Abstract
Rationale: Mxi1 is regarded as a potential tumor suppressor protein that antagonizes the transcriptional activity of proto-oncogene Myc. However, the clinical significances and underlying mechanisms by which Mxi1 is regulated in lung cancer remain poorly understood.
Methods: Mass spectrometry analysis and immunoprecipitation assay were utilized to detect the protein-protein interaction. The phosphorylation of Mxi1 was evaluated by in vitro kinase assays. Poly-ubiquitination of Mxi1 was examined by in vivo ubiquitination assay. Lung cancer cells stably expressing wild-type Mxi1 or Mxi1-S160A were used for functional analyses. The expression levels of Mxi1 and S6K1 were determined by immunohistochemistry in lung cancer tissues and adjacent normal lung tissues.
Results: We found that Mxi1 is downregulated and correlated with poor prognosis in lung cancer. Using tandem affinity purification technology, we provided evidence that β-Trcp E3 ubiquitin ligase interacts with and promotes the ubiquitination and degradation of Mxi1. Furthermore, we demonstrated that Mxi1 is phosphorylated at S160 site by the protein kinase S6K1 and subsequently degraded via the ubiquitin ligase β-Trcp. Moreover, a phosphorylation mutant form of Mxi1 (Mxi1-S160A), which cannot be degraded by S6K1 and β-Trcp, is much more stable and efficient in suppressing the transcriptional activity of Myc and radioresistance in lung cancer cells. More importantly, a strong inverse correlation between S6K1 and Mxi1 expression was observed in human lung cancer tissues.
Conclusion: Our findings not only establish a crosstalk between the mTOR/S6K1 signaling pathway and Myc activation, but also suggest that targeting S6K1/Mxi1 pathway is a promising therapeutic strategy for the treatment of lung cancer.
Keywords: Mxi1, S6K1, β-Trcp, phosphorylation, lung cancer
Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide, and its incidence is rapidly rising in many developing countries 1, 2. Non-small cell lung cancer (NSCLC), including adenocarcinoma, squamous cell carcinoma and large cell carcinoma, comprises about 85% of the total cases 3. Despite recent advances in molecular diagnosis and targeted therapies, the clinical outcomes of this disease remain poor, with the 5-year survival rate of less than 15% 4. Local relapses and distant metastases are still the major obstacles to the improvement of patient survival. Thus, there is an urgent need to elucidate the mechanisms underlying tumorigenesis and identify novel therapeutic targets for lung cancer patients.
Ribosomal protein S6 kinase 1, also known as S6K1, is a serine-threonine kinase that functions as the critical downstream effector of the mTOR signaling pathway 5, 6. A growing body of evidence demonstrate that S6K1 plays critical roles in multiple key cellular processes, including cell cycle progression, protein synthesis, cell metabolism, adipogenesis and tumorigenesis 7-9. S6K1 exhibits these functions mainly through its ability to phosphorylate downstream substrates and most of its substrates contain a S6K1 consensus phosphorylation motif (K/RXRXXS/T), as previously described 10. For example, Dr. Sahin and colleagues showed that S6K1-mediated phosphorylation of BMAL1 regulates protein synthesis 11, while phosphorylation of EPRS by S6K1 has been reported to contribute to adiposity in mice 12. Additionally, a previous study showed that phosphorylation of PDCD4 by S6K1 is required for cell growth and cell cycle progression 13. Moreover, the involvement of S6K1-mediated Gli1 phosphorylation in tumorigenesis of esophageal adenocarcinoma has also been documented 14. These and many other findings strongly suggest that S6K1 is a potentially promising drug target for cancer treatment.
Myc, a well-known oncoprotein, has been studied extensively in the past decades. As a transcription factor, Myc dimerizes with MAX to bind to promoters and therefore controls downstream gene expression 15. Deregulation of Myc contributes to many altered cellular processes, which include cell proliferation, differentiation, apoptosis, metabolism, angiogenesis, genomic instability and tumorigenesis 16, 17. MAX interactor 1 (Mxi1), a member of the MAD family, functions as an antagonist of Myc by competing for Max and recruiting transcriptional repressor 18, 19. In this regard, Mxi1 is considered as a negative regulator of Myc through repressing the Myc-dependent transcriptional activity. Mxi1 gene localizes at chromosome 10q24‑q25, a cancer hotspot which has been linked to the development of prostate cancer, glioblastoma and lung cancer 19. Accumulating evidence suggest that Mxi1 has crucial roles in cell growth, differentiation, cell cycle control and tumorigenesis 18. For instance, Mxi1 has been shown to suppress the growth of prostate cancer and glioblastoma cells 20, 21. Moreover, Mxi1-/- mice exhibit increased susceptibility to tumorigenesis 22. These data indicate that Mxi1 may be a potential tumor-suppressor. However, exactly how Mxi1 is regulated in lung cancer remains poorly understood.
In the current study, we reported that Mxi1 is a novel substrate of S6K1 protein kinase and β-Trcp ubiquitin ligase. We showed that the S6K1-dependent phosphorylation of Mxi1 at Ser160 site acts to recruit the β-Trcp ubiquitin ligase, which facilitates the ubiquitination and degradation of Mxi1. Furthermore, we demonstrated that S6K1 and β-Trcp-dependent degradation of Mxi1 is crucial for Myc-dependent transcriptional activation and radioresistance in lung cancer cells. Importantly, a strong correlation between S6K1 overexpression and Mxi1 downregulation was observed in human lung cancer tissues, suggesting that S6K1 phosphorylation-dependent degradation of Mxi1 by β-Trcp ubiquitin ligase may suppress Mxi1 expression and therefore promote Myc transcriptional activity and radioresistance in lung cancer.
Materials and Methods
Cell lines and transfection
Human lung cancer cell lines (A549, H1299, H292, HCC827 and H1975), HBE, HeLa and HEK293T cell lines were obtained from American Type Culture Collection and cultured in DMEM medium containing 10% FBS and 100 μg/mL penicillin and streptomycin at 37 °C in 5% CO2. Plasmid transfection was carried out using the polyethylenimine reagent or Lipofectamine 2000 (Invitrogen).
Constructs
The Mxi1 plasmid was generated by polymerase chain reaction (PCR) and subcloned into pDNOR201 entry vector and then transferred to gateway-compatible destination vector with indicated SFB or HA tag using Gateway Technology (Invitrogen). S6K1, Mad1, Mad3 and Mad4 plasmids were purchased from DF/HCC DNA Resource Core. HA-S6K1-CA (F5A-T389E-R3A), HA-S6K1-KD (T229A), HA-AKT1, Myc-β-Trcp1, Myc-β-Trcp1-R474A and Myc-β-Trcp2 were gifts from Dr. Wenyi Wei (Harvard Medical School, USA). Mutations were introduced by using the KOD Hot Start DNA Polymerase (Novagen), and all mutations were verified by DNA sequencing.
Antibodies and reagents
The following antibodies in our study were used for immunoblotting: Phospho-Mxi1 antibody, which was raised against peptide RIRMDS(p)IGSTISS by immunizing rabbits; anti-Mxi1 antibodies (sc-1042, Santa Cruz Biotechnology and HPA035319, Sigma-Aldrich); anti-S6K1 (sc-230), anti-Myc (sc-40), anti-Cdc20 (sc-13162), and anti-GAPDH (sc-166574) antibodies (Santa Cruz Biotechnology); anti-phospho-S6K1(Thr389) (#9234), anti-β-Trcp1 (#4394), anti-Cyclin B1(#4138), anti-Ubiquitin (#3933) and anti-HA (#3724) antibodies (Cell Signaling Technology); anti-Max (ab199489) , anti-β-Trcp2 (ab137770) and anti-Myc (ab32072) antibodies (Abcam); anti-RSK2 (23762-1-AP) and anti-GAPDH (60004-1-Ig) antibodies (Proteintech); anti-Flag (F1804), anti-Actin (A5441) and anti-Tubulin (T-5168) antibodies (Sigma-Aldrich). MG132 (474790, a proteasome inhibitor) was purchased from Millipore, Hydroxyurea (H8627) and CHX (01810, Cycloheximide, a protein synthesis inhibitor) were purchased from Sigma-Aldrich.
Lentivirus Packaging and generation of stable cell lines
Lentiviral supernatants were generated by transient transfection of HEK293T cells with Mxi1 plasmid and packaging plasmids pSPAX2 and pMD2G (kindly provided by Dr. Zhou Songyang, Baylor College of Medicine) and harvested 48 h after transfection. Supernatants were filtered and used to infect cells with the addition of 10 μg/mL Polybrene for 48 h. Stable cell lines were selected with media containing 2 μg/mL puromycin and confirmed by Western blotting.
RNAi treatment
The sequences of oligonucleotides targeting S6K1 and β-Trcp were as follows: 5-GGACAUGGCAGGAGUGUUU-3 and 5-AAGUGGAAUUUGUGGAACAUC-3, which has been described previously 13, 23, 24. RSK siRNA-1: 5′- AGCGCTGAGAATGGACAGCAA-3′; RSK siRNA-2: 5′-GGGAGGAGAUUUGUUUACACGCUUA-3′, which has been described previously 25, 26. A549, H1299 or HeLa cells were seeded into each well of a 6-well tissue culture dish the day before transfection. SiRNAs transfection was performed according to the manufacturer's instructions using Lipofectamine RNAiMAX transfection reagent (Invitrogen).
Tandem affinity purification of SFB-tagged Mxi1
Tandem affinity purification of SFB-tagged Mxi1 was performed as previously described 27. Briefly, HEK293T cells stably expressing exogenous SFB-tagged Mxi1 were collected after the MG132 treatment for 4 h. Cells were lysed with NETN buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl, and 0.5 % Nonidet P-40) for 20 min. The lysates were centrifuged (20,817g) and then incubated with streptavidin-conjugated beads (Amersham) for 2 h at 4ºC. After being washed three times, beads-bound proteins were eluted twice with 2 mg/mL biotin (Sigma-Aldrich) diluted in NETN buffer for 2 h. The elutes were further incubated with S protein agarose (Novagen) for 1 h. The immunocomplexes were again washed three times and resolved by SDS-PAGE. Protein bands were identified and analyzed by mass spectrometry.
Western blotting and immunoprecipitation
Cells were lysed in NETN buffer, and the clarified lysates were resolved by SDS-PAGE for Western blotting analysis. Alternatively, the clarified supernatants were first incubated with S protein beads (Novagen) overnight, and the precipitates were washed five times with NETN buffer. To investigate the endogenous interaction, the clarified supernatants were first incubated with Mxi1, S6K1 or β-Trcp antibody for 2 h at 4°C. Protein A/G-agaroses were then added overnight and the precipitates were washed five times with NETN buffer and analyzed by Western blotting.
In vitro kinase assays
To construct plasmids that express His-Mxi1-WT and His-Mxi1-160A proteins, the coding sequences of Mxi1-WT and Mxi1-160A were subcloned into pET22b (+) vector. Each expression plasmid was transformed into E. coli BL21, and the recombinant proteins were purified by His Sepharose Ni (GE Healthcare). HA-S6K1-WT or HA-S6K1-KD was obtained by immunoprecipitation using anti-HA agarose from the lysates of Hela cells overexpressing HA-S6K1-WT or HA-S6K1-KD. The samples were then incubated with 5 μg of His-Mxi1-WT or His-Mxi1-160A in 50 μL kinase buffer (Cell Signaling Technology) containing 10 mM ATP for 30 min at 30°C. Subsequently, the samples were dissolved in SDS-PAGE loading buffer and subjected to Western blotting.
λ-PPase treatment
SFB-Mxi1 was immunoprecipitated with anti-Flag agarose from lysates of cells overexpressing SFB-Mxi1, resuspended in λ-PPase buffer containing 200 U of enzyme (New England BioLabs), and incubated for 1 h at 30°C. The reaction was stopped by heat inactivation at 65°C for 1 h.
In vivo ubiquitination assay
HeLa cells were cotransfected with the indicated plasmids for 24 h. MG132 (10 μM) was added for 4 h prior to collection, and cells were lysed in NETN buffer. The clarified supernatants were first incubated with S protein beads (Novagen) for 2 h and then subjected to western blotting.
Real-time quantitative PCR
This assay was carried out as described previously 28. Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized using the qPCR RT Master Mix (Toyobo). The primers were as follows: ODC1: F 5′-GATGACTTTTGATAGTGAAGTTGAGTTGA-3′; R 5′-GGCACCGAATTTCACACTGA-3′; CCND2: F 5′-GGCACCGAATTTCACACTGA-3′; R 5′-GTTGCAGATGGGACTTCGGA-3′; eIF2a: F 5′-ATGGGACCTTGTTTGCCTGG-3′; R 5′-CCACGTTGCCAGGACAGTAT-3′; Mxi1: F 5′- TGGCTACGCCTCTTCATTCC -3′; R 5′- CTGTTGGCAGTGCTGGTGTT-3′; GAPDH: F 5′-AATCCCATCACCATCTTCCAG-3′; R 5′-GAGCCCCAGCCTTCTCCAT-3.
Clonogenic cell survival assay
The A549 or H1299 stable cells were plated in triplicate in six-well plates and exposed to irradiation with indicated doses. After cultured for two weeks, cell clones were washed and fixed in methanol for 30 min, then dyed with crystal violet for 30 min. Afterward, the dye was washed off and colonies that contained more than 50 cells were counted.
Immunohistochemical (IHC) assay
A lung cancer tissue microarray was purchased from Shanghai Outdo Biotech (Shanghai, China), which contained 90 carcinoma tissues and paired para-carcinoma tissues. All patients had been pathologically diagnosed with lung adenocarcinoma. IHC of Mxi1 and S6K1 were performed using anti-Mxi1 (HPA035319, Sigma-Aldrich) and anti-S6K1 (sc-230, Santa Cruz Biotechnology) antibody respectively. The IHC analysis was performed as previously described 29-32. In brief, the tissue sections were dewaxed and endogenous peroxidase was blocked by 1% hydrogen peroxide. After incubated with primary antibody against Mxi1 or S6K1 overnight at 4°C and being washed, tissue sections were treated with biotinylated secondary antibody for 1 h at room temperature. Finally, tissue sections were reacted with 3,3-diaminobenzidine and counterstained with hematoxylin. The total score (values 0-12) of protein expression was calculated by multiplying the percentage of immunopositively areas (0-25% = 1, 26-50% = 2, 51-75% = 3, >75% = 4) and immunostaining intensity (negative = 0, weak = 1, moderate = 2, or strong = 3). The score ≥ 6 defined high expression, while score < 6 defined low expression.
Statistical analysis
An unpaired Student's t test was used for statistical comparison. The Kaplan-Meier method was used to estimate the overall survival, correlation between S6K1 and Mxi1 expression was analyzed by using the Pearson's chi-squared test. Statistically significant differences were defined when P values were at least below 0.05.
Results
Mxi1 is downregulated and predicts poor prognosis in lung cancer patients
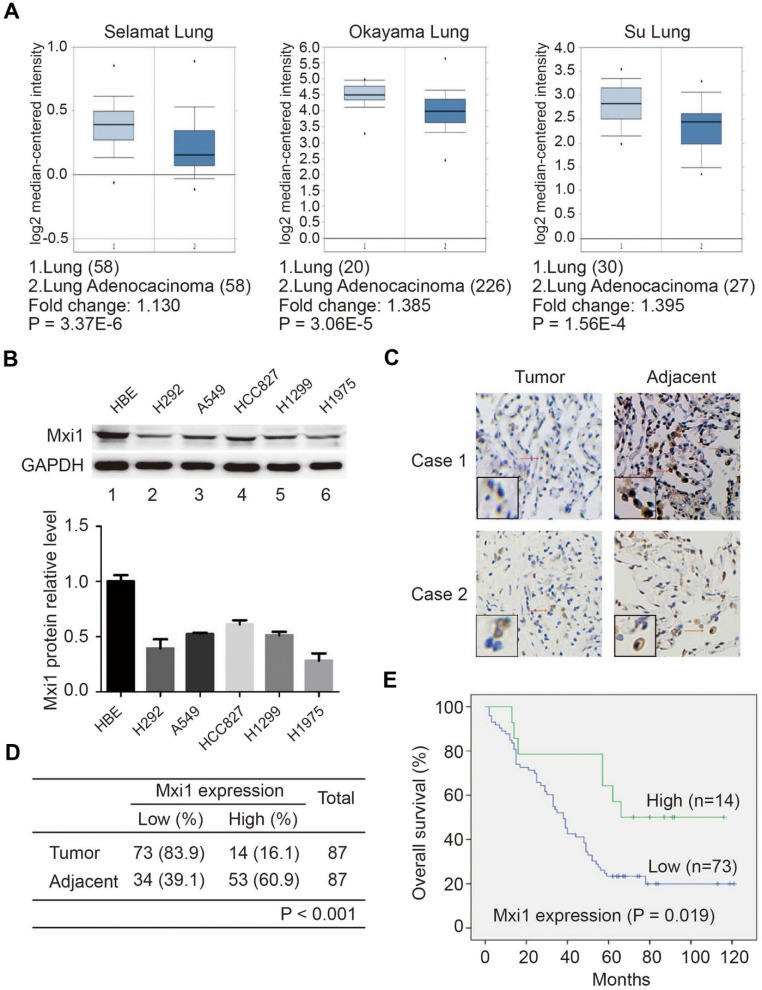
Mxi1 potentially functions as tumor suppressor due to its ability to counteract Myc-dependent oncogenic activities. To investigate Mxi1 expression in lung cancer, we first analyzed Mxi1 mRNA level in lung adenocarcinoma and normal lung tissue using the Oncomine database. As shown in Figure 1A, Mxi1 mRNA level was significantly downregulated in lung cancer. To further determine whether Mxi1 expression is indeed reduced in tumors, we first analyzed Mxi1 protein levels in five human lung cancer cell lines (H292, A549, HCC827, H1299, and H1975) and found that Mxi1 was remarkably downregulated in lung cancer cell lines compared with that in normal human lung epithelial cell HBE (Figure 1B). Next, we quantified Mxi1 protein level in lung adenocarcinoma tissue arrays by immunohistochemistry assay. Our data showed that Mxi1 localized in the nucleus of cancer cells but Mxi1 protein level was low in the lung cancer tissues compared with that in adjacent lung tissues (Figure 1C, D). To elucidate whether Mxi1 expression is associated with clinical outcome, overall survival data of patients with lung adenocarcinoma were divided into two groups based on the intensity and the percentage of Mxi1 positive cells (low and high). Kaplan-Meier survival analysis demonstrated that low Mxi1 level was significantly associated with poor overall survival (P < 0.05) (Figure 1E). Therefore, our findings suggest that Mxi1 may be a tumor suppressor and its downregulation contributes to the development and progression of lung cancer.
Figure 1.
Mxi1 is downregulated and predicts poor prognosis in lung cancer patients. A. Box plots show the Mxi1 mRNA levels are down-regulated in lung adenocarcinoma tissue in comparison to normal lung tissue. B. Upper panel: Western blotting analysis of Mxi1 expression in five human lung cancer cell lines and one normal human bronchial epithelial cell line (HBE). Lower panel: quantification of Mxi1 protein expression (n=3). C. Representative immunohistochemical staining images for Mxi1 in lung adenocarcinoma tissue and adjacent lung tissue. Scale bar, 100 μm. D. Statistical analysis of immunohistochemical staining for Mxi1 in lung adenocarcinoma tissue microarray. E. OS (overall survival) curve shows that low expression of Mxi1 predicts poor prognosis in lung adenocarcinoma patients.
β-Trcp binds to Mxi1 and promotes the degradation of Mxi1
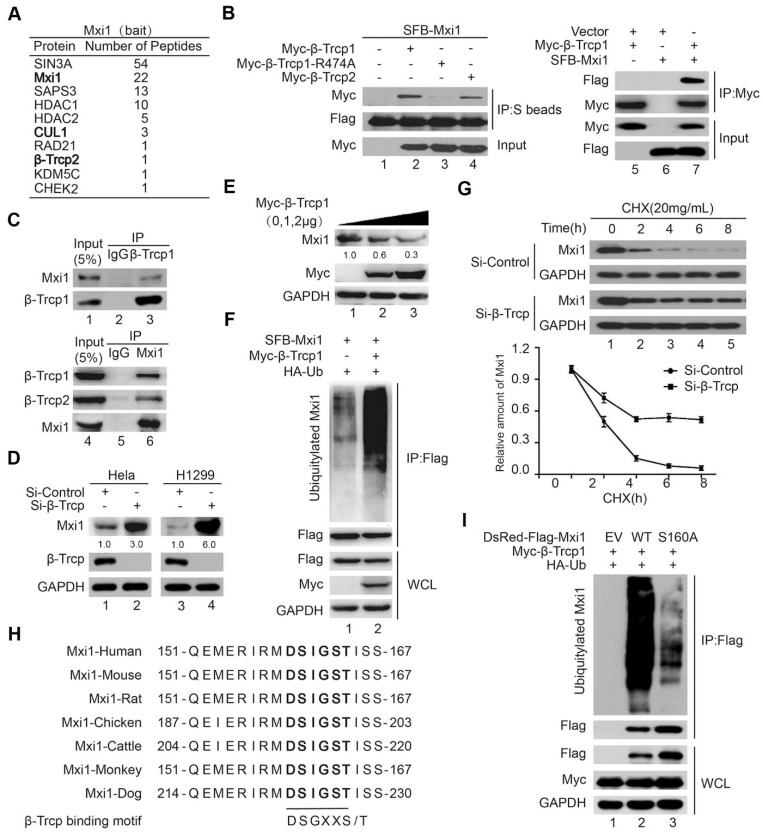
It has been reported that Myc was involved in G0/S progression 33, so we examined the abundance of Mxi1, which is considered an antagonist of Myc during cell cycle. Interestingly, the level of Mxi1 was higher when cells were arrested at early S phase after hydroxyurea stimulation and then started to decrease (Figure S1), indicating Mxi1 may be a cell cycle-regulated and labile protein. To explore how Mxi1 is regulated, we performed an unbiased tandem affinity purification (TAP) and mass spectrometry (MS) technology to identify Mxi1-interacting proteins in cells. As shown in Figure 2A, a number of previously reported Mxi1-interacting proteins, including SIN3A, SAPS3, HDAC1 and HDAC2, were identified. Intriguingly, we noticed that ubiquitin ligase complex Cullin 1-β-Trcp2 is a candidate Mxi1 binding partner. There are two distinct paralogs of β-Trcp protein β-Trcp1 and β-Trcp2, which share identical biological and biochemical traits 34. Therefore, we will use the term β-Trcp to refer to both. To validate our mass spectrometry results, co-immunoprecipitation experiments were carried out with β-Trcp since it is the substrate specificity subunit of the Cullin complexes and binds directly to substrates. As shown in Figure 2B, exogenous β-Trcp1 or β-Trcp2 associated with Mxi1, and vice versa. The R474 site of β-Trcp1 is required for mediating its association with most of its substrates 35. Notably, the interaction between Mxi1 and β-Trcp1-R474A mutant was dramatically reduced, further supporting that Mxi1 is likely a substrate of β-Trcp. Furthermore, we also found that the endogenous Mxi1 interacted with β-Trcp1 and β-Trcp2 (Figure 2C). More importantly, β-Trcp depletion using siRNAs targeting both β-Trcp1 and β-Trcp2 led to Mxi1 accumulation in two different cancer cell lines (Figure 2D). Additionally, overexpression of β-Trcp1 reduced endogenous Mxi1 levels in a dose-dependent manner (Figure 2E). These results indicate that β-Trcp negatively controls the stability of Mxi1. To further examine whether Mxi1 is a ubiquitination substrate of β-Trcp, we performed in vivo ubiquitination assay and showed that β-Trcp ligase could polyubiquitinate Mxi1 in cells (Figure 2F). In line with this notion, the half-life of Mxi1 was markedly extended after β-Trcp depletion (Figure 2G). These data indicate that Mxi1 is a novel substrate of β-Trcp which ubiquitinates and targets it for degradation.
Figure 2.
β-Trcp binds to Mxi1 and promotes the ubiquitination and degradation of Mxi1. A. Mxi1-associated proteins in HEK293T cells identified by tandem affinity purification and mass spectrometry analysis were presented. The recovered peptide numbers for a given protein were listed as indicated. B. Exogenously expressed β-Trcp interacts with Mxi1 and vice versa. HEK293T cells were transfected with indicated constructs. Cell lysates were subjected to pulldown using S protein beads or immunoprecipitation (IP) using anti-Myc beads and then analyzed by Western blotting using indicated antibodies (n=3). C. Endogenous β-Trcp binds to Mxi1 and vice versa. HeLa cell lysates were subjected to immunopreciptation using IgG or anti-β-Trcp antibodies and then analyzed by Western blotting as indicated (n=3). D. β-Trcp knockdown leads to accumulation of endogenous Mxi1. HeLa and H1299 cells were transfected with control or β-Trcp siRNA for 48 h and then analyzed by Western blotting as indicated (n=3). The ratio shows relative Mxi1 protein expression normalized for GAPDH (control, set at 1). E. Overexpression of β-Trcp1 decreases the levels of Mxi1 protein. HeLa cells were transfected with increasing amounts of β-Trcp1, followed by Western blotting with indicated antibodies (n=3). The ratio shows relative Mxi1 protein expression normalized for GAPDH (control, set at 1). F. HeLa cells were transfected with indicated constructs for 24 h and treated with MG132 (10 μM) for another 4 h. The samples were subjected to immunopreciptation (IP) using anti-Flag beads and then analyzed by Western blotting using indicated antibodies (n=3). WCL: whole cell lysate. G. Upper panel: HeLa cells transfected with indicated siRNA for 48 h were treated with CHX (20 mg/mL) for the indicated times and then analyzed by Western blotting using indicated antibodies. Lower panel: Quantification of the Mxi1 band intensities over time (n=3). H. Alignment of the candidate phosphodegron sequence in Mxi1 from different species. I. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were transfected with indicated constructs and then treated with MG132 (10 μM) for another 4 h. The samples were analyzed as described in (E) (n=3).
It has been well documented that β-Trcp ligase binds to its substrates by recognizing a characterized phosphodegron motif DSGXXS 34. we noticed that the C-terminus of Mxi1 contains a conserved 159DSIGST164 sequence, which is similar to a canonical DSGXXS motif (Figure 2H). To investigate whether β-Trcp ubiquitinates Mxi1 via this motif, we generated serine160 to alanine mutant (S160A) and found that the S160A mutant was less ubiquitinated than wild-type Mxi1 in the presence of β-Trcp (Figure 2I), suggesting that Mxi1 phosphodegron motif (DSIGST) determines ubiquitination and degradation of Mxi1 by β-Trcp ligase.
S6K1 is involved in the regulation of Mxi1 stability
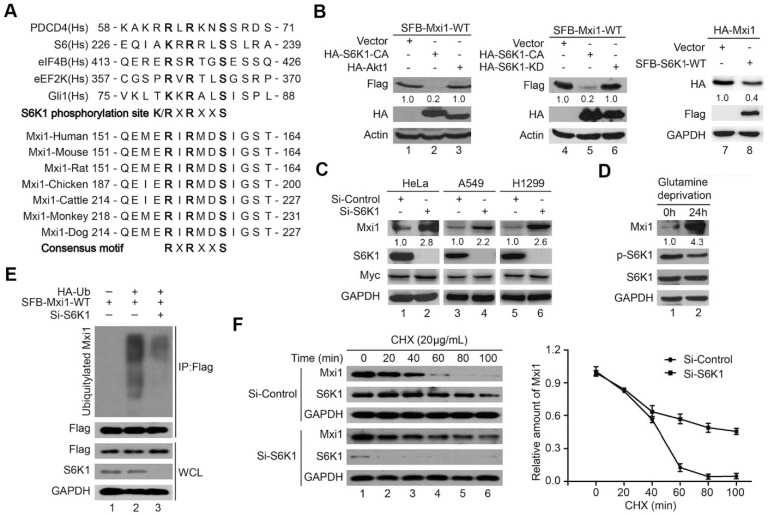
Emerging evidence have shown that β-Trcp recognizes downstream substrates with phosphorylated Ser residue in the phosphodegron motif DSGXXS 34. In addition, it has also been reported that several protein kinases including CK1, GSK3β and S6K1 often participate in β-Trcp-mediated ubiquitination and degradation 36. We therefore sought to identify the upstream kinase that phosphorylates Mxi1 and triggers its destruction. Excitingly, the β-Trcp-binding degron in Mxi1 is near to a canonical RXRXXS phosphorylation consensus motif for S6K1 (Figure 3A) 10. Thus, we suspected that Mxi1 may be a downstream substrate of S6K1 kinase. To test this hypothesis, we examined the Mxi1 expression in cells transfected with S6K1 or AKT1 and found that overexpression of S6K1-CA (constitutively active) and S6K1-WT (wild-type), but not that of AKT1, resulted in decreased Mxi1 expression (Figure 3B). Importantly, S6K1-KD (kinase dead) mutant did not affect Mxi1 protein level (Figure 3B). RSK, another AGC kinase, has been reported to be capable of recognizing RXRXXS consensus motif. However, we did not detect the accumulation of Mxi1 protein in RSK-depleted cells (Figure S2). Therefore, all of these results suggest that S6K1 but not AKT1 and RSK may be responsible for regulating Mxi1 stability. Consistent with this notion, knockdown of S6K1 stabilized endogenous Mxi1 but not Myc in three different cancer cell lines (Figure 3C). Additionally, accumulation of Mxi1 was also observed after glutamine deprivation in which the activity of S6K1 was inhibited (Figure 3D). These results strongly suggest that Mxi1 downregulation in cancer cells may be due to S6K1 overexpression and/or activation.
Figure 3.
Mxi1 protein level is negatively regulated by S6K1. A. Upper panel: alignment of the amino acid sequences encompassing the S6K1 consensus phosphorylation motif in previously reported S6K1 substrates. Lower panel: alignment of S6K1/AKT phosphodegron motifs in Mxi1 from different species. B. S6K1 but not AKT1 overexpression results in decreased Mxi1 protein level. HEK293T cells were transfected with indicated constructs. Cells were collected and analyzed as in (B) (n=3). The ratio shows relative Mxi1 protein expression normalized for GAPDH or Actin (control, set at 1). C. Knockdown of S6K1 stabilizes endogenous Mxi1. HeLa, A549 and H1299 cells were transfected with indicated siRNAs for 48 h. Cell lysates were collected and immunoblots were performed with the indicated antibodies (n=4). The ratio shows relative Mxi1 protein expression normalized for GAPDH (control, set at 1). D. HeLa cells were treated with glutamine deprivation for 24 h. Cells were collected and analyzed as in (B) (n=3). The ratio shows relative Mxi1 protein expression normalized for GAPDH (control, set at 1). E. HeLa cells were transfected with indicated constructs after cells were transfected with S6K1 siRNA for 24 h. Cells were collected after treatment with MG132 (10 μM) for 4 h. The samples were subjected to immunoprecipitation using anti-Flag beads and then analyzed by Western blotting as indicated (n=3). F. Left panel: H1299 cells transfected with indicated siRNA for 48 h were treated with CHX (20 mg/mL) for the indicated times and then analyzed by Western blotting using indicated antibodies. Right panel: Quantification of the Mxi1 band intensities over time (n=3).
Considering that Mxi1 is degraded via a proteasome-dependent pathway, we hypothesized that S6K1 may negatively regulate the stability of Mxi1 by promoting its ubiquitination. To test this hypothesis, we performed in vivo ubiquitination assay and showed that the ubiquitination of wild-type Mxi1 was dramatically reduced when S6K1 was silenced (Figure 3E). Likewise, S6K1 depletion also prolonged the half-life of Mxi1 (Figure 3F). These findings indicate that S6K1 negatively controls the stability of Mxi1 through ubiquitin proteasome pathway.
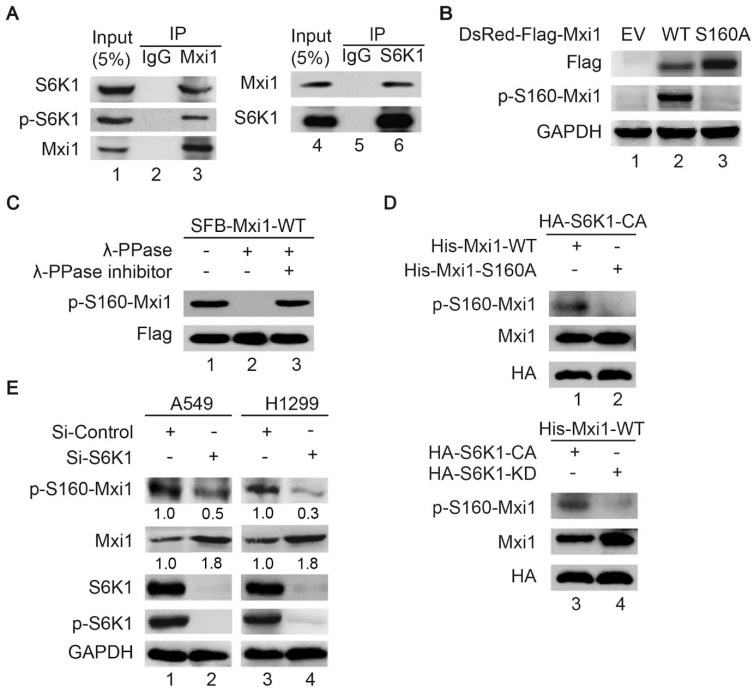
S6K1 interacts with Mxi1 and phosphorylates Mxi1 at Ser160 in vitro and in vivo
Given that the abundance of Mxi1 is controlled by S6K1 and S6K1 is a serine/threonine kinase, we asked whether Mxi1 is a direct substrate of S6K1. First, we examined whether S6K1 could associate with Mxi1. As shown in Figure 4A, endogenous S6K1 and p-S6K1 was present in Mxi1 immunoprecipitates and vice versa, indicating that S6K1 binds to Mxi1 in cells. We noted a highly conserved S6K1 phosphorylation motif in Mxi1 (RXRXXS160) (Figure 3A). To examine whether S6K1 regulates Mxi1 through phosphorylation at Ser160, we generated a rabbit polyclonal antibody that specifically recognizes phosphorylated Ser160 of Mxi1 (p-S160-Mxi1). Indeed, this antibody specifically recognized the ectopically expressed wild-type Mxi1, but not the Mxi1 S160A mutant in which the serine residue at position 160 was replaced with alanine (Figure 4B). Additionally, the anti-p-S160-Mxi1 antibody was able to recognize wild-type Mxi1, but not the same Mxi1 that was pre-treated with λ-PPase (Figure 4C). These results demonstrated that the anti-p-S160-Mxi1 antibody is highly specific for recognizing Ser160 phosphorylated Mxi1 and Mxi1 is phosphorylated at this residue in vivo. Using in vitro kinase assay, we confirmed again that S6K1 could phosphorylate wild-type Mxi1, but not the S160A mutant (Figure 4D). Only the catalytically active version of S6K1 (S6K1-CA), but not the catalytically dead version of S6K1 (S6K1-KD), was able to phosphorylate Mxi1 (Figure 4D). Moreover, we showed that phosphorylation of endogenous Mxi1 at Ser160 decreased when S6K1 was depleted in two different lung cancer cell lines (Figure 4E). Taken together, these results strongly suggest that S6K1 directly phosphorylates Mxi1 at Ser160 in vitro and in vivo.
Figure 4.
S6K1 binds and phosphorylates Mxi1 at Ser160 in vitro and in vivo. A. Endogenous S6K1 binds to Mxi1 and vice versa. HeLa cell lysates were subjected to immunoprecipitation using IgG, anti-Mxi1, or anti-S6K1 antibodies and then analyzed by Western blotting using indicated antibodies (n=3). B. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were lysed and then analyzed by Western blotting using indicated antibodies (n=4). C. Lysates prepared from HeLa cells transfected with SFB-Mxi1 were treated with λ-PPase with or without λ-PPase inhibitor and then analyzed by Western blotting as indicated (n=3). D. Upper panel: His-Mxi1-WT or S160A protein were incubated in vitro with immunoprecipitates isolated from HEK293T cells transfected with HA-S6K1 construct and then analyzed by Western blotting using indicated antibodies. Lower panel: Proteins immunoprecipitated using anti-HA beads from HEK293T cells transfected with constructs encoding HA-S6K1 or HA-S6K1-KD were incubated with His-Mxi1-WT protein in vitro separately and then analyzed by Western blotting using indicated antibodies (n=3). E. A549 or H1299 cells were transfected with indicated S6K1 siRNA for 48 h and cell lysates were analyzed by Western blotting using antibodies as indicated (n=3). The ratio shows relative p-S160-Mxi1 or Mxi1 protein expression normalized for GAPDH (control, set at 1).
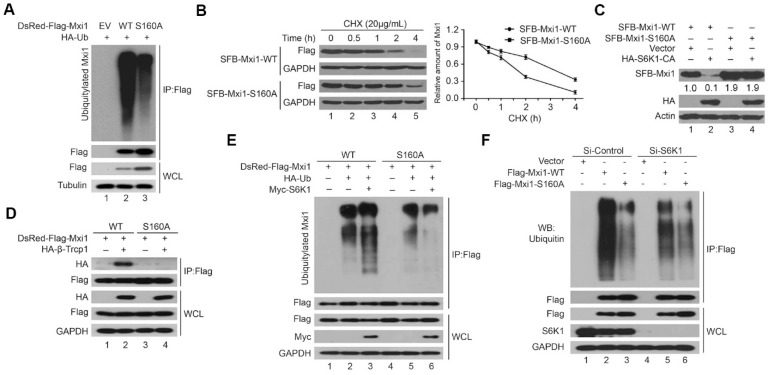
Phosphorylation of Mxi1 by S6K1 at S160 site promotes its binding to β-Trcp and ubiquitin-mediated degradation
The above findings demonstrate that S6K1 and β-Trcp negatively regulates the stability of Mxi1 through ubiquitin proteasome pathway. To further determine whether Mxi1 degradation is dependent on its phosphorylation by S6K1, we compared ubiquitination of wild-type Mxi1 with that of Mxi1 S160A mutant. As shown in Figure 5A, the polyubiquitination of Mxi1 S160A mutant was significantly reduced. Consistent with this finding, the half-time of Mxi1 S160A mutant was longer than that of wild-type Mxi1 (Figure 5B). These data indicate that phosphorylation of Mxi1 at Ser-160 promotes polyubiquitination of Mxi1. Additionally, we showed that wild-type Mxi1, but not the S160A mutant of Mxi1, can be degraded by S6K1 (Figure 5C), further supporting that phosphorylation at S160 is critical for the stability of Mxi1.
Figure 5.
S6K1 phosphorylates Mxi1 at Ser160 site to promote β-Trcp-mediated ubiquitination and degradation of Mxi1. A. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were transfected with indicated construct for 24 h and then treated with MG132 (10 μM) for another 4 h. Cell lysates were analyzed as described in Figure 2F (n=3). B. Upper panel: HeLa cells transfected with constructs encoding SFB-Mxi1-WT or SFB-Mxi1-S160A were treated with CHX (20 mg/mL) for the indicated times and then analyzed as described in Figure 2F. Lower panel: Quantification of the Mxi1 band intensities over time (n=3). C. HEK293T cells were transfected with constructs as indicated. Cell lysates were analyzed by Western blotting as indicated (n=4). The ratio shows relative Mxi1 protein expression normalized for Actin (control, set at 1). D. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were transfected with indicated construct for 24 h and then subjected to immunopreciptation using anti-Flag beads. The samples then were analyzed by Western blotting using indicated antibodies (n=3). E. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were transfected with indicated constructs and then treated with MG132 (10 μM) for another 4 h. The samples were analyzed as described in (A) (n=3). F. HeLa cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were transfected with indicated siRNAs for 48 h and then treated with MG132 (10 μM) for another 4 h. The samples were analyzed as described in (E) (n=3).
It has been reported that most, if not all, β-Trcp substrates must be phosphorylated before they can be recognized by β-Trcp. Therefore, we examined whether Ser160 phosphorylation of Mxi1 would be required for its binding to β-Trcp. As shown in Figure 5D, unlike wild-type Mxi1, Mxi1 S160A mutant failed to associate with β-Trcp. Consistent with this finding, the S160A mutant was less ubiquitinated than wild-type Mxi1 (Figure 5E). More importantly, S6K1 overexpression could accelerate the ubiquitination of wild-type Mxi1 but not S160A mutant, whereas knockdown of S6K1 exhibited an opposite effect (Figure 5E, F). These results indicate that Mxi1 phosphorylation at Ser160 site by S6K1 facilitates its binding and degradation by β-Trcp. Collectively, these results suggest that S6K1-dependent Mxi1 phosphorylation at Ser160 site promotes the binding of Mxi1 to β-Trcp and triggers its polyubiquitination and degradation.
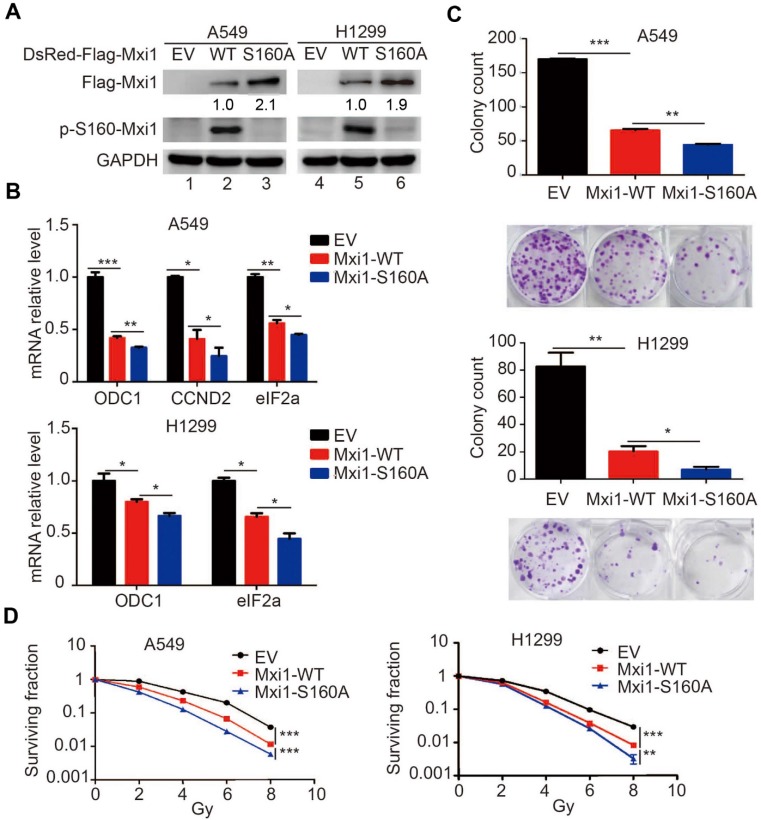
Phosphorylation of Mxi1 at S160 by S6K1 promotes Myc activation and radioresistance in lung cancer cells
To determine the biological consequence of S6K1-dependent phosphorylation of Mxi1 at Ser160, we generated A549 and H1299 derivative cell lines stably transfected with retrovirus expressing wild-type or S160A mutant of Mxi1 (Figure 6A). Notably, we demonstrated that the mRNA levels of Mxi1 in Mxi1-WT and Mxi1-S160A stably expressed A549 and H1299 cells are equally transcribed (Figure S3), further supporting the notion that the regulation of Mxi1 by S6K1 occurs at the post-translational level. It has been reported that Mxi1 is a well-known Myc inhibitory protein; we then examined whether Mxi1 phosphorylation would affect Myc-dependent gene transcription. Indeed, the mRNA levels of ODC1, CCND2, and eIF2a, three well-established Myc-target genes, were decreased in cells stably expressing wild-type Mxi1, when compared to control cells (Figure 6B). Importantly, cells stably expressing the Mxi1 S160A mutant showed even stronger inhibition of these Myc-target genes (Figure 6B). These results suggest that Mxi1 phosphorylation in lung cancer cells promotes Myc-mediated transcription.
Figure 6.
Phosphorylation of Mxi1 at Ser160 by S6K1 promotes Myc activation and cell radioresistance in lung cancer. A. A549 and H1299 cells stably expressing empty vector (EV), wild-type Mxi1, or Mxi1 S160A mutant were collected and analyzed by Western blotting using indicated antibodies (n=4). The ratio shows relative Mxi1 protein expression normalized for GAPDH (control, set at 1). B. The mRNA levels of the indicated genes were analyzed by Real time-PCR in A549 and H1299 cells stably expressing wild-type or mutant Mxi1. * P < 0.05, ** P < 0.01, *** P <0.001 (n=4). C. A549 and H1299 cells stably expressing wild-type or mutant Mxi1 were seeded and cultured for two weeks. The colonies were stained by crystal violet and then counted. Representative pictures are shown. * P < 0.05, ** P < 0.01, *** P <0.001 (n=3). D. A549 and H1299 cells stably expressing wild-type or mutant Mxi1 were irradiated with indicated doses. After two weeks, the colonies contained more than 50 cells were counted. ** P < 0.01, *** P <0.001 (n=3).
Myc proto-oncogene has been shown to be involved in cell proliferation in various human cancers. Therefore, we speculated that phosphorylation of Mxi1 at Ser160 by S6K1 may affect the cell proliferation in lung cancer. As expected, cells with wild-type Mxi1 expression showed a reduction in cell colony formation comparing with control cells (Figure 6C), which is consistent with previous reports suggesting that Mxi1 may function as a tumor suppressor. Importantly, we found that cells expressing Mxi1 S160A mutant displayed reduced colony formation than those cells expressing wild-type Mxi1 (Figure 6C). Furthermore, we demonstrated that cells expressing the S160A mutant of Mxi1 had fewer colonies than cells expressing wild-type Mxi1 following irradiation (Figure 6D), suggesting that Mxi1 phosphorylation at Ser160 may promote cancer cell radioresistance. Together, these results indicate that Mxi1 phosphorylation in lung cancer cells promotes Myc-mediated transcription and cell proliferation in normal condition as well as in response to DNA damage.
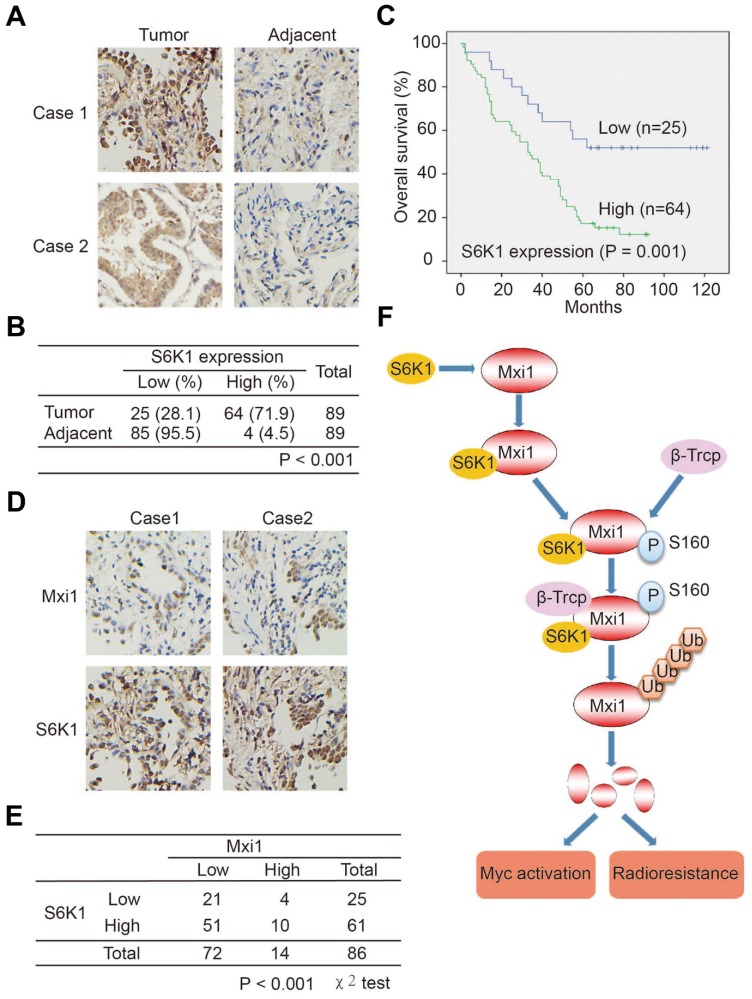
Correlation between S6K1 overexpression and Mxi1 downregulation in human lung cancer tissues
Our data above strongly suggest Mxi1 downregulation in cancer may be dependent on S6K1 overexpression and/or activation, which has been implicated in several types of cancer 37, 38. Thus, we speculated that S6K1 may have oncogenic activity in lung cancer via its ability to suppress Mxi1 expression. We first detected the levels of S6K1 and p-S6K1 in a panel of lung cancer cell lines and found that p-S6K1 expression varied dramatically and total S6K1 protein levels were enriched (Figure S4). We then performed IHC staining of lung adenocarcinoma tissue arrays to illustrate the protein level of S6K1 in 89 pairs of lung adenocarcinomas tissue and adjacent normal lung tissue. We observed that S6K1 expression was detected in both the nucleus and cytoplasm of cancer cells, and it was significantly upregulated in lung adenocarcinoma tissue (Figure 7A, B). Moreover, Kaplan-Meier analysis showed that patients with high S6K1 level displayed dramatically worse overall survival (P = 0.001) than those with low S6K1 level (Figure 7C). Therefore, S6K1 overexpression has a potential role in lung cancer development and correlates with poor outcome for lung cancer patients. Given that S6K1 negatively regulates Mxi1 protein level in lung cancer cells (Figure 2), we further explored the correlation between S6K1 and Mxi1 expression in lung adenocarcinoma tissue microarrays. Interestingly, there was an inverse correlation between the protein levels of S6K1 and Mxi1 (P < 0.001, χ2 tests; Figure 7D, E), suggesting that S6K1 overexpression may contribute to Mxi1 down-regulation in lung adenocarcinoma.
Figure 7.
Overexpression of S6K1 correlates with Mxi1 downregulation and predicts poor prognosis in lung cancer. A. Representative immunohistochemical staining images for S6K1 in lung adenocarcinoma tissue and adjacent lung tissue. B. Statistical analysis of immunohistochemical staining for S6K1 in lung adenocarcinoma tissue microarray. C. OS (overall survival) curve shows that high expression of S6K1 associates with poor prognosis in lung adenocarcinoma patients. D. Representative immunohistochemical staining images for Mxi1 and S6K1 in two lung adenocarcinoma tissues. E. Statistical analysis of immunohistochemical staining for Mxi1 and S6K1 shows Mxi1 low expression correlates with S6K1 overexpression in lung adenocarcinoma tissue microarray. F. A proposed model shows mechanistically how Mxi1 is regulated by S6K1-mediated phosphorylation and β-Trcp-induced ubiquitination and degradation. The phosphorylation of Mxi1 at Ser160 by S6K1 leads to the binding of β-Trcp and results in Mxi1 degradation, which ultimately results in Myc activation and promotes cell radioresistance in lung cancer.
Discussion
Accumulating evidence support the role of Mxi1 as a tumor suppressor. In this study, we showed that S6K1 is a key kinase responsible for Mxi1 phosphorylation and downregulation. Our data support a working hypothesis in which S6K1 phosphorylates Mxi1 at Ser160 site to promote its binding to the β-Trcp ubiquitin ligase complex. This interaction subsequently leads to Mxi1 ubiquitination and degradation by β-Trcp. Moreover, the S6K1-dependent phosphorylation of Mxi1 regulates Myc-dependent transcription activity and cell radioresistance. Importantly, S6K1 overexpression correlates with Mxi1 downregulation and serves as a poor prognosis marker in human lung cancer samples, implying that targeting S6K1/Mxi1 pathway is a promising therapeutic strategy for lung cancer treatment.
As a key negative regulator of Myc oncogenic activities, Mxi1 is tightly regulated at multiple levels. Previous studies have demonstrated that several transcriptional factors could regulate Mxi1 at transcriptional level. For instance, Sp1, AP2 and HIF-1 induce Mxi1 at mRNA level 39, 40. In addition, some microRNAs are involved in the regulation of Mxi1 at translational level. For example, miR-191 controls erythroid enucleation by directly targeting Mxi1 41. Mxi1 is also considered as a direct target of miR-155 and suppresses glioma cell proliferation 42. In this study, we demonstrated that Mxi1 is also regulated at post-translational level. We observed that inhibition of kinase activity or protein level of S6K1 led to upregulation of Mxi1 at protein level. We demonstrated that Mxi1 is a bona fide substrate of S6K1 and the S6K1-dependent phosphorylation of Mxi1 is a major post-translational modification of Mxi1 responsible for Mxi1 degradation. Importantly, our functional analysis showed that phosphorylation of Mxi1 at Ser160 is critical for Mxi1 function in Myc transcriptional activation and cell radioresistance. Notably, previous study has reported that S6K1 and RSK could phosphorylate Mad1, a member of Mad family, at Ser145 and promote Mad1 degradation 43. It has long been known that Mad family consists of four related proteins: Mad1, Mxi1(also termed Mad2), Mad3 and Mad4, which function to inhibit the transcriptional activity of Myc 18. Our further study clearly demonstrated that the other three Mad family proteins (Mad1, Mad3 and Mad4) were also negatively controlled by S6K1 (Figure S5). Intriguingly, we observed that all four Mad proteins contain S6K1 binding motif (Figure S5). Accordingly, our data, combined with previous reports, strongly suggest that S6K1 overexpression promotes the transcriptional activation of Myc by destabilizing all Mad family proteins. Collectively, our work establishes an exciting crosstalk between S6K1 signaling pathway and Myc activation via the regulation of Mad family members, which occurs without affecting Myc protein level.
Moreover, our data also support a role of β-Trcp in this process. β-Trcp is a critical substrate-recognition subunit of the Cullin E3 ubiquitin ligase, which targets substrates for ubiquitination and degradation 44-46. It is well recognized that β-Trcp binds to its substrates only after they are properly phosphorylated 46. Our data demonstrate that S6K1-dependent phosphorylation of Mxi1 at Ser160 site results in its subsequent ubiquitination and degradation via β-Trcp E3 ligase, suggesting that S6K1 acts cooperatively with β-Trcp to control the stability of Mxi1. Given that all four Mad proteins contain β-Trcp binding motif (Figure S5), we speculated that other Mad family members (Mad1, Mad3 and Mad4) may also be recognized and degraded by β-Trcp ligase after phosphorylation by S6K1. Additionally, it is worth noting that the poly-ubiquitination of Mxi1-S160A was also detected (Figure 5A), indicating that there may be additional ubiquitin ligases that are also involved in the degradation of Mxi1 in this setting, and this requires further investigation.
Mxi1 is a potential tumor suppressor since it antagonizes the activity of Myc oncogene. However, very little is known about Mxi1 expression in cancers. Our work showed that Mxi1 is downregulated in lung cancer, which predicts poor prognosis. More importantly, we demonstrated a strong correlation between Mxi1 downregulation and overexpression of S6K1, an oncogene known to promote tumorigenesis 47, 48. In addition, we also found 21 tumors that contained both low levels of S6K1 and Mxi1, suggesting that other molecules, in addition to S6K1, may be involved in Mxi1 downregulation. Our findings clearly showed that β-Trcp binds to Mxi1 and promotes the degradation of Mxi1, suggesting the abundance of Mxi1 is also negatively regulated by β-Trcp. It has been reported that β-Trcp is overexpressed in multiple types of human cancer 49, 50, including lung cancer 51. Therefore, all of these results indicate that S6K1 acts cooperatively with β-Trcp to contribute to the decreased level of Mxi1 in lung cancer tissues. Collectively, our study provides a mechanism by which Mxi1 is downregulated in cancer, as proposed in Figure 7F. These data also provide important insights into the crosstalk between the mTOR/S6K1 signaling pathway and Myc activation. Several S6K1 inhibitors, such as PF-4708671, LY-2779964 and AD80, have been suggested to be effective in some advanced cancers 52, 53. In particular, the compound LY-2779964 has undergone a phase I clinical trial in patients with advanced cancers 54. Our work provides a strong rationale for the development of S6K1 inhibitors as anti-cancer agents in human lung cancer. Given that S6K1 is overexpressed in lung cancer tissues, we reason that targeting the S6K1/Mxi1 signaling pathway may be effective for treatment of lung cancers.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81301719 to S.X), the National Natural Science Foundation of China (81672979 to GW), National Key R&D Program of China (2016YFC1303800 to L.L), National Key R&D Program of China (2016YFC0106700 to G.W and S.Z), National Key R&D Program of China (2017YFC0909904 to Y.Z, S.S and S.X), the National Natural Science Foundation of China (81301732 and 81772821 to K.H), Guangdong Natural Science Foundation (2017A030313471 to K.H) and Department of Defense Era of Hope research scholar award (W81XWH-09-1-0409 to J.C).
Author contributions
S.X conceived and designed the study, S.X, G.W and J. C supervised the study, S.X, Y.H, K.H and S.Z performed experiments and analyzed the data, X.D, Z.Y, R.M, Y.Z, X.D, T.Z, K.Y, L.L, K.H, S.S and Y.Z provided advice and technical assistance, S.X wrote the manuscript. All authors have contributed to and approved the final manuscript.
Abbreviations
- Mxi1
MAX interactor 1
- NSCLC
non-small cell lung cancer
- PCR
polymerase chain reaction
- IHC
immunohistochemistry
- TAP
tandem affinity purification
- MS
mass spectrometry
- IP
immunoprecipitation
- OS
overall survival.
References
- 1.Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Fillmore CM, Hammerman PS. et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–46. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton TR, Gout IT. Functions and regulation of the 70kDa ribosomal S6 kinases. Int J Biochem Cell Biol. 2011;43:47–59. doi: 10.1016/j.biocel.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochemical J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 9.Tavares MR, Pavan IC, Amaral CL. et al. The S6K protein family in health and disease. Life Sci. 2015;131:1–10. doi: 10.1016/j.lfs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 11.Lipton JO, Yuan ED, Boyle LM. et al. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161:1138–51. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arif A, Terenzi F, Potdar AA. et al. EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature. 2017;542:357–61. doi: 10.1038/nature21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorrello NV, Peschiaroli A, Guardavaccaro D. et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Ding Q, Yen CJ. et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer cell. 2012;21:374–87. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stine ZE, Walton ZE, Altman BJ. et al. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–39. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–74. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Cascon A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res. 2012;72:3119–24. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 20.Taj MM, Tawil RJ, Engstrom LD. et al. Mxi1, a Myc antagonist, suppresses proliferation of DU145 human prostate cells. Prostate. 2001;47:194–204. doi: 10.1002/pros.1063. [DOI] [PubMed] [Google Scholar]
- 21.Manni I, Tunici P, Cirenei N. et al. Mxi1 inhibits the proliferation of U87 glioma cells through down-regulation of cyclin B1 gene expression. Br J Cancer. 2002;86:477–84. doi: 10.1038/sj.bjc.6600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber-Agus N, Meng Y, Hoang T. et al. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–7. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 23.Zeng X, Kinsella TJ. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res. 2008;68:2384–90. doi: 10.1158/0008-5472.CAN-07-6163. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Matsumoto M, Kitagawa K. et al. Chk1 phosphorylates the tumour suppressor Mig-6, regulating the activation of EGF signalling. EMBO J. 2012;31:2365–77. doi: 10.1038/emboj.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dehan E, Bassermann F, Guardavaccaro D. et al. betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y, Yamada N, Tanaka T. et al. Crucial roles of RSK in cell motility by catalyzing serine phosphorylation of EphA2. Nat Commun. 2015;6:7679. doi: 10.1038/ncomms8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, Li X, Gong Z. et al. Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol Cell Proteomics. 2014;13:2986–3000. doi: 10.1074/mcp.M113.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S, Wu Y, Chen Q. et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013;23:423–35. doi: 10.1038/cr.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Ma J, Lu Y. et al. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36:5321–5330. doi: 10.1038/onc.2017.161. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, Ma J, Li Y. et al. CDP138 silencing inhibits TGF-β/Smad signaling to impair radioresistance and metastasis via GDF15 in lung cancer. Cell Death Dis. 2017;8:e3036. doi: 10.1038/cddis.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Feng Z, Zhang M. et al. hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene. 2011;30:2219–29. doi: 10.1038/onc.2010.596. [DOI] [PubMed] [Google Scholar]
- 32.Kang T, Wei Y, Honaker Y. et al. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell. 2008;13:36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker W, Zhou ZQ, Ota S. et al. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J Cell Biol. 2005;169:405–13. doi: 10.1083/jcb.200411013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G, Xu G, Schulman BA. et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–56. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Liu P, Inuzuka H. et al. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahin F, Kannangai R, Adegbola O. et al. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–5. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 38.Maruani DM, Spiegel TN, Harris EN. et al. Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation. Oncogene. 2012;31:5073–80. doi: 10.1038/onc.2011.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benson LQ, Coon MR, Krueger LM. et al. Expression of MXI1, a Myc antagonist, is regulated by Sp1 and AP2. J Biol Chem. 1999;274:28794–802. doi: 10.1074/jbc.274.40.28794. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Gao P, Fukuda R. et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Flygare J, Wong P. et al. miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 2011;25:119–24. doi: 10.1101/gad.1998711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Wang W, Gao Z. et al. MicroRNA-155 promotes glioma cell proliferation via the regulation of MXI1. PLoS One. 2013;8:e83055. doi: 10.1371/journal.pone.0083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription byphosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci U S A. 2008;105:6584–9. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Dai X, Zhong J. et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao D, Lu X, Wang G. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542:484–8. doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura JL, Garcia E, Pieper RO. S6K1 plays a key role in glial transformation. Cancer Res. 2008;68:6516–23. doi: 10.1158/0008-5472.CAN-07-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ip CK, Wong AS. Exploiting p70 S6 kinase as a target for ovarian cancer. Expert Opin Ther Targets. 2012;16:619–30. doi: 10.1517/14728222.2012.684680. [DOI] [PubMed] [Google Scholar]
- 49.Ougolkov A, Zhang B, Yamashita K. et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96:1161–70. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 50.Shaik S, Nucera C, Inuzuka H. et al. SCF(beta-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J Exp Med. 2012;209:1289–307. doi: 10.1084/jem.20112446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J, Zhou W, Yang F. et al. The beta-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filbin MG, Dabral SK, Pazyra-Murphy MF. et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat Med. 2013;19:1518–23. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Feng X, Ennis KN. et al. Pharmacologic Targeting of S6K1 in PTEN-Deficient Neoplasia. Cell Rep. 2017;18:2088–95. doi: 10.1016/j.celrep.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolcher A, Goldman J, Patnaik A. et al. A phase I trial of LY2584702 tosylate, a p70 S6 kinase inhibitor, in patients with advanced solid tumours. Eur J Cancer. 2014;50:867–75. doi: 10.1016/j.ejca.2013.11.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.