholar]
- 13.Kraus S, Nabiochtchikov I, Shapira S, Arber N. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347:15–21. doi: 10.1016/j.canlet.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Zhang Y, Niu Y, Li K, Liu X, Chen H, Gao C. A systematic review and meta-analysis of diagnostic and prognostic serum biomarkers of colorectal cancer. PLoS One. 2014;9:e103910. doi: 10.1371/journal.pone.0103910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. J Clin Oncol. 2009;27:e279–280. doi: 10.1200/JCO.2009.25.6156. [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler TM, Johnson-camacho K, Peto M, Wang NJ, Macey TA, Korkola JE, Koppie TM, Corless CL, Gray JW, Spellman PT. Exome sequencing of cell-free DNA from metastatic cancer patients identifies clinically actionable mutations distinct from primary disease. PLoS One. 2015;10:e0136407. doi: 10.1371/journal.pone.0136407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husain H, Velculescu VE. Cancer DNA in the circulation: The liquid biopsy. Jama. 2017;318:1272–1274. doi: 10.1001/jama.2017.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, Shovlin M, Jaffe ES, Staudt LM, Lai C, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015;16:541. doi: 10.1016/S1470-2045(15)70106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu S, Lou F, Wu Y, Sun DQ, Zhang JB, Chen W, Ye H, Liu JH, Wei S, Zhao MY, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett. 2016;370:324–331. doi: 10.1016/j.canlet.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanie Mortimer: Early, molecular detection of cancer utilizing circulating cell-free DNA assay with ultra high accuracy and sensitivity. ASCO. 2016 [Google Scholar]
- 24.Zhou J, Chang L, Guan Y, Yang L, Xia X, Cui L, Yi X, Lin G. Application of circulating tumor DNA as a non-invasive tool for monitoring the progression of colorectal cancer. PLoS One. 2016;11:e0159708. doi: 10.1371/journal.pone.0159708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdomo S, Anantharaman D, Mckay J, Brennan P. Abstract 5230: Circulating tumor DNA as a ‘liquid biopsy’ in head and neck cancer. Cancer Res. 2015;75:5230–5230. doi: 10.1158/1538-7445.AM2015-5230. [DOI] [Google Scholar]
- 26.Aravanis AM, Lee M, Klausner RD. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 27.Tie J, Wang Y, Kinde I, Steel M, Elsaleh H, Singh MS, Singh MS, Turner NH, Tran B, Strausberg R, et al. Circulating tumor DNA (ctDNA) in nonmetastatic colorectal cancer (CRC): Potential role as a screening tool. J Clin Oncol. 2017;3:518. [Google Scholar]
- 28.Morgans A, Allen F. Getting ethics committee approval for research: A beginners guide. Australasian J Paramedicine. 2015;3 [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup: The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller C, Mardis ER, Ding L, Wilson RK. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R: A Language and Environment for Statistical Computing, R Core Team. R Foundation for Statistical Computing; Vienna, Austria: [Google Scholar]
- 33.Hari DM, Leung AM, Lee JH, Sim MS, Vuong B, Chiu CG, Bilchik AJ. AJCC Cancer Staging Manual 7th edition criteria for colon cancer: Do the complex modifications improve prognostic assessment? J Am Coll Surg. 2013;217:181–190. doi: 10.1016/j.jamcollsurg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay Li Q, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 36.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras and p53-alternative genetic pathways to colorectal cancer; Proc Natl Acad Sci USA; 2002; pp. 9433–9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing; Proc Natl Acad Sci USA; 2012; pp. 14508–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]