Abstract
MicroRNA-32 (miR-32) is associated with tumor progression and poor prognosis in certain malignant tumors. However, the function and clinical relevance of miR-32 in human hepatocellular carcinoma (HCC) has not yet been elucidated. The present study aimed to investigate the expression and prognostic value of miR-32 from liver samples in patients with HCC. The expression of miR-32 was analyzed in HCC and healthy tissues using Gene Expression Omnibus datasets. Reverse transcription-quantitative polymerase chain reaction was used to analyze the levels of miR-32 mRNA in 154 HCC liver samples, 33 of which were paired with adjacent non-tumor tissues. The overall survival (OS) rate in patients with HCC was evaluated using Kaplan-Meier survival analysis, and the factors that may affect the prognosis and survival of patients with HCC were analyzed using univariate (log-rank test) and multivariate Cox proportional hazard models. The present results demonstrated that miR-32 expression levels were significantly upregulated in HCC liver biopsies compared with normal tissues (P<0.05). miR-32 expression was significantly associated with the number of foci and tumor diameter (P<0.05). In addition, Kaplan-Meier analysis revealed that patients with low miR-32 expression had longer OS and disease-free survival compared with those with high miR-32 expression (P<0.01). Altogether, to the best our knowledge, the present study is the first study to indicate the association between increased miR-32 expression with HCC progression and poor prognosis in patients. This suggests that miR-32 may have potential prognostic value and may be used as a tumor biomarker for the diagnosis of patients with HCC.
Keywords: microRNA-32 expression, hepatocellular carcinoma, overall survival, Gene Expression Omnibus, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver malignancy (1). Currently, HCC ranks as the third leading cause of cancer-associated mortality worldwide and the sixth leading cause of human cancer (2–4). During the past few decades, a number of HCC treatments have been developed. However, the therapeutic effects are not optimal due to tumor recurrence, metastasis and, in particular, a low survival rate (5). Therefore, novel treatment options based on an improved understanding of the molecular mechanisms of HCC need to be investigated.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules (20–24 nucleotides), processed as miRNA precursors with a hairpin structure (6–8). The specific involvement of miRNAs in different tumor types and associations with prognosis are of interest. miRNAs are highly conserved and tissue-specific. Previous studies have shown that miRNA expression varies in different types of cancer, indicating that miRNAs may be involved not only in tissue function and cell specificity, but also in complex gene regulation (9–11).
In recent years, a plethora of studies have suggested that dysregulation of miRNA-32 (miR-32) is associated with tumor progression and poor prognosis in different types of tumors. miR-32 has been shown to be upregulated in colorectal cancer (CRC) (12), renal cell carcinoma (13) and prostate cancer (14), and downregulated in non-small cell lung cancer (NSCLC) (15), osteosarcoma (16), gastric cancer (17) and oral squamous cell carcinoma (18). Yan et al (19) reported that miR-32 levels were upregulated in HCC tissue, and cell proliferation, invasion and migration were upregulated by targeting phosphatase and tensin homolog (PTEN). These results indicate that miR-32 may be a potential biomarker for HCC diagnosis. However, the clinical relevance and function of miR-32 in HCC remains elusive.
The present study aimed to investigate the expression levels of miR-32 in HCC tissues by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). In addition, the association of miR-32 and patient clinicopathological characteristics was analyzed.
Materials and methods
Ethics statement
The present study was approved by the Ethics Committee of Jilin University (approval no. 20151101; Jilin, China). Each patient provided written informed consent prior to participation in the present study.
miRNA expression in HCC from Gene Expression Omnibus (GEO) datasets
miRNA expression levels were investigated in HCC tissues and normal tissue samples in the GEO datasets using the NCBI Platform (http://www.ncbi.nlm.nih.gov/). A total of three original datasets were downloaded (GEO accession no. GSE31383, GSE21362 and GSE22058), and differentially expressed miRNAs in HCC samples and adjacent non-tumor tissue were analyzed. Fold-change (FC) ≥2 or ≤0.5 and P<0.01 were used as basic screening parameters for cluster analysis. Hierarchical clustering was performed using the Multiple Experiment Viewer (MeV) 4.7.1 software program (http://www.tm4.org/).
Clinical specimens
A total of 154 HCC primary tumor tissues obtained from patients with HCC (123 male, 30 female and 1 unknown) with a median age of 65.4 years (range, 45–81 years), who underwent surgical resection between July 2004 and October 2013 at China-Japan Union Hospital, Jilin University (Changchun, China), were analyzed. In total, 33 HCC samples were paired with adjacent non-tumor tissues. No patient received chemotherapy or radiotherapy prior to surgery. Follow-up data and statistics were recorded for all patients until September 2014.
RNA extraction and RT-qPCR for miRNA
Total RNA was isolated from HCC and normal tissue specimens using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. RNA concentration was determined using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.), and purity was identified in 1.5% denaturing agarose gels. TaqMan probe-based qPCR was carried out using TaqMan MicroRNA Reverse Transcription kit (no. 4366597) and Universal Master Mix II (no. 4440048) (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the protocol of the manufacturer. The specific primers are as follows: for hsa-miR-32, forward, 5′-GCACATTCATCATACACGCCG-3′ and reverse, 5′-GACCACTGAGGTTAGAGCCA-3′; for U6, forward, 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse, 5′-ACGCTTCACGAATTTGCGTGTC-3′. Thermocycling conditions for RT reaction were as follows: Initial reaction at 16°C for 30 min, followed by 42°C for 30 min, 85°C for 5 min, with a final reaction at 4°C. Thermocycling conditions for PCR were as follows: Initial denaturation at 94°C for 10 min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, with a final extension at 72°C for 10 min. Each reaction was independently tested in duplicate a minimum of three times. U6 small nuclear RNA was used as an internal control, and the 2−ΔΔCq method (20) was used to analyze miR-32 expression levels.
Statistical analysis
All statistical analyses were performed using the IBM SPSS statistical software (version 22.0; IBM SPSS, Armonk, NY, USA). χ2 test and t-test were used to examine the associations between clinical characteristics and miR-32 expression. Overall survival (OS) curves were constructed using Kaplan-Meier survival analysis, and results were compared using the log-rank test. In order to estimate independent prognostic factors associated with survival, univariate and multivariate survival analyses were performed using the Cox regression model. P<0.05 was considered to indicate a statistically significant difference.
Results
Cluster analysis of miRNA expression in HCC using MEV4.7.1 software
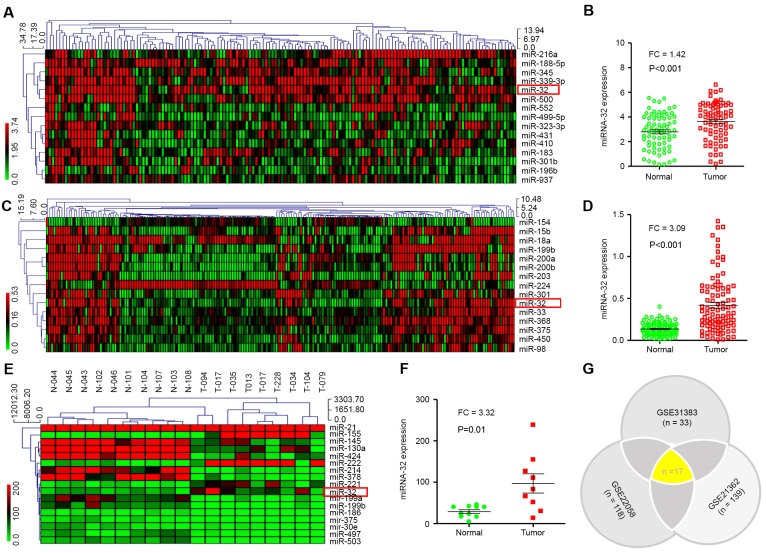
miRNA expression was investigated in HCC and normal tissues (n=73). Raw data were retrieved using the search terms ‘GSE#21362’ in the GEO dataset. A total of 15 different miRNAs with P<0.01 and FC ≥1.4 were identified. The MEV4.7.1 clustering software was used to analyze 15 different miRNAs (Fig. 1A). The analysis indicated that miR-32 expression in HCC tissues was significantly higher compared with non-tumor tissues (P<0.001; Fig. 1B).
Figure 1.
Analysis of miR-32 expression in patients with HCC was performed using GEO datasets, and MEV4.7.1 clustering software was used to analyze different miRNAs. (A) Clustering analysis was performed using the MEV4.7.1 based on 15 dysregulated miRNAs using FC ≥1.4 and P<0.01. (B) miR-32 expression levels in HCC tissues vs. normal tissues were analyzed using the GEO database (GEO accession no. GSE21362). (C) Clustering analysis was performed with the MEV4.7.1 software. A total of 15 dysregulated miRNAs were screened using FC ≥3 or ≤0.3 and P<0.01. (D) The expression levels of miR-32 in HCC (n=96) and normal tissues (n=96) derived from the GEO database (GEO accession no. GSE22058) were analyzed. (E) Clustering analysis was performed using MEV4.7.1 based on 17 dysregulated miRNAs and screened using FC ≥3 or ≤0.3 and P<0.01. (F) miR-32 expression in HCC tissues (n=9) was significantly higher compared with non-tumor tissues (n=10). Datasets derived from the GEO database (GEO accession no. GSE22058). (G) A Venn diagram was generated using three GEO datasets. A total of 17 common miRNAs were identified, including miR-32. HCC, hepatocellular carcinoma; miR, microRNA; FC, fold-change; GEO, Gene Expression Omnibus; miRNA, microRNA.
Next, the raw data were downloaded from the GEO database (GEO accession no. GSE22058). In the present study, the genome-wide expression profiles of miRNAs from paired tumor and adjacent non-tumor tissues from a cohort of 96 patients with HCC in Hong Kong were evaluated. A total of 15 dysregulated miRNAs were screened out using FC ≥3 or ≤0.3 and P<0.01 (Fig. 1C). It was revealed that miR-32 expression was also significantly higher in HCC compared with non-tumor tissues (P<0.001; Fig. 1D).
The same method was then used to analyze the ‘GSE31383’ GEO dataset (Fig. 1E). Approximately 57 miRNAs were tested in HCC samples (n=9) and normal tissue (n=10). A total of 17 dysregulated miRNAs were screened with P<0.01. miR-32 expression in HCC tissue was significantly higher (P=0.01) compared with non-tumor tissue (Fig. 1F).
Altogether, differentially expressed miRNAs were screened using three different raw datasets (GEO accession no. GSE31383, GSE22058 and GSE21362). Notably, 17 miRNAs were identified, including miR-32, which were common in all datasets (P<0.01; Fig. 1G). These results indicated that miR-32 is increased in HCC compared with non-tumor tissues, and therefore miR-32 may be a potential biomarker for diagnosis of HCC.
Increased expression of miR-32 in HCC
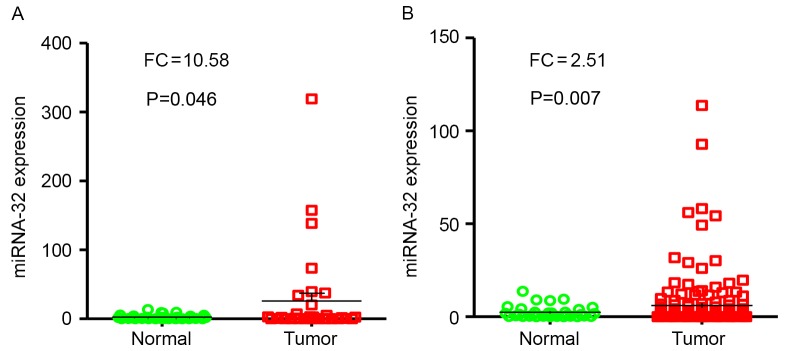
miR-32 expression was evaluated in tumor samples (n=33) compared with adjacent non-tumor tissues (n=33) by RT-qPCR. Results of the present study revealed that miR-32 expression levels were significantly higher in HCC tumor specimens compared with adjacent non-neoplastic tissues (P<0.05; FC, 10.58; Fig. 2A).
Figure 2.
miR-32 expression in normal liver and HCC tissues. (A) miR-32 expression levels in HCC vs. paired adjacent non-tumor tissues (n=33). (B) miR-32 expression levels in 154 HCC samples and 33 non-tumor tissues. miR, microRNA; HCC, hepatocellular carcinoma.
miR-32 expression levels were examined in tumor samples (n=154), and levels of expression were compared with paired adjacent non-tumor tissues (n=33) from patients with HCC using RT-qPCR. Notably, results demonstrated that the levels of miR-32 expression were significantly higher in HCC tumor specimens compared with normal tissues (P=0.007; FC, 2.51; Fig. 2B).
Association between miR-32 expression and clinicopathological characteristics of patients with HCC
The association between miR-32 expression levels and the individual clinicopathological characteristics were evaluated in patients with HCC. As shown in Table I, miR-32 expression was positively associated with the number and diameter of foci (P<0.05). However, no significant association was observed between miR-32 expression and other clinical characteristics, including age, sex and tumor differentiation (P>0.05).
Table I.
miR-32 expression and clinicopathological characteristics of patients with hepatocellular carcinoma.
| miR-32 expression | Overall survival, months | ||||||
|---|---|---|---|---|---|---|---|
| Factor | n | Low | High | P-value | Mean | 95% CI | P-value |
| Age, years | |||||||
| ≥60 | 84 | 42 | 42 | 0.996 | 45.98 | 39.11–52.84 | 0.472 |
| <60 | 70 | 35 | 35 | 37.97 | 32.42–43.52 | ||
| Sex | |||||||
| Male | 123 | 61 | 62 | 0.957 | 41.33 | 37.14–47.54 | 0.101 |
| Female | 30 | 15 | 15 | 42.46 | 31.48–51.44 | ||
| Unknown | 1 | 1 | 0 | ||||
| Tumor differentiation | |||||||
| Poor | 11 | 4 | 7 | 0.541 | 20.91 | 5.77–36.04 | 0.015 |
| Good | 125 | 65 | 60 | 44.42 | 39.30–49.55 | ||
| Unknown | 18 | 8 | 10 | ||||
| Tumor diameter, cm | |||||||
| ≥5 | 58 | 21 | 37 | 0.012 | 36.51 | 30.79–42.22 | 0.009 |
| <5 | 96 | 56 | 40 | 46.01 | 39.63–52.39 | ||
| Number of foci | |||||||
| Multiple | 81 | 32 | 49 | 0.022 | 34.41 | 27.86–40.96 | 0.014 |
| Single | 67 | 39 | 28 | 51.96 | 46.57–57.34 | ||
| Unknown | 6 | 6 | 0 | – | – | ||
CI, confidence interval; miR-32, microRNA-32.
Association between clinical characteristics and HCC prognosis
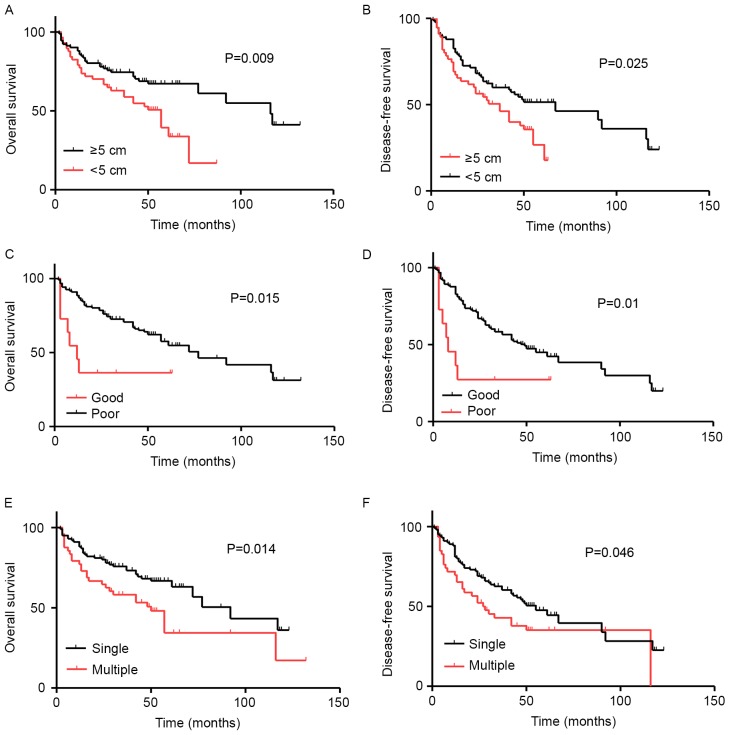
In order to analyze whether clinical factors, including sex, age, diameter, tumor differentiation and number of foci affect HCC prognosis, Kaplan-Meier survival curves were plotted and compared using a log-rank test (Table I). It was observed that tumor diameter was significantly associated with decreased duration of OS (P=0.009; Fig. 3A) and disease-free survival (DFS) (P=0.025; Fig. 3B) in patients with HCC. In addition, tumor differentiation was significantly associated with decreased duration of OS (P=0.015; Fig. 3C) and DFS (P=0.01; Fig. 3D) in patients with HCC. Similar results were obtained regarding the number of foci and duration of OS (P=0.014; Fig. 3E) and DFS (P=0.046; Fig. 3F).
Figure 3.
Univariate survival analysis of different clinical parameters with OS and DFS in patients with HCC. Kaplan-Meier plots indicating the associations between: (A) OS and HCC tumor diameter; (B) DFS and HCC tumor diameter; (C) OS and tumor differentiation; (D) DFS and tumor differentiation; (E) OS and number of foci and (F) DFS and number of foci. HCC, hepatocellular carcinoma; OS, overall survival; DFS, disease-free survival.
As shown in Table II, univariate analysis using the Cox regression model revealed that miR-32 expression levels [hazard ratio (HR)=2.51; confidence interval (CI): 1.48–4.27; P=0.001], number of foci (HR=36.78; CI: 8.98–150.68; P<0.001) and tumor diameter (HR=1.95; CI: 1.17–3.24; P=0.011) were positively associated with poor prognosis (P<0.05). However, clinicopathological characteristics, including age, sex and tumor differentiation exhibited no association with HCC prognosis (P>0.05).
Table II.
Cox regression model analysis for OS based on the clinicopathological characteristics of patients with hepatocellular carcinoma.
| A, Univariate analysis | |||
|---|---|---|---|
| Factor | HR | 95% CI | P-value |
| Sex (male vs. female) | 0.526 | 0.24–1.16 | 0.111 |
| Age, years (≥60 vs. <60) | 1.21 | 0.42–1.08 | 0.481 |
| Tumor differentiation (poorly vs. moderately/well) | 0.9 | 0.60–1.36 | 0.632 |
| Tumor diameter, cm (≥5 vs. <5) | 1.95 | 1.17–3.24 | 0.011 |
| Number of foci (multiple vs. single) | 36.78 | 8.98–150.68 | <0.001 |
| miR-32 expression (high vs. low) | 2.51 | 1.48–4.27 | 0.001 |
| B, Multivariate analysis | |||
| Factor | HR | 95% CI | P-value |
| Sex | |||
| Age, years | |||
| Tumor differentiation | |||
| Tumor diameter, cm | 4.47 | 2.13–9.35 | <0.001 |
| Number of foci | 4.63 | 2.20–9.76 | <0.001 |
| miR-32 expression | |||
HR, hazard ratio; CI, confidence interval; miR-32, microRNA-32.
miR-32 upregulation is a prognostic marker for survival in patients with HCC
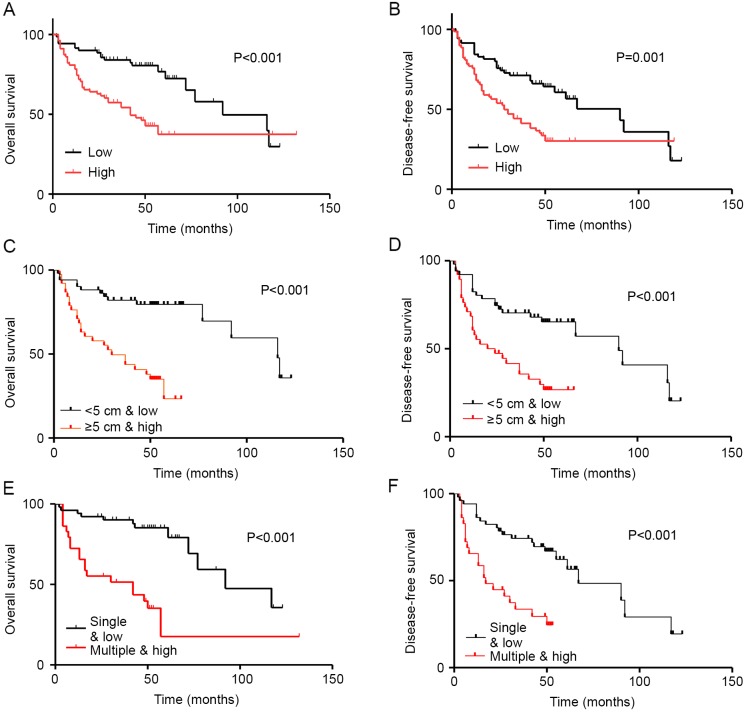
To determine the prognostic value of miR-32 expression in HCC, Kaplan-Meier survival analysis was used to evaluate the associations between miR-32 expression and OS and DFS. The results revealed that high miR-32 expression was associated with poorer OS, whereas low miR-32 mRNA levels were associated with increased OS. Therefore, miR-32 expression was significantly associated with decreased duration of OS (P<0.001; Fig. 4A) and DFS (P=0.001; Fig. 4B) in patients with HCC.
Figure 4.
Survival analysis of miR-32 expression and clinical parameters in patients with HCC. Univariate survival analysis of (A) OS and (B) DFS in HCC as determined by Kaplan-Meier plots based on miR-32 expression. Multivariate analysis of (C) OS and (D) DFS by Kaplan-Meier survival analysis was based on diameter and miR-32 expression. Multivariate analysis of (E) OS and (F) DFS by Kaplan-Meier survival analysis was based on number of foci and miR-32 expression. HCC, hepatocellular carcinoma; OS, overall survival; DFS, disease-free survival; miR-32, microRNA-32.
Kaplan-Meier survival analysis indicated that patients with HCC with high miR-32 expression and large tumor size (≥5 cm) had significantly decreased duration of Kaplan-Meier survival analysis indicated that patients with HCC with high miR-32 expression and large tumor size (≥5 cm) had significantly decreased duration of OS (P<0.001; Fig. 4C) and DFS (P=0.001; Fig. 4D). In addition, low miR-32 expression and multiple tumor foci were associated with poorer OS and DFS (Fig. 4E and F).
Discussion
HCC ranks as the sixth most common type of cancer worldwide. HCC treatment includes radical surgery or chemotherapy, but the postoperative recurrence rate is high (21). Therefore, the prognosis of HCC currently remains unsatisfactory. The toxicity of chemotherapeutic drugs and cancer-associated tumor resistance are matters of concern, and improvements in therapy are required. In the past few years, the development of new molecular biology techniques have enabled targeted HCC therapy (22). miRNAs are regarded as ideal biomarkers, since the diagnostic and therapeutic sensitivity of miRNAs is associated with clinical outcome compared with other biomarkers, including metabolites, antibodies and nucleic acids (23–25).
Recently, a study confirmed that the differences in miRNA expression levels in HCC tissues compared with normal tissues are not only statistically significant, but also associated with HCC diagnosis, prognosis and treatment (26). Li et al (27) reported that six different miRNAs were significantly upregulated in HCC samples compared with non-tumor tissues. Notably, it was revealed that miR-375 alone exhibited diagnostic value for HCC (27). Sukata et al (28) demonstrated that miR-98, let-7f and let-7a are potential early diagnostic markers for liver cancer. Pineau et al (29) revealed that downregulation of miR-221 in HCC performs an important role in tumorigenesis and drug resistance by inducing apoptosis.
miR-32 is able to function as a tumor suppressor and as a tumor promoter in different types of cancer (30). Due to the tissue specificity of miR-32, its biological function has been thoroughly studied. miR-32 functions as a tumor suppressor in osteosarcoma and NSCLC (16,31). In another study, Wu et al (12,32) reported that miR-32 is also involved in development of CRC, in part, due to suppression of PTEN. Furthermore, upregulation of miR-32 was associated with specific clinicopathological characteristics of patients with CRC. Therefore, miR-32 is considered to be a putative molecular marker of poor prognosis in patients with CRC.
Next, raw datasets from the GEO database were analyzed. In the raw datasets, it was revealed that miR-32 expression levels were significantly higher in HCC tumor specimens compared with non-neoplastic tissues (33–35). Indeed, using FC ≥2 or ≤0.5 and P<0.01 for screening in cluster analysis revealed that miR-32 was a common denominator of the three datasets. In a series of in vitro experiments performed by Yan et al (19), it was reported that miR-32 induces cell migration, proliferation and invasion in HCC by targeting PTEN. However, the clinical relevance between miR-32 and HCC currently remains unknown.
In the present study, it was revealed that miR-32 expression in HCC samples was significantly higher compared with normal tissues. The association between miR-32 and the clinical parameters was also examined in patients with HCC. High miR-32 expression was associated with tumor diameter and the number of foci, indicating that the upregulation of miR-32 has a crucial role in the progression of liver cancer. In order to investigate the potential prognostic value of miR-32, the association between the expression of miR-32 and OS in patients with HCC was analyzed. Kaplan-Meier survival analysis revealed that patients with high miR-32 expression levels had decreased OS time. Furthermore, Cox analysis demonstrated that miR-32 was an independent prognostic indicator of HCC. Therefore, levels of miR-32 expression may be an optimal indicator and risk factor for reduced OS and DFS in patients with HCC.
In summary, the present study demonstrated the clinical and prognostic significance of miR-32 in HCC. High miR-32 expression may be an optimal indicator of poor HCC prognosis. Altogether, the present analysis indicated that high miR-32 expression may be a potential biomarker for patients with HCC.
Acknowledgements
This study was supported partly by grants from the National Natural Science Foundation of China (grant nos. 81201535, 81302065, 81472202, 81772932, 81472209 and 81702243), Jilin Provincial Science and Technology Department (grant no. 20140414061GH), Shanghai Natural Science Foundation (grant nos. 12ZR1436000 and 16ZR1428900) and Shanghai Municipal Commission of Health and Family Planning (grant nos. 201440398 and 201540228). The funding agencies had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Wang Y, Ma Y, Fang Y, Wu S, Liu L, Fu D, Shen X. Regulatory T cell: A protection for tumor cells. J Cell Mol Med. 2012;16:425–436. doi: 10.1111/j.1582-4934.2011.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. Ca Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y, Fu D, Tang WQ, Cai Y, Ma D, Wang HJ, Xue RY, Liu TT, Huang XW, Dong L, et al. Ubiquitin C-terminal hydrolase 37, a novel predictor for hepatocellular carcinoma recurrence, promotes cell migration and invasion via interacting and deubiquitinating PRP19. Biochim Biophys Acta. 2013;1833:559–572. doi: 10.1016/j.bbamcr.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Chen YJ, Ma YS, Fang Y, Wang Y, Fu D, Shen XZ. Power and promise of ubiquitin carboxyl-terminal hydrolase 37 as a target of cancer therapy. Asian Pac J Cancer Prev. 2013;14:2173–2179. doi: 10.7314/APJCP.2013.14.4.2173. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu LL, Fu D, Ma Y, Shen XZ. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011;20:2023–2030. doi: 10.1089/scd.2011.0012. [DOI] [PubMed] [Google Scholar]
- 8.Hou LK, Ma YS, Han Y, Lu GX, Luo P, Chang ZY, Xie RT, Yang HQ, Chai L, Cai MX, et al. Association of microRNA-33a molecular signature with non-small cell lung cancer diagnosis and prognosis after chemotherapy. PLoS One. 2017;12:e0170431. doi: 10.1371/journal.pone.0170431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Cho WC. OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 1007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30. doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Fu D, Shen XZ. The potential role of ubiquitin C-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Jänne OA, Seppälä J, Lähdesmäki H, Tammela TL, Visakorpi T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31:4460–4471. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y, Wang YL, Yao WJ, Guo L, Xi HF, Li SY, Zhao BS. Expression of miR-32 in human non-small cell lung cancer and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol. 2015;8:824–829. [PMC free article] [PubMed] [Google Scholar]
- 16.Xu JQ, Zhang WB, Wan R, Yang YQ. MicroRNA-32 inhibits osteosarcoma cell proliferation and invasion by targeting Sox9. Tumor Biol. 2014;35:9847–953. doi: 10.1007/s13277-014-2229-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Kuai X, Song M, Chen X, Yu Z, Zhang H, Mao Z. microRNA32 inhibits the proliferation and invasion of the SGC-7901 gastric cancer cell line in vitro. Oncol Lett. 2014;7:270–274. doi: 10.3892/ol.2013.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Ni Z, Xu X. Decreased microRNA-32 in oral squamous cell carcinoma. Med Sci Monit. 2014;20:2527–2535. doi: 10.12659/MSM.892636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan SY, Chen MM, Li GM, Wang YQ, Fan JG. miR-32 induces cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting PTEN. Tumor Biol. 2015;36:4747–4755. doi: 10.1007/s13277-015-3124-9. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Fang Y, Mu J, Ma Y, Ma D, Fu D, Shen X. The interaction between ubiquitin C-terminal hydrolase 37 and glucose-regulated protein 78 in hepatocellular carcinoma. Mol Cell Biochem. 2012;359:59–66. doi: 10.1007/s11010-011-0999-7. [DOI] [PubMed] [Google Scholar]
- 22.Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie RT, Yang HQ, Chang ZY, Sun R, Chai L, et al. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896–11905. doi: 10.18632/oncotarget.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu SD, Ma YS, Fang Y, Liu LL, Fu D, Shen XZ. Role of the microenvironment in hepatocellular carcinoma development and progression. Cancer Treat Rev. 2012;38:218–225. doi: 10.1016/j.ctrv.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Ge S, Huang D. Systemic therapies for hepatocellular carcinoma. Drug Discov Ther. 2015;9:352–362. doi: 10.5582/ddt.2015.01047. [DOI] [PubMed] [Google Scholar]
- 25.Bartels CL, Tsongalis GJ. MicroRNAs: Novel biomarkers for human cancer. Ann Biol Clin (Paris) 2010;68:263–272. doi: 10.1684/abc.2010.0429. (In French) [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Hou L, Yu F, Lu G, Qin S, Xie R, Yang H, Wu T, Luo P, Chai L, et al. Incidence and physiological mechanism of carboplatin-induced electrolyte abnormality among patients with non-small cell lung cancer. Oncotarget. 2017;8:18417–18423. doi: 10.18632/oncotarget.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 28.Sukata T, Sumida K, Kushida M, Ogata K, Miyata K, Yabushita S, Uwagawa S. Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett. 2011;200:46–52. doi: 10.1016/j.toxlet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigebesis; Proc Natl Acad Sci USA; 2010; pp. 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Zhu Y, Ma Y, Wang J, Zhang F, Xia Q, Fu D. The role of cancer stem cells in cancer metastasis: New perspective and progress. Cancer Epidemiol. 2013;37:60–63. doi: 10.1016/j.canep.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Zhu D, Chen H, Yang X, Chen W, Wang L, Xu J, Yu L. miR-32 functions as a tumor suppressor and directly targets SOX9 in human non-small cell lung cancer. Onco Targets Ther. 2015;8:1773–1783. doi: 10.2147/OTT.S72457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Wu W, Yang P, Feng X, Wang H, Qiu Y, Tian T, He Y, Yu C, Yang J, Ye S, Zhou Y. The relationship between and clinical significance of MicroRNA-32 and phosphatase and tensin homologue expression in colorectal cancer. Genes Chromosomes Cancer. 2013;52:1133–1140. doi: 10.1002/gcc.22108. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Xu T, Jiang Y, Xu H, Yan Y, Fu D, Chen J. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia. 2015;17:239–255. doi: 10.1016/j.neo.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S, Shimizu K. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6:e16435. doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]