Abstract
Yes-associated protein 1 (YAP1) is a transcriptional regulator of the Hippo pathway, which regulates the development and progression of a number of types of cancer, including that of the colon. In the present study, the expression levels of Hippo pathway genes and their clinical significance were investigated in 458 patients with colon adenocarcinoma (COAD), the most frequently diagnosed neoplastic disease globally, using data obtained from The Cancer Genome Atlas database. Notably, mRNA expression of YAP1 was higher in COAD than in other types of gastrointestinal tract cancer. Expression of YAP1 mRNA was higher in COAD than in normal colon samples and was significantly higher in Tumor-Node-Metastasis (TNM) stages III–IV than in stages I–II. YAP1 protein levels, a protein primarily localized in the nucleus, was greater in TNM stages III–IV than in stages I–II. The level of pYAP1, which is inactive and localized in the cytoplasm, was significantly higher in TNM stages III–IV than in stages I–II. However, the YAP1/pYAP1 ratio, which is representative of activity, was higher in TNM stages III–IV than in stages I–II. High mRNA expression of YAP1, TAZ and TEAD4 was associated with a poor prognosis in patients with COAD. Bioinformatics analysis revealed that YAP1 was associated with DNA duplication, cell proliferation and development. Wnt signaling and transforming growth factor-β signaling were significantly higher in the high-YAP1 group, according to data from Gene Set Enrichment Analysis. Taken together, the results indicate that the subcellular distribution of YAP1 and high mRNA expression of YAP1, TAZ and TEAD4 may be associated with poorer overall survival rates in patients with COAD.
Keywords: yes-associated protein 1, colon adenocarcinoma, The Cancer Genome Atlas, Hippo signaling pathway, overall survival
Introduction
Yes-associated protein 1 (YAP1) and its transcriptional co-activator with PDZ-binding domain taffazin (TAZ) form the backbone of the Hippo pathway kinase cascade, which is involved in regulation of tissue homeostasis, organ size, regeneration and tumorigenesis (1). In mammals, the Hippo pathway is comprised of the core kinase complexes mammalian Ste2-like kinase 1 and 2 (MST1 and MST2) and large tumor suppressor kinase 1 and 2 (LATS1 and LATS2) (2). Upon activation of the Hippo pathway, the inhibitory MST/LSTS kinases phosphorylate YAP1 and TAZ. This phosphorylation leads to the nuclear exclusion of YAP1 and TAZ, which are then sequestered and undergo ubiquitin-mediated proteasomal degradation in the cytoplasm to suppress the expression of YAP1- and TAZ-regulated genes (3,4). If molecular events, including crosstalk with oncogenic signaling pathways, trigger the dysregulation of the Hippo pathway, YAP1/TAZ are translocated into the nucleus (5). There, they interact with four transcriptional enhancer associated domain (TEAD) transcription factors, TEAD1-4, and promote cell proliferation and inhibit apoptosis (6).
The Hippo pathway was first hypothesized to be important in human cancer when tissue overgrowth was observed in Drosophila melanogaster flies with mutations in the Hippo pathway (7–9). A number of studies have indicated that human tumors use these biological properties of YAP1 to foster their own proliferation, progression and metastasis (4,10,11). Increased activation of YAP1 has been identified in a broad range of carcinomas including lung cancer, colorectal cancer, ovarian cancer, prostate cancer, melanoma and glioblastoma, and has often been associated with poor prognosis (12–18). However, the exact function of YAP1 in the tumorigenesis of certain types of cancer remains obscure, despite its oncogenic behavior in several types of cancer. In breast cancer, nuclear YAP1 expression does not differ significantly between normal breast and ductal carcinoma breast tissues (14). In hematological malignancies, including lymphoma and multiple myeloma, the expression of YAP1 is markedly downregulated (19).
In colon cancer, several studies reported that YAP1 is overexpressed, contributes to tumorigenesis and is associated with poor prognosis (20–22). However, Yuen et al (23) demonstrated that TAZ was the only prognostic marker in colorectal cancer. The functions of YAP1 and Hippo pathway genes have not yet been fully investigated in a large cohort of patients with colon cancer. Therefore, in the present study, the expression levels of Hippo pathway-associated genes including YAP1, TAZ, TEAD1, TEAD2, TEAD3, TEAD4, MST1, MST2, LATS1 and LATS2 were investigated and their clinical significance evaluated in a population of 458 patients with colon adenocarcinoma (COAD) using data obtained from The Cancer Genome Atlas (TCGA) research network (https://cancergenome.nih.gov/).
Materials and methods
Gene expression profiling
Level 3 mRNA expression data from 41 normal samples and 458 COAD samples were obtained from TCGA database (https://portal.gdc.cancer.gov/). Raw data were initially analyzed using R software (v.3.2.5) (24). Chip data were normalized using the RankNormalize module in GenePattern (https://genepattern.broadinstitute.org). GeneNeighbors and ClassNeighbors, modules programmed in GenePattern, were used to select genes closely associated with YAP1 (25). cBioportal (http://www.cbioportal.org/) and Firebrowse (http://firebrowse.org) were used to analyze mRNA expression and alterations in Hippo pathway genes.
Functional enrichment analysis
Differentially expressed genes were imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (26) for Gene Ontology (GO)-based functional enrichment analysis. Gene Set Enrichment Analysis (GSEA) was utilized to identify mRNAs predicted to associate with pathways in C2 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene sets (27,28). GO categories encompass three domains: Biological processes, cellular components and molecular functions.
Survival analysis
Cutoff Finder (http://molpath.charite.de/cutoff) was used to determine threshold values in mRNA and protein expression of COAD using log-rank tests (29). Cumulative event (mortality) rate was calculated using the Kaplan-Meier method, with the time to the first event as the outcome variable. The probability of recurrence and calculated risk for recurrence were determined by actuarial analysis. The criteria for statistical analysis were the date of surgery and the date of mortality.
Statistical analysis
The distributions of characteristics between the two groups were compared using unpaired Student's t-test for continuous variables (or the Kolmogorov-Smirnov test when the expected frequency within any cell was <5) and the χ2 test (or Fisher's exact test when the expected frequency within any cell was <5) for categorical variables. The distributions of characteristics between >3 groups were compared using analysis of variance and Newman-Keuls post-hoc test. Survival curves were compared by the log-rank test for various recurrence factors and Cox's model for multivariate analysis. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using the GraphPad Prism software (version 5.0; GraphPad Prism Software, La Jolla, CA, USA) and the Statistical Package for Social Sciences v.13.0 for Windows (SPSS, Inc., Chicago, IL, USA).
Results
Cross-cancer mRNA expression of YAP1
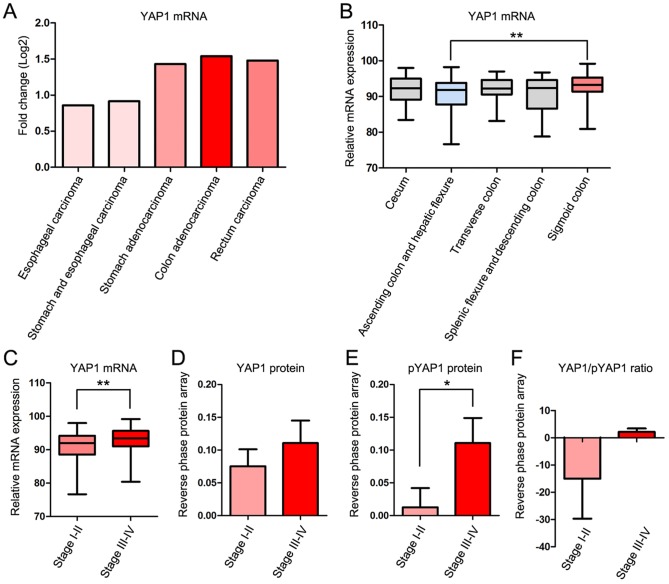
The fold change of YAP1 mRNA expression compared with 41 normal control samples in cases of COAD was higher than that in four other gastrointestinal cancer types; Esophageal carcinoma, stomach and esophageal carcinoma, stomach adenocarcinoma and rectal carcinoma in TCGA database. Clinicopathological information for the patients is presented in Table I. YAP1 mRNA expression was increased in COAD, stomach adenocarcinoma and rectal carcinoma, but decreased in esophageal carcinoma and ‘stomach and esophageal carcinoma’ combined compared with normal samples (Fig. 1A).
Table I.
Clinicopathological information of the patients with colon adenocarcinoma.
| Feature | Patients, n (%) |
|---|---|
| Number | 458 (100.0) |
| Sex | 458 (100.0) |
| Female | 216 (47.2) |
| Male | 242 (52.8) |
| Age, years | 458 (100.0) |
| ≤65 | 189 (41.3) |
| >65 | 269 (58.7) |
| Anatomic subdivision | 442 (96.5) |
| Ascending colon | 87 (19.0) |
| Cecum | 108 (24.0) |
| Descending colon | 20 (4.4) |
| Hepatic flexure | 27 (5.9) |
| Rectosigmoid junction | 1 (0.2) |
| Sigmoid colon | 152 (33.2) |
| Splenic flexure | 7 (1.5) |
| Transverse colon | 40 (8.7) |
| Histological type | 453 (98.9) |
| Colon adenocarcinoma | 391 (85.4) |
| Colon mucinous adenocarcinoma | 62 (13.5) |
| Vital status | 458 (100.0) |
| Alive | 356 (77.7) |
| Dead | 102 (22.3) |
| Postoperative Treatment | 390 (85.1) |
| Yes | 147 (32.1) |
| No | 243 (53.0) |
| Pathologic stage (TNM) | 448 (97.8) |
| I | 76 (16.6) |
| II | 178 (38.9) |
| III | 129 (28.2) |
| IV | 65 (14.2) |
| Lymphatic invasion | 414 (90.4) |
| Absent | 250 (54.6) |
| Present | 164 (35.8) |
| Perineural invasion | 179 (39.0) |
| Absent | 133 (29.0) |
| Present | 46 (10.0) |
| Venous invasion | 397 (86.8) |
| Absent | 301 (65.7) |
| Present | 96 (21.1) |
TNM, Tumor-Node-Metastasis.
Figure 1.
mRNA and protein expression of YAP1 in various TNM stages of COAD. (A) Fold change of mRNA expression of YAP1 in gastrointestinal cancers including esophageal carcinoma, stomach and esophageal carcinoma combined, stomach adenocarcinoma, colon adenocarcinoma and rectum carcinoma. (B) mRNA expression of YAP1 in various locations of colon cancer. (C) mRNA expression of YAP1 between TNM stage I–II and III–IV. (D) Protein level of YAP1 between TNM stage I–II and III–IV. (E) Phosphorylated YAP1 (pYAP1) between TNM stage I–II and III–IV. (F) YAP1/pYAP1 ratio between TNM stage I–II and III–IV. mRNA microarray data of YAP1 in colon adenocarcinoma obtained from The Cancer Genome Atlas data portal. *P<0.05 and **P<0.01. YAP1, yes-associated protein 1; pYAP, phosphorylated form of YAP1; TNM, Tumor-Node-Metastasis; COAD, colon adenocarcinoma.
mRNA expression of Hippo pathway genes in COAD
The Hippo pathway genes included in the present study were YAP1, TAZ, TEAD1, TEAD2, TEAD3, TEAD4, MST1, MST2, LATS1 and LATS2 (Table II). The fold change in mRNA expression in COAD compared with normal control samples identified that YAP1, TAZ, TEAD1, TEAD2, TEAD4, MST1 and MST2 were highly expressed in COAD compared with normal control samples, with TEAD4 and MST1 exhibiting the largest shifts. However, TEAD3, LATS1 and LATS2 exhibited lower expression in COAD than in normal control samples.
Table II.
Hippo pathway genes in colon adenocarcinoma.
| Symbol | Gene name | Chromosome location | Fold change, Log | Alteration, % |
|---|---|---|---|---|
| YAP1 | Yes associated protein 1 | 11q22.1 | 1.54 | 7.0 |
| TAZ | Tafazzin | Xq28 | 1.78 | 4.0 |
| TEAD1 | TEA domain transcription factor 1 | 11p15.3 | 1.11 | 4.0 |
| TEAD2 | TEA domain transcription factor 2 | 19q13.33 | 1.52 | 7.0 |
| TEAD3 | TEA domain transcription factor 3 | 6p21.31 | 0.87 | 5.0 |
| TEAD4 | TEA domain transcription factor 4 | 12p13.33 | 4.91 | 6.0 |
| MST1 | Macrophage stimulating 1 | 3p21.31 | 4.04 | 1.6 |
| MST2 | STK3, serine/threonine kinase 3 | 8q22.2 | 1.97 | 20.0 |
| LATS1 | Large tumor suppressor kinase 1 | 6q25.1 | 0.89 | 7.0 |
| LATS2 | Large tumor suppressor kinase 2 | 13q12.11 | 0.83 | 10.0 |
mRNA and protein expression of YAP1 in various Tumor-Node-Metastasis (TNM) stages of COAD
To examine the association between YAP1 expression and the location and progression of COAD, the expression of YAP1 was studied according to the location and TNM stage of COAD (30). YAP1 mRNA expression was significantly increased in the sigmoid colon compared with the ascending colon and hepatic flexure (Fig. 1B) and in TNM stages III–IV compared with stages I–II (Fig. 1C). Protein expression of YAP1 was markedly increased in TNM stages III–IV compared with stages I–II (Fig. 1D). The serine-1217-phosphorylated form of YAP1 (pYAP), which is inactive and localized in the cytoplasm, was significantly increased in TNM stages III–IV compared with stages I–II (Fig. 1E). However, the YAP/pYAP ratio, which represents YAP1 activity, was increased in TNM stages III–IV compared with stages I–II (Fig. 1F).
YAP1 mRNA and protein expression in T, N and M stages
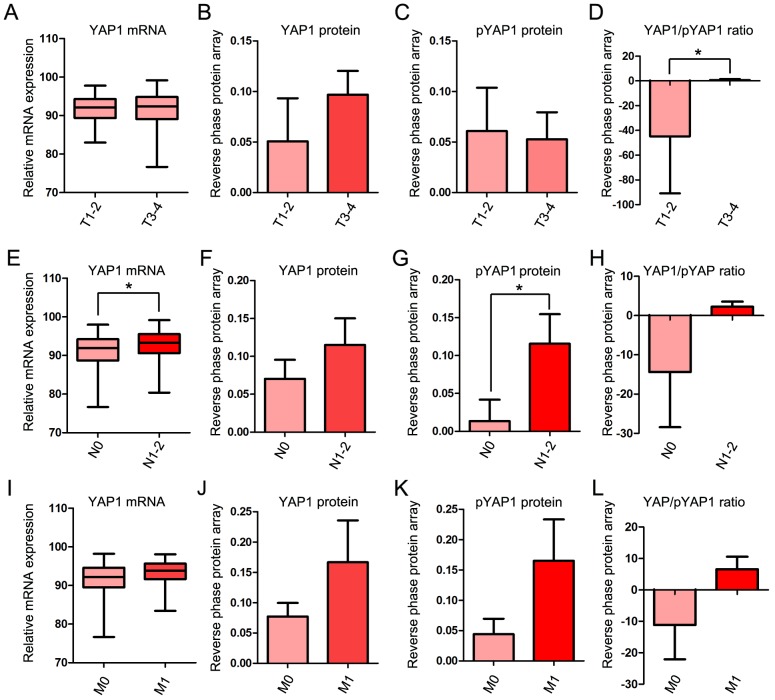
To investigate the association between YAP1 expression and progression of COAD in more detail, the mRNA and protein expression of YAP1 was examined in each of the T, N and M stages. In the T stage, YAP1/pYAP was significantly increased in T3-4 compared with T1-2 stages (Fig. 2A-D). In the M stage, YAP1 mRNA and protein expression were significantly increased in N1-2 compared with N0 (Fig. 2E-H). Although mRNA expression and protein expression of YAP1 were higher in M1 than in M0, the differences were not significant (Fig. 2I-L).
Figure 2.
YAP1 mRNA and protein expression in TNM stages. (A) Relative mRNA expression of YAP1, (B) protein level of YAP1, (C) protein level of pYAP and (D) YAP1/pYAP ratio between T1-2 and T3-4. (E) Relative mRNA expression of YAP1, (F) protein level of YAP1, (G) protein level of pYAP and (H) YAP1/pYAP ratio between N0 and N1-2. (I) Relative mRNA expression of YAP1, (J) protein level of YAP1, (K) protein level of pYAP and (L) YAP1/pYAP ratio between M0 and M1. mRNA and protein expression data of YAP1 in COAD obtained from The Cancer Genome Atlas data portal. *P<0.05. YAP1, yes-associated protein 1; pYAP, phosphorylated form of YAP1; TNM, Tumor-Node-Metastasis; COAD, colon adenocarcinoma.
GeneNeighbors analysis of YAP1
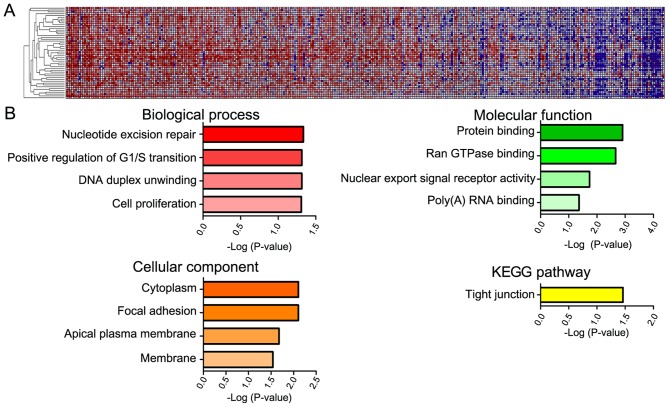
The 100 genes most closely associated with YAP1 were selected using the GeneNeighbors program (Fig. 3) and were classified using DAVID. The genes were sorted into three groups: GO terms that differed significantly served functions in: i) Biological processes, ii) cellular components, and iii) molecular functions. Genes that were highly expressed in COAD and associated with biological processes were mainly associated with DNA duplication (positive regulation of G1/S transition, nucleotide excision repair and DNA duplex unwinding) (Fig. 3B). As for cellular components, highly expressed genes in COAD were primarily associated with the cytoplasm and membrane (focal adhesion, apical plasma membrane and membrane). Regarding molecular functions, genes that were expressed at a high level in COAD were primarily associated with protein binding (Ran GTPase-binding and poly (A) RNA binding) and nuclear export. In addition, when genes were analyzed according to cell signaling pathway (KEGG), the tight junction pathway was the most significant.
Figure 3.
GeneNeighbors of YAP1 in 458 COAD samples. (A) Hierarchical clustering of YAP1 GeneNeighbors in colon adenocarcinoma. COAD samples are arranged in decreasing order of YAP1 mRNA expression. Colors in the heat map represent expression relative to the mean expression value, with red indicating higher expression and blue indicating lower expression. GeneNeighbors of YAP1 are presented in the column. (B) GeneNeighbors were characterized as biological processes, cellular components, molecular function and KEGG pathway-associated. COAD, colon adenocarcinoma; YAP1, yes-associated protein 1; pYAP, phosphorylated form of YAP1; KEGG, Kyoto Encyclopedia of Genes and Genomes.
ClassNeighbors analysis of YAP1
Analysis using ClassNeighbors divided the COAD samples in to two classes: Class A contained the most marked 10% of YAP1-upregulated COAD samples and class B contained the most marked 10% of YAP1-downregulated COAD samples (Fig. 4A). Of the 20,502 probe sets, the 150 genes that were most significantly associated and most highly expressed in classes A and B were selected (Table III). DAVID analysis classified these genes into groups based on the GO terms: i) Biological processes, ii) cellular components and iii) molecular functions, as well as iv) KEGG pathways (Fig. 4). Genes highly expressed in class A were mainly associated with development (multicellular organism development, somatic stem cell population maintenance and anatomical structure morphogenesis) and pathways (Wnt signaling pathway, pathway restricted SMAD protein phosphorylation) in biological processes; the plasma membrane (apical plasma membrane, integral component of plasma membrane and cytoplasmic side of plasma membrane) in cellular components; activity (growth factor activity and transcriptional activity) and binding [sequence-specific DNA binding and transforming growth factor-β (TGF-β) receptor binding] in molecular function; and signaling pathways (Wnt signaling, TGF-β signaling) in KEGG pathways. Genes highly expressed in class B were mostly associated with ions (cellular response to zinc ion and cadmium ion, and negative regulation of growth) in biological process; integral component of plasma membrane and pronuclear region of cytoplasm in cellular components; activity (phospholipase A2 activity and prostaglandin E receptor activity) in molecular function, mineral absorption in KEGG pathways.
Figure 4.
ClassNeighbors of YAP1 in 458 colon adenocarcinoma samples. Hierarchical clustering of differentially expressed genes (top 10%) upregulated and downregulated in colon adenocarcinoma cases according to Pearson distance. (A) Colors in the heat map represent expression relative to the mean expression value, with red indicating higher expression and blue indicating lower expression. (B) Genes in class A were divided into biological processes, cellular components, molecular function and KEGG pathway-associated. (C) Genes in class B were divided into biological processes, cellular components, molecular function and KEGG pathway-associated. YAP1, yes-associated protein 1; pYAP, phosphorylated form of YAP1; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table III.
DAVID analysis of ClassNeighbors.
| A, Class A | |||
|---|---|---|---|
| Term | Count | % | P-value |
| Biological process | |||
| GO: 0030178-Negative regulation of Wnt signaling pathway | 6 | 4.5 | <0.01 |
| GO: 0001942-Hair follicle development | 4 | 3.0 | <0.01 |
| GO: 0001580-Detection of chemical stimulus involved in sensory perception of bitter taste | 4 | 3.0 | <0.01 |
| GO: 0007275-Multicellular organism development | 11 | 8.2 | <0.01 |
| GO: 0090090-Negative regulation of canonical Wnt signaling pathway | 6 | 4.5 | <0.01 |
| GO: 0016055-Wnt signaling pathway | 6 | 4.5 | 0.01 |
| GO: 0035019-Somatic stem cell population maintenance | 4 | 3.0 | 0.01 |
| GO: 0060279-Positive regulation of ovulation | 2 | 1.5 | 0.01 |
| GO: 0046882-Negative regulation of follicle-stimulating hormone secretion | 2 | 1.5 | 0.02 |
| GO: 0007411-Axon guidance | 5 | 3.7 | 0.02 |
| GO: 0009653-Anatomical structure morphogenesis | 4 | 3.0 | 0.02 |
| GO: 0070858-Negative regulation of bile acid biosynthetic process | 2 | 1.5 | 0.03 |
| GO: 0030154-Cell differentiation | 8 | 6.0 | 0.04 |
| GO: 0001755-Neural crest cell migration | 3 | 2.2 | 0.04 |
| GO: 0042423-Catecholamine biosynthetic process | 2 | 1.5 | 0.04 |
| GO: 0046881-Positive regulation of follicle-stimulating hormone secretion | 2 | 1.5 | 0.04 |
| GO: 0010862-Positive regulation of pathway-restricted SMAD protein phosphorylation | 3 | 2.2 | 0.04 |
| GO: 0009072-Aromatic amino acid family metabolic process | 2 | 1.5 | 0.05 |
| GO: 0021516-Dorsal spinal cord development | 2 | 1.5 | 0.05 |
| GO: 0000122-Negative regulation of transcription from RNA polymerase II promoter | 10 | 7.5 | 0.05 |
| GO: 0010470-Regulation of gastrulation | 2 | 1.5 | 0.05 |
| Cellular component | |||
| GO: 0005576-Extracellular region | 24 | 17.9 | <0.01 |
| GO: 0005615-Extracellular space | 21 | 15.7 | <0.01 |
| GO: 0016324-Apical plasma membrane | 7 | 5.2 | 0.01 |
| GO: 0071944-Cell periphery | 3 | 2.2 | 0.03 |
| GO: 0005887-Integral component of plasma membrane | 17 | 12.7 | 0.03 |
| GO: 0009898-Cytoplasmic side of plasma membrane | 3 | 2.2 | 0.03 |
| Molecular function | |||
| GO: 0008083-Growth factor activity | 7 | 5.2 | <0.01 |
| GO: 0043565-Sequence-specific DNA binding | 10 | 7.5 | 0.01 |
| GO: 0003700-Transcription factor activity, sequence-specific DNA binding | 14 | 10.4 | 0.01 |
| GO: 0016714-Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen | 2 | 1.5 | 0.03 |
| GO: 0000981-RNA polymerase II transcription factor activity, sequence-specific DNA binding | 5 | 3.7 | 0.03 |
| GO: 0005160-Transforming growth factor beta receptor binding | 3 | 2.2 | 0.03 |
| GO: 0000978-RNA polymerase II core promoter proximal region sequence-specific DNA binding | 7 | 5.2 | 0.03 |
| KEGG | |||
| hsa04310: Wnt signaling pathway | 5 | 3.7 | 0.02 |
| hsa04350: TGF-β signaling pathway | 4 | 3.0 | 0.02 |
| hsa04970: Salivary secretion | 4 | 3.0 | 0.02 |
| hsa04530: Tight junction | 4 | 3.0 | 0.07 |
| hsa04550: Signaling pathways regulating pluripotency of stem cells | 4 | 3.0 | 0.07 |
| hsa04151: PI3K-Akt signaling pathway | 6 | 4.5 | 0.09 |
| B, Class B | |||
| Term | Count | % | P-value |
| Biological process | |||
| GO: 0071294-Cellular response to zinc ion | 7 | 5.0 | <0.01 |
| GO: 0045926-Negative regulation of growth | 7 | 5.0 | <0.01 |
| GO: 0071276-Cellular response to cadmium ion | 5 | 3.6 | <0.01 |
| GO: 0051453-Regulation of intracellular pH | 4 | 2.9 | <0.01 |
| GO: 0015701-Bicarbonate transport | 4 | 2.9 | <0.01 |
| GO: 0007586-Digestion | 4 | 2.9 | 0.01 |
| GO: 2001225-Regulation of chloride transport | 2 | 1.4 | 0.01 |
| GO: 0036018-Cellular response to erythropoietin | 2 | 1.4 | 0.01 |
| GO: 0032849-Positive regulation of cellular pH reduction | 2 | 1.4 | 0.03 |
| GO: 1902476-Chloride transmembrane transport | 4 | 2.9 | 0.03 |
| GO: 0006029-Proteoglycan metabolic process | 2 | 1.4 | 0.05 |
| GO: 0007189-Adenylate cyclase-activating G protein-coupled receptor signaling pathway | 3 | 2.1 | 0.05 |
| Cellular component | |||
| GO: 0005887-Integral component of plasma membrane | 19 | 13.6 | 0.01 |
| GO: 0048471-Perinuclear region of cytoplasm | 11 | 7.9 | 0.01 |
| GO: 0042589-Zymogen granule membrane | 2 | 1.4 | 0.08 |
| GO: 0005886-Plasma membrane | 38 | 27.1 | 0.08 |
| Molecular function | |||
| GO: 0008499-UDP-galactose:β-N-acetylglucosamine β-1,3-galactosyltransferase activity | 3 | 2.1 | <0.01 |
| GO: 0004623-Phospholipase A2 activity | 3 | 2.1 | 0.02 |
| GO: 0004957-Prostaglandin E receptor activity | 2 | 1.4 | 0.04 |
| GO: 0008270-Zinc ion binding | 15 | 10.7 | 0.04 |
| GO: 0005254-Chloride channel activity | 3 | 2.1 | 0.06 |
| GO: 0046983-Protein dimerization activity | 4 | 2.9 | 0.09 |
| GO: 0004089-Carbonate dehydratase activity | 2 | 1.4 | 0.10 |
| KEGG | |||
| hsa04978: Mineral absorption | 7 | 5.0 | <0.01 |
| hsa04972: Pancreatic secretion | 6 | 4.3 | <0.01 |
| hsa04975: Fat digestion and absorption | 4 | 2.9 | <0.01 |
| hsa04924: Renin secretion | 3 | 2.1 | 0.09 |
| hsa00830: Retinol metabolism | 3 | 2.1 | 0.10 |
DAVID, database for annotation, visualization and integrated discovery; KEGG, Kyoto Encyclopedia of Genes and Genomes; PI3K, phosphoinositide 3-kinase; Akt, RAC serine/threonine protein kinase; SMAD, mothers against decapentaplegic homolog.
GSEA analysis
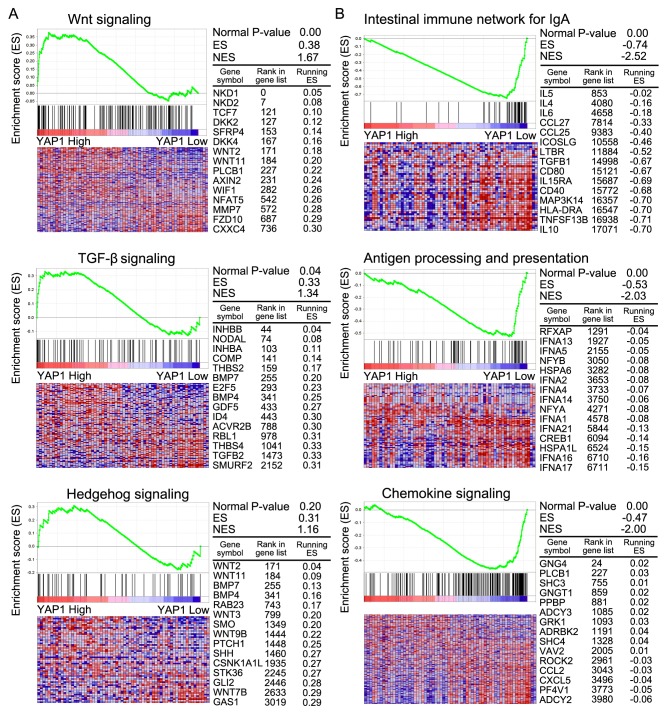
GSEA was conducted to compare more specifically the significantly enriched pathways between classes A and B (Table IV). In class A, pathways involving Wnt signaling, TGF-β signaling and Hedgehog signaling were significantly enriched compared with class B. In class B, pathways were mainly involved in the intestinal immune network for immunoglobulin A, antigen processing and presentation and chemokine signaling. In class A, Wnt and TGF-β signaling were associated with cancer progression (Fig. 5A). Immune-associated signaling pathways were associated with class B (Fig. 5B).
Table IV.
Gene set enrichment analysis of class A and class B.
| A, Class A | ||||
|---|---|---|---|---|
| KEGG pathway | Size | ES | NES | NOM P-value |
| RNA polymerase | 29 | 0.54 | 1.76 | <0.01 |
| Melanoma | 71 | 0.42 | 1.69 | <0.01 |
| Wnt signaling pathway | 150 | 0.38 | 1.67 | <0.01 |
| Basal cell carcinoma | 55 | 0.41 | 1.53 | 0.01 |
| Basal transcription factors | 35 | 0.40 | 1.37 | 0.10 |
| TGF-β signaling pathway | 85 | 0.33 | 1.34 | 0.04 |
| Homologous recombination | 26 | 0.40 | 1.30 | 0.11 |
| ECM receptor interaction | 83 | 0.30 | 1.22 | 0.11 |
| Hedgehog signaling pathway | 56 | 0.31 | 1.16 | 0.20 |
| Adherens junction | 73 | 0.30 | 1.15 | 0.20 |
| Spliceosome | 114 | 0.26 | 1.10 | 0.23 |
| B, Class B | ||||
| Name | Size | ES | NES | NOM P-value |
| Intestinal immune network for IgA production | 46 | −0.74 | −2.52 | <0.01 |
| Hematopoietic cell lineage | 84 | −0.66 | −2.50 | <0.01 |
| Allograft rejection | 35 | −0.71 | −2.30 | <0.01 |
| Primary immunodeficiency | 35 | −0.70 | −2.27 | <0.01 |
| Antigen processing and presentation | 81 | −0.53 | −2.03 | <0.01 |
| Chemokine signaling pathway | 188 | −0.47 | −2.00 | <0.01 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; ES, enrichment score; NES, normalized enrichment score; NOM, nominal; TGF-β, transforming growth factor-β; IgA, immunoglobulin A.
Figure 5.
Gene Set Enrichment Analysis. (A) Gene Set Enrichment Analysis of Class A and B. WNT and TGF-β signaling were significantly enriched in Class A. (B) Intestinal immune network for IgA and Antigen processing and presentation pathway were significantly enriched in Class B. TGF-β, transforming growth factor-β; IgA, immunoglobulin A.
Survival analysis
To determine the prognostic significance of Hippo pathway genes in patients with COAD, the association between mRNA expression of Hippo pathway genes and overall survival was evaluated using Kaplan-Meier curves (Fig. 6). High mRNA expression of YAP1, TAZ, TEAD4 and LATS2 was significantly associated with poor prognosis in COAD.
Figure 6.
Kaplan-Meier analysis of the association between Hippo pathway genes and overall survival. In order to investigate the Hippo pathway, survival was studied against levels of (A) YAP1 mRNA, (B) YAP1 protein and (C) pYAP1 protein. The mRNA levels of (D) TAZ, (E) TEAD1, (F) TEAD2, (G) TEAD3, (H) TEAD4, (I) MST1, (J) MST2, (K) LATS1 and (L) LATS2 were also assessed. Cutoff Finder was used to determine cut-off values in mRNA and protein expression of colon adenocarcinoma using log-rank tests. mRNA of YAP1, TAZ, TEAD4 and LATS2 exhibited the poor prognosis. YAP1, yes-associated protein 1; pYAP, phosphorylated form of YAP1; TAZ, taffazin; TEAD, transcriptional enhancer associated domain; MST, macrophage stimulating; LATS, large tumor suppressor kinase.
Discussion
In the present study, the expression of YAP1 mRNA in cases of COAD was identified to be higher than that in other types of gastrointestinal tract cancer. YAP1 mRNA expression was significantly increased in the sigmoid colon compared with the ascending colon and hepatic flexure, and in advanced TNM stages. YAP1 protein was highly expressed in advanced TNM stages, and pYAP1 levels were high; however, YAP1/pYAP1 was also higher in the advanced TNM stages. When YAP1 expression was compared separately for each of the TNM stages, YAP1/pYAP1 was only significantly elevated in advanced T stages, but was markedly higher in the advanced N and M stages. YAP1 mRNA and pYAP1 levels were significantly elevated in the advanced N stage. YAP1 was mainly associated with cell proliferation and development. WNT and TGF-β signaling were significantly enriched in the high YAP1-expression group, as assessed by GSEA. Finally, YAP1, TAZ, TEAD4 and LATS2 mRNA expression were associated with poor overall survival rates in patients with COAD.
Evidence indicates that the right and left sides of the colon exhibit significantly different histological and molecular characters (31–34). At the molecular level, genes are significantly differentially expressed between right- and left-sided colon cancer (33). In the present study, expression of YAP1 mRNA in COAD was significantly higher in the sigmoid colon than in the ascending colon and hepatic flexure. Although levels of pYAP1 were increased in advanced TNM stages, the YAP1/pYAP1 ratio, which represents the nuclear activity of YAP1, was consistently higher in advanced TNM stages, particularly in the advanced T stage.
Increases in the expression of YAP1 and other Hippo pathway genes have been investigated in multiple types of cancer, including cancer of the liver, colon, lung and prostate (12–14,35,36). In liver cancer, YAP1 was identified as an independent prognostic marker for overall and disease-free survival (36). In ovarian cancer, YAP1 has a marked association with poor prognosis (16). In colon cancer, several studies have reported that Hippo pathway genes were overexpressed and associated with poor prognosis (20–23). Wang et al (21) reported that co-overexpression of YAP1 and TAZ is an independent predictor of prognosis for patients. Liang et al (22) demonstrated that YAP1 and TEAD1 were upregulated and MST1 and LATS2 were down regulated in colorectal cancer. Yuen et al (23) reported that TAZ exhibited greater prognostic value than YAP1 in colorectal cancer. In accordance with previous findings, the present study demonstrated that YAP1 and TAZ were highly expressed and associated with poor overall survival in COAD (Fig. 6). In addition, the present study identified that TEAD4 was significantly associated with poor prognosis (P=0.038; Fig. 6H). Among upstream components, MST1/2 was highly expressed, whereas LATS1/2 was expressed at a low level. Although LATS2 exhibited low expression in the present study, LATS2 was associated with poor overall survival. In the hippo pathway, YAP1, TAZ, TEAD4 and LATS2 genes may be able to serve as molecular markers in COAD.
To verify the function and mechanism of YAP1 in COAD, bioinformatics analysis was conducted. GeneNeighbors analysis revealed that cell proliferation and protein binding-associated genes were associated with YAP1 in 458 samples from patients with COAD (Fig. 3). Additionally, ClassNeighbors analysis classified YAP1-expressing COAD samples into class A, which expresses genes associated with development, stem cell maintenance and growth factor activity, and class B, which expresses genes associated with the negative regulation of growth, cellular response to ions and mineral absorption. Class A genes enhance development and cell growth-associated functions, whereas class B genes enhance the suppression of cell growth and mineral interaction-associated functions. GSEA was performed to compare pathways that were enriched between classes A and B. In class A, pathways involved in tight junction, Wnt signaling, TGF-β signaling and adherens junctions pathways exhibited greater activity than those in class B. In class B, pathways involved in primary immunodeficiency, intestinal immune network for immunoglobulin A production and regulation of autophagy were enriched. The Hippo pathway is able to interact with other oncogenic signaling pathways, including Wnt, TGF-β, Sonic hedgehog, Notch and epidermal growth factor receptor/KRAS proto-oncogene, GTPase pathways, to modify more downstream components (37,38). In the present study, YAP1 expression was associated with Wnt signaling and TGF-β signaling, which are associated with cancer progression (1,39). Additionally, GSEA also identified that YAP1 was associated with RNA polymerase, basal transcription factors, ECM receptor interaction and adherens junction in COAD.
In conclusion, the expression and clinical significance of Hippo pathway genes in COAD was investigated using a cohort of 458 patients obtained from TGCA. YAP1 mRNA was highly expressed in sigmoid colon cancer. YAP1 activity was consistently higher in advanced TNM stages, particularly in the advanced T stage. YAP1 was associated with proliferation and development, and was significantly associated with Wnt and TGF-β signaling, as indicated by bioinformatics analysis. High mRNA expression of YAP1 and its associated genes, TAZ, TEAD4 and LATS2, was significantly associated with poor patient prognosis in COAD. However, further study is required to confirm the prognostic value of TAZ, TEAD4 and LATS2, and the underlying molecular mechanisms of their functions in COAD.
References
- 1.Ikushima H, Miyazono K. TGFbeta signalling: A complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 2.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson R, Halder G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Santucci M, Vignudelli T, Ferrari S, Mor M, Scalvini L, Bolognesi ML, Uliassi E, Costi MP. The Hippo pathway and YAP/TAZ-TEAD protein-protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58:4857–4873. doi: 10.1021/jm501615v. [DOI] [PubMed] [Google Scholar]
- 7.Justice N, Roegiers F, Jan LY, Jan YN. Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr Biol. 2003;13:778–783. doi: 10.1016/S0960-9822(03)00288-4. [DOI] [PubMed] [Google Scholar]
- 8.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 9.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/S0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 10.Cui ZL, Han FF, Peng XH, Chen X, Luan CY, Han RC, Xu WG, Guo XJ. YES-associated protein 1 promotes adenocarcinoma growth and metastasis through activation of the receptor tyrosine kinase Axl. Int J Immunopathol Pharmacol. 2012;25:989–1001. doi: 10.1177/039463201202500416. [DOI] [PubMed] [Google Scholar]
- 11.Pei T, Li Y, Wang J, Wang H, Liang Y, Shi H, Sun B, Yin D, Sun J, Song R, et al. YAP is a critical oncogene in human cholangiocarcinoma. Oncotarget. 2015;6:17206–17220. doi: 10.18632/oncotarget.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, AOCS Study group. Bowtell DD, Harvey KF. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 16.Hall CA, Wang R, Miao J, Oliva E, Shen X, Wheeler T, Hilsenbeck SG, Orsulic S, Goode S. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70:8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nallet-Staub F, Marsaud V, Li L, Gilbert C, Dodier S, Betaille V, Sudol M, Herlyn M, Mauviel A. Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol. 2014;134:123–132. doi: 10.1038/jid.2013.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, ten Hacken E, Bertilaccio MT, Antonini E, Neri A, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20:599–606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Xie C, Li Q, Xu K, Wang E. Clinical and prognostic significance of Yes-associated protein in colorectal cancer. Tumour Biol. 2013;34:2169–2174. doi: 10.1007/s13277-013-0751-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, Wen W, Zhu A. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8:e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liang K, Zhou G, Zhang Q, Li J, Zhang C. Expression of hippo pathway in colorectal cancer. Saudi J Gastroenterol. 2014;20:188–194. doi: 10.4103/1319-3767.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuen HF, McCrudden CM, Huang YH, Tham JM, Zhang X, Zeng Q, Zhang SD, Hong W. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One. 2013;8:e54211. doi: 10.1371/journal.pone.0054211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team, corp-author. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 25.Golub TR, Slonim DK, Tamayo P, Huard C, Baasenbeek M, Mersirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hari DM, Leung AM, Lee JH, Sim MS, Vuong B, Chiu CG, Bilchik AJ. AJCC cancer staging manual 7th edition criteria for colon cancer: Do the complex modifications improve prognostic assessment? J Am Coll Surg. 2013;217:181–190. doi: 10.1016/j.jamcollsurg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benedix F, Kube R, Meyer F, Schmidt U, Gastinger I, Lippert H, Colon/Rectum Carcinomas (Primary Tumor) Study Group Comparison of 17,641 patients with right- and left-sided colon cancer: Differences in epidemiology, perioperative course, histology and survival. Dis Colon Rectum. 2010;53:57–64. doi: 10.1007/DCR.0b013e3181c703a4. [DOI] [PubMed] [Google Scholar]
- 32.Papagiorgis P, Oikonomakis I, Karapanagiotou I, Wexner SD, Nikiteas N. The impact of tumor location on the histopathologic expression of colorectal cancer. J Buon. 2006;11:317–321. [PubMed] [Google Scholar]
- 33.Glebov OK, Rodriguez LM, Nakahara K, Jenkins J, Cliatt J, Humbyrd CJ, DeNobile J, Soballe P, Simon R, Wright G, et al. Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiol Biomarkers Prev. 2003;12:755–762. [PubMed] [Google Scholar]
- 34.Azzoni C, Bottarelli L, Campanini N, Di Cola G, Bader G, Mazzeo A, Salvemini C, Morari S, Di Mauro D, Donadei E, et al. Distinct molecular patterns based on proximal and distal sporadic colorectal cancer: Arguments for different mechanisms in the tumorigenesis. Int J Colorectal Dis. 2007;22:115–126. doi: 10.1007/s00384-006-0093-x. [DOI] [PubMed] [Google Scholar]
- 35.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Znder L, Lowe SW, Poon RT, Luk JM. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irvine KD. Integration of intercellular signaling through the Hippo pathway. Semin Cell Dev Biol. 2012;23:812–817. doi: 10.1016/j.semcdb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B, Li L, Guan KL. Hippo signaling at a glance. J Cell Sci. 2010;123:4001–4006. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]