Abstract
Hearing loss may place a heavy burden on the patient and patient's family. Given the high incidence of hearing loss among newborns and the huge cost of treatment and care (including cochlear implantation), prenatal diagnosis is strongly recommended. Termination of the fetus may be considered as an extreme outcome to the discovery of a potential deaf fetus, and therefore preimplantation genetic diagnosis has become an important option for avoiding the birth of affected children without facing the risk of abortion following prenatal diagnosis. In one case, a couple had a 7-year-old daughter affected by non-syndromic sensorineural hearing loss. The affected fetus carried a causative compound heterozygous mutation c.919-2 A>G (IVS7-2 A>G) and c.1707+5 G>A (IVS15+5 G>A) of the solute carrier family 26 member 4 gene inherited from maternal and paternal sides, respectively. The present study applied multiple displacement amplification for whole genome amplification of biopsied trophectoderm cells and next-generation sequencing (NGS)-based single nucleotide polymorphism haplotyping on an Ion Torrent Personal Genome Machine. One unaffected embryo was transferred in a frozen-thawed embryo transfer cycle and the patient was impregnated. To conclude, to the best of our knowledge, this may be the first report of NGS-based preimplantation genetic diagnosis (PGD) for non-syndromic hearing loss caused by a compound heterozygous mutation using an Ion Torrent Personal Genome Machine. NGS provides unprecedented high-throughput, highly parallel and base-pair resolution data for genetic analysis. The method meets the requirements of medium-sized diagnostics laboratories. With decreased costs compared with previous techniques (such as Sanger sequencing), this technique may have potential widespread clinical application in PGD of other types of monogenic disease.
Keywords: congenital deafness, non-syndromic hearing loss, preimplantation genetic diagnosis, next-generation sequencing
Introduction
Congenital deafness is one of the most common birth defects and its incidence rate is ~1-3‰ worldwide in 2003 (1). Ministry of Health figures from China identify 115,000 children under the age of 7 years with severe-to-profound deafness and 30,000 babies born each year with hearing impairment (2). Overall, hearing loss is usually categorized into 3 subgroups; sensorineural, conductive or mixed. The primary cause of hearing loss is attributed to genetic or environmental factors (3). Genetic transmission accounts for 50% of cases of congenital deafness, and of these, ~30% are syndromic and 70% are non-syndromic (4). Syndromic hearing loss (SHL) or non-(N) SHL refers to hearing loss with or without clinical symptoms, respectively (5). In total, ~77% of NSHL cases are due to autosomal recessive inheritance (6), 10–20% are autosomal dominant, 1% are X-linked and <1% are due to mitochondrial inheritance (7). In the Chinese population, extensive studies of deafness molecular epidemiology have demonstrated that a number of NSHL cases are caused by multiple mutated genes, including the gap junction protein β-2 gene (GJB2 gene), solute carrier family 26 member 4 (SLC26A4) gene (PDS gene) and mitochondrial gene, mitochondrially encoded 12S RNA (6).
There is frequent co-morbidity following cochlear implantation and the child/person remains handicapped (7). Cochlear implants are costly and therefore burden the affected families and patients (8). Given the increased incidence of hearing loss among newborns (1) and the huge cost of treatments and care, preventing this birth defect is strongly recommended. Pregnancy termination may be considered an extreme action taken for a deaf fetus, and therefore preimplantation genetic diagnosis (PGD) has become an important option for avoiding the birth of affected children without risking abortion following prenatal diagnosis (9,10). PGD is not only used for single gene disorders (SGDs) (8) but is also used for human leukocyte antigen matching (10) and inherited types of cancer (11). The primary goal of PGD is to aid parents who are attempting to conceive healthy offspring.
Current methods of PGD for SGDs are direct mutation detection using polymerase chain reaction (PCR) or Sanger sequencing; however, PGD accuracy and diagnostic efficiency have been limited due to amplification failures, allelic drop-out (ADO), mosaicism and contamination (12). Linkage analysis, including mutation detection combined with short tandem repeat (STR) identifier analysis, increases the efficiency of PGD diagnosis and confirms the genetic diagnosis (13). Linkage analysis based on STR is usually associated with choosing 3–8 genetic markers within 2 Mb of the mutation. Compared with STR analysis, karyomapping provides the analysis of more single nucleotide polymorphism (SNP) markers and has been used to diagnose SGDs in the clinic with high efficiency, accuracy and reliability (14–17). However, the dependence on DNA from family members may limit its application (18). If detailed information on affected relatives is not available, karyomapping may be performed in parallel with conventional PCR methods for direct detection of the mutation to increase the accuracy of the genetic diagnosis (19). With certain genes that have decreased SNP coverage, performing PCR testing (including STR analysis) or direct mutation detection in parallel is necessary (20).
Next-generation sequencing (NGS) is a rapidly developing technology that produces enormous amounts of data with a wide range of applications (21). PGD with NGS provides more informative genetic markers in high throughput (22), and whole genome amplification (WGA) provides novel possibilities for diagnosis and parameters for evaluation of SGDs. NGS also allows for analysis of aneuploidy or translocation of all chromosomes and mutations responsible for any single-gene disease, using one biopsy and one process (23). Furthermore, in the absence of suitable affected family members in SGD cases, NGS-based linkage analysis may still correctly diagnose the embryos by using the affected embryo as the proband.
The development of different NGS platforms and decreased costs enable their introduction into PGD (21). In the present study, Ion Torrent technology was used, which incorporates the use of non-optical, single-nucleotide, semiconductor-based sequencing on an Ion Torrent Personal Genome Machine (PGM). This technology combines aspects of parallel sequencing, including bead-based emulsion PCR but employs a complementary metal oxide semiconductor chip with microwells that serve as pH-sensitive pixels to detect the release of a hydrogen ion (registered as an electrical signal) when a nucleotide is incorporated during sequencing by synthesis (24). This approach suggests the requirement for chemiluminescent dyes, serial optical image acquisition, a motorized camera stage and extensive storage of preanalytic files for subsequent processing (25).
To the best of our knowledge, this is the first report of an NGS-based PGD case on an Ion Torrent PGM platform for non-syndromic sensorineural hearing loss caused by the SLC26A4 mutation.
Materials and methods
Patient information and ethics
A couple (maternal age, 30; paternal age, 31) who had a 7-year-old daughter affected by non-syndromic sensorineural hearing loss attended the Reproductive Medicine Center, Department of Obstetrics and Gynecology in the First Affiliated Hospital of Anhui Medical University (Anhui, China) for PGD. The result of target NGS revealed that the daughter carried a causative compound heterozygous mutation c.919-2 A>G (IVS7-2 A>G) and c.1707+5 G>A (IVS15+5 G>A) in intron 7 and 15 of the SLC26A4 gene. DNA testing of the couple using Sanger sequencing of SLC26A4 confirmed the presence of a maternal splicing mutation c.919-2 A>G and a paternal splicing mutation c.1707+5 G>A that were inherited by the child.
The couple was counseled about PGD and signed informed consent forms for intracytoplasmic sperm injection (ICSI) treatment and PGD. The Ethics Committee of Anhui Medical University approved the present study.
Pre-clinical test: NGS-based haplotyping of the family
Haplotyping for the pre-analytic testing of the family was performed using genomic DNA extracted from peripheral blood of the mother, father and child in order to establish the mutation-associated haplotype based on informative SNPs. This was achieved by using a magnetic genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China). A total of 160 high-frequency SNPs located 3 Mb upstream, 3 Mb downstream of SLC26A4 and 11 high-frequency SNP markers, including mutation sites in the SLC26A4 gene, were selected for NGS-based SNP haplotyping. These SNPs were then submitted to a primer design website (www.ampliseq.com). Following DNA purification (Agencourt AMPure XP; Beckman Coulter, Inc., Brea, CA, USA) the target region was amplified using multiplex PCR. The reaction contained 10 µl 2X Ion AmpliSeq™ Primer Pool, 4 µl 5X Ion AmpliSeq™ HiFi Master mix, 6 µl 10 ng DNA and nuclease-free water, and an Ion AmpliSeq™ Library kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used. The target area was amplified according to the following procedure: 99°C for 2 min for 1 cycle, 99°C for 15 sec and 60°C for 4 min for 21 cycles. Samples were then incubated at 4°C for the next step. The Amplicon Library Preparation protocol (using Ion AmpliSeq™ Library kits) was used as recommended by the supplier (Thermo Fisher Scientific, Inc.). Template preparation was performed using an Ion One Touch 2 system and an Ion One Touch Enrichment System in accordance to the manufacturers protocol (version 2.0; Ion Onetouch 200 Template kit; Thermo Fisher Scientific, Inc.). The template positive Ion Sphere Particles were sequenced on an Ion Torrent PGM (Thermo Fisher Scientific, Inc.) using the 318 chip following the protocol provided by the Ion Sequencing kit (version 2.0; Thermo Fisher Scientific, Inc.) (26). Informative SNPs located 3 Mb upstream and 2.5 Mb downstream of the SLC26A4 gene [Minor Allele Frequency, (MAF)>0.2] in the genomes of Han Chinese in Beijing and the Southern Han Chinese from the 1,000 Genomes Project were selected for NGS-based haplotyping.
ICSI procedure, embryo biopsy and vitrification
A standard pituitary downregulation protocol (26) was used for ovarian stimulation This was performed with Gonal-F (Merck Serono, Darmstadt, Germany) in a long-protocol cycle. When ≥2 follicles were >18 mm in diameter, 250 µg recombinant human chorionic gonadotropin (Ovitrelle; Merck Serono) was administered via hypodermic injection (27) and the oocytes were retrieved 36 h later, guided by transvaginal ultrasound. ICSI was performed for insemination of mature oocytes to avoid contamination by extraneous sperm. Fertilization was assessed 16–18 h after ICSI. A ~14 µm hole was made in the zona pellucida of the embryos using a ZILOS-tk laser (Hamilton Thorne, Inc., Beverly, MA, USA) on the morning of day 3 after ICSI. Blastocyst biopsy was performed on day 5 or 6 using the laser when the trophectoderm cells herniated out of the zona pellucida. The quality of blastocysts were scored between 1 and 6 according to their expansive degree and development condition of inner cell mass and trophectoderm outlined by Gardner's grading system (28). Categorization was as follows; 1, an early blastocyst with a blastocoels cavity <50% of the embryo volume; 2, a blastocyst with a blastocoels that is ≥50% of the embryo volume; 3, a blastocyst with a blastocoel that is 100% of the whole embryo; 4, an expanded blastocyst with a blastocoel filling the embryo and a thinning zona pellucida; 5, a hatching blastocyst with the trophectoderm beginning to extrude from the zona pellucida; and 6, a hatched blastocyst which has completely escaped from the zona pellucida. Additionally, according to the development condition of the inner cell mass and trophectoderm, the blastocyst inner cell mass and trophectoderm were graded (A-C). The inner cell mass was graded as follows: A, numerous cells packed tightly; B, several cells loosely grouped; and C, only very few cells. The trophectoderm was graded as follows: A, large numbers of cells forming a cohesive epithelium; B, few cells forming a loose epithelium; and C, very few large cells. Biopsied blastocysts were then vitrified using a Kitazato vitrification kit (Kitazato, Tokyo, Japan). The biopsied cells and fragments were each placed in 2 µl PBS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in 0.2 ml PCR tubes and multiple displacement amplification (MDA) was performed.
Multiple displacement amplification and NGS
MDA was performed using a REPLI-g single cell kit (Qiagen GmbH, Hilden, Germany). A total of 4 µl trophectoderm cells (supplied with PBS) were placed into a microcentrifuge tube and 3 µl prepared Buffer D2 (provided by the kit) was added. Following incubation at 65°C for 10 min, 3 µl stop solution was added. For each reaction, 40 µl master mix (9 µl H2Osc, 29 µl REPLI-g Reaction Buffer and 2 µl REPLI-g DNA Polymerase, from the REPLI-g single cell kit) was added to 10 µl obtained solution and incubated at 30°C for 8 h. The reaction was stopped by incubation at 65°C for 3 min to inactivate the Phi29 polymerase (REPLI-g sc DNA polymerase) and amplified DNA was stored at −20°C until required.
The following steps were similar to the construction of the haplotype map for the family. Library preparation was then performed and followed by chip loading for PGM (Thermo Fisher Scientific, Inc.) sequencing. The procedure was performed according to the manufacturer's protocol (Thermo Fisher Scientific, Inc.). The data from the PGM sequencing were analyzed by Beijing Jia Renhe Medical Technology Co., Ltd. (Beijing, China). Amniocentesis was performed at 19 weeks of gestation.
Results
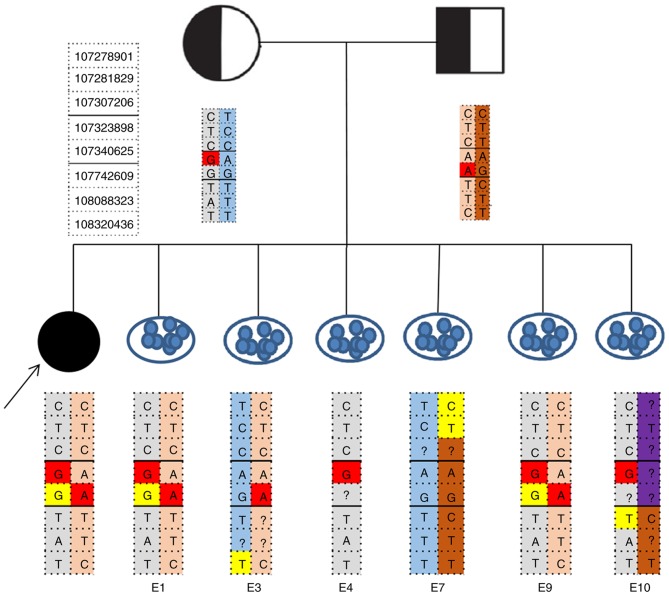
Haplotyping for the pre-analytic testing of the family was successfully performed. A total of 60 informative SNPs including mutation sites were identified in the family of NSHL. Following controlled ovarian stimulation, 14 oocytes were retrieved and 11 of them were suitable for ICSI; however, only 7 fertilized oocytes developed into hatched blastocysts, the remaining 4 were arrested. The 7 hatched blastocysts were biopsied successfully on day 5 or 6 of culture. Biopsied trophectoderm cells (5–10 cells) from embryo 1, 3, 4, 7, 9 and 10 were successfully amplified using MDA, whereas the biopsies from embryo 6 failed to amplify. All embryos were then analyzed by NGS. On the basis of the embryonic SLC26A4 genotypes results (Figs. 1 and 2), embryo 7 was genotypically normal, embryo 3 revealed a carrier pattern with the normal maternally inherited allele. Two embryos (embryo 1 and 9) were affected, inheriting affected parental haplotypes. Embryo 4, which inherited only the maternally affected haplotype, was diagnosed as monosomic. Paternal recombination occurred in embryo 10 and the diagnosis was ambiguous. The quality of the blastocyst was assessed based on Gardner's grading system (Table I) (28). No external DNA contamination events occurred in any tested embryos. Three months after oocyte retrieval, embryo 7, which was diagnosed as being normal, was transferred back to the mother in frozen-thawed embryo transfer cycle, resulting in a single ongoing pregnancy. Prenatal diagnosis by amniocentesis at 19 weeks demonstrated concordance of the embryo. Remaining embryos were cryopreserved.
Figure 1.
NGS-based SNP haplotyping for NSHL diagnosis. Gray and blue represented pathogenic and normal haplotype of the mother, respectively. Pink and dark orange represented pathogenic and normal haplotype of the father, respectively. The arrow indicated the proband that represented the daughter of the couple affected by non-syndromic sensorineural hearing loss. Gene mutation sites were marked in red. Allele drop-out sites were marked in yellow. Embryo 7 was genotypically normal, embryo 3 demonstrated a carrier pattern and embryos 1 and 9 were affected. Embryo 4 was diagnosed as monosomic. Paternal recombination occurred in embryo 10 (marked in purple). NSHL, non-syndromic hearing loss; NGS, next generation sequencing; SNP, single nucleotide polymorphism; E, embryo; ?, site was not detected.
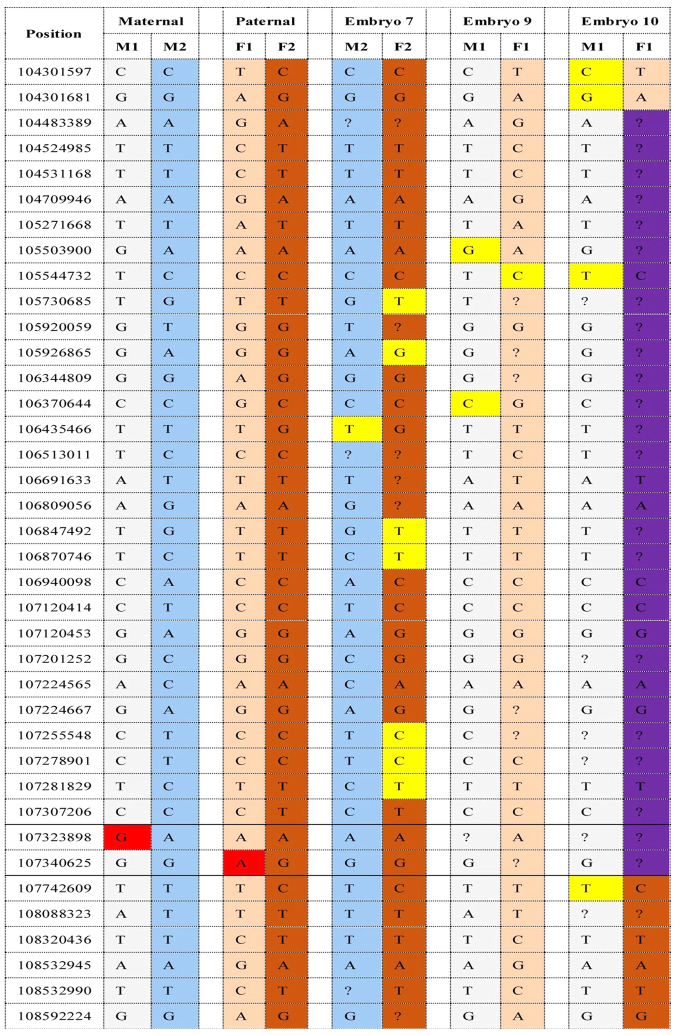
Figure 2.
Next gene sequencing-based single nucleotide polymorphism haplotyping for embryos 7, 9 and 10. Gray and blue represent pathogenic and normal haplotype of the mother, respectively. Pink and dark orange represented pathogenic and normal haplotype of the father, respectively. Embryo 7 was genotypically normal and embryo 9 was affected. Paternal recombination occurred in embryo 10 (marked in purple). Allele drop-out sites are marked in yellow and gene mutation sites are marked in red.
Table I.
The quality of biopsied blastocysts and result of haplotype analysis.
| Embryo | Quality of blastocystsa | Result |
|---|---|---|
| 1 | 5BB | Pathogenic |
| 3 | 5BB | Carrier |
| 4 | 6BB | Pathogenic, monosomic |
| 6 | 5BB | Amplification failure |
| 7 | 5BA | Normal |
| 9 | 5BB | Pathogenic |
| 10 | 5CC | Recombination |
Using Gardner's grading system.
Discussion
The present study may provide the first successful application of NGS-based PGD using a semiconductor technology, an Ion Torrent platform, for nonsyndromic sensorineural hearing loss caused by compound heterozygous mutation in SLC26A4.
Although hearing loss is not life-threatening, children with hearing loss may encounter multiple issues, given that spoken language is the predominant form of communication and social interaction (29). Adequate auditory stimulation and sufficient language exposure in early childhood are critical in their subsequent linguistic acquisition, cognitive development and psychosocial functioning (29). Hearing loss causes hearing disability, affects mental health and places a heavy burden on the patient's family and society. The 2006 National Sample Survey in China (30) demonstrated that hearing impairment affected ~20 million people, accounting for 24.16% of all people with disabilities. In 2011, ~30,000 babies are born with congenital hearing impairment annually in China and nearly 1/2 of these cases of congenital deafness are estimated to be associated with genetic factors (31). Children affected with nonsyndromic deafness using cochlear implants require more hospitalization for postoperative complications, including device extrusion requiring further surgery and wound infection (32).
Given the high incidence of hearing loss among newborns and huge cost of treatments and care (8), the medical genetic services in China strongly recommend to prevent this birth defect (1,30,31). PGD is now clinically established globally for preventing the birth defect. It may detect birth defects at the embryo stage, avoiding the trauma of prenatal diagnosis and the possibility of terminating affected pregnancies, which may be appropriate for deafness since abortion for deafness is viewed as a nonacceptable option for the majority of the population (9).
PGD for nonsyndromic deafness based on a single cell protocol (33,34) has been reported. However, the single cell protocol for PGD failed to avoid the misdiagnosis caused by ADO. PGD for nonsyndromic deafness based on Sanger sequencing and STR linkage analysis has been applied (9,35); however, the number of STR markers used for linkage analysis are limited and when a crossover event occurred, low density STR markers may still result in misdiagnosis.
To overcome increased ADO rates due to small amounts of genetic material and improve the accuracy of diagnosis, newly developed methods involving WGA and high-resolution methods, including karyomapping may be used in SGD-PGD.
Karyomapping, which uses genome-wide linkage to reveal the inheritance of genetic disease loci present in one or both parents, is efficient, accurate and reliable (17). However, it also has certain limitations. For example, the dependence on DNA samples from family members may limit its application (18). It is not appropriate for cases when detailed information of affected relatives is not available. In addition, when certain genes have decreased SNP coverage, PCR testing including STR analysis or direct mutation detection is necessary and need to be performed in parallel (18). Although karyomapping has the potential for providing a simultaneous identification of aneuploidy, it has not been validated for microdeletions (19) and cannot detect sequence-identical chromosome duplication that may result from malsegregation of chromosomes during the early cleavage divisions of the embryo (12). In addition, the cost of consumables required for karyomapping is significantly increased compared with that of the reagents required for conventional PGD methods.
Previously, NGS provided unprecedented high-throughput, highly parallel and base-pair resolution data for genetic analysis (36). It provides powerful application not only to molecular diagnostics but may also help advance research, due to an interest in long-term solutions, including prevention of disease/disabilities in future family members and advanced rehabilitation and therapeutics based on research outcome (37). Treff et al (36) applied NGS-based SNP haplotyping in PGD for single-gene disorders and provided blastocyst PGD results with consistency using established methodologies. Chen et al (22) reported PGD for the patient affected by congenital contractual arachnodactyly and spinal and bulbar muscular atrophy, illustrating the reliability of NGS-based SNP haplotyping. In the present study, mutations and high-frequency SNP markers were selected for haplotyping using NGS. The unaffected blastocyst was transferred to the patient who became pregnant.
Compared with karyomapping, NGS-based haplotying may still correctly diagnose the embryos using the affected embryo as a proband under the condition of the absence of affected family members (20). Comprehensive aneuploidy screening of blastocysts may also be performed simultaneously and NGS has the advantage of detecting chromosome microdeletion and microduplication (38,39).
To conclude, to the best of our knowledge, this is the first study in which NGS-based haplotying was combined with WGA and applied to PGD for nonsyndromic sensorineural hearing loss caused by compound heterozygous mutation of the SLC26A4 gene using an Ion Torrent platform. NGS combined with WGA applied in PGD may potentially offer a powerful means to prevent genetic transmission and therefore may benefit the family and society's health care system.
Acknowledgements
The authors would like to thank laboratory technicians, Lixian Xing and Yaohua Zhu, at Peking Jabrehoo Med Tech., Ltd. (Beijing, China) for their assistance with sequencing and data analysis. The present study was supported by the Key Science and Technology Project of Anhui Province (grant no. 1604a0802077) and the College Natural Science Project of Anhui Province (grant no. KJ2015A057).
References
- 1.White KR. Early hearing detection and intervention programs: Opportunities for genetic services. Am J Med Genet A. 2004;130A:1–36. doi: 10.1002/ajmg.a.30048. [DOI] [PubMed] [Google Scholar]
- 2.Liang Q, Mason B. Enter the dragon-China's journey to the hearing world. Cochlear Implants Int. 2013;14(Suppl 1):S26–S31. doi: 10.1179/1467010013Z.00000000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RJ, Bale JF, Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 4.Taneja MK. Preimplantation genetic diagnosis: Its role in prevention of deafness. Indian J Otolaryngol Head Neck Surg. 2014;66:1–3. doi: 10.1007/s12070-014-0711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahboubi H, Dwabe S, Fradkin M, Kimonis V, Djalilian HR. Genetics of hearing loss: Where are we standing now? Eur Arch Otorhinolaryngol. 2012;269:1733–1745. doi: 10.1007/s00405-011-1910-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu RM, Liu HJ, Cong JL, Sun AL, Du JD, Sun CM. Genetic characteristics of the couple with non-syndromic sensorineural hearing loss and fertility guidance. Int J Clin Exp Med. 2015;8:21746–21754. [PMC free article] [PubMed] [Google Scholar]
- 7.Park MK, Sagong B, Lee JD, Bae SH, Lee B, Choi KS, Choo YS, Lee KY, Kim UK. A1555G homoplasmic mutation from A1555G heteroplasmic mother with Pendred syndrome. Int J Pediatr Otorhinolaryngol. 2014;78:1996–1999. doi: 10.1016/j.ijporl.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Nadège C, Valérie G, Laura F, Hélène DB, Vanina B, Olivier D, Bernard F, Laurent M. The cost of cochlear implantation: A review of methodological considerations. Int J Otolaryngol. 2011;2011:210838. doi: 10.1155/2011/210838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altarescu G, Eldar-Geva T, Brooks B, Zylber-Haran E, Varshaver I, Margalioth EJ, Levy-Lahad E, Renbaum P. Preimplantation genetic diagnosis (PGD) for nonsyndromic deafness by polar body and blastomere biopsy. J Assist Reprod Genet. 2009;26:391–397. doi: 10.1007/s10815-009-9335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuliev A, Rechitsky S, Tur-Kaspa I, Verlinsky Y. Preimplantation genetics: Improving access to stem cell therapy. Ann N Y Acad Sci. 2005;1054:223–227. doi: 10.1196/annals.1345.028. [DOI] [PubMed] [Google Scholar]
- 11.Drüsedau M, Dreesen JC, Derks-Smeets I, Coonen E, van Golde R, van Echten-Arends J, Kastrop PM, Blok MJ, Gómez-García E, Geraedts JP, et al. PGD for hereditary breast and ovarian cancer: The route to universal tests for BRCA1 and BRCA2 mutation carriers. Eur J Hum Genet. 2013;21:1361–1368. doi: 10.1038/ejhg.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kou S. Preimplantation genetic diagnosis: An update on current technologies and ethical considerations. Reprod Med Biol. 2016;15:69–75. doi: 10.1007/s12522-015-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qubbaj W, Al-Swaid A, Al-Hassan S, Awartani K, Deek H, Coskun S. First successful application of preimplantation genetic diagnosis and haplotyping for congenital hyperinsulinism. Reprod Biomed Online. 2011;22:72–79. doi: 10.1016/j.rbmo.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, Coonen E, Dreesen JC, Stevens SJ, Paulussen AD, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014;16:838–845. doi: 10.1038/gim.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giménez C, Sarasa J, Arjona C, Vilamajó E, Martínez-Pasarell O, Wheeler K, Valls G, Garcia-Guixé E, Wells D. Karyomapping allows preimplantation genetic diagnosis of a de-novo deletion undetectable using conventional PGD technology. Reprod Biomed Online. 2015;31:770–775. doi: 10.1016/j.rbmo.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw MA, Griffin DK. Karyomapping: A universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47:651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 17.Thornhill AR, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, Harton G, Cliffe K, Affara N, Konstantinidis M, et al. Karyomapping - a comprehensive means of simultaneous monogenic and cytogenetic PGD: Comparison with standard approaches in real time for Marfan syndrom. J Assist Reprod Genet. 2015;32:347–356. doi: 10.1007/s10815-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natesan SA, Handyside AH, Thornhill AR, Ottolini CS, Sage K, Summers MC, Konstantinidis M, Wells D, Griffin DK. Live birth after PGD with confirmation by a comprehensive approach (karyomapping) for simultaneous detection of monogenic and chromosomal disorders. Reprod Biomed Online. 2014;29:600–605. doi: 10.1016/j.rbmo.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Konstantinidis M, Prates R, Goodall NN, Fischer J, Tecson V, Lemma T, Chu B, Jordan A, Armenti E, Wells D, Munné S. Live births following Karyomapping of human blastocysts: Experience from clinical application of the method. Reprod Biomed Online. 2015;31:394–403. doi: 10.1016/j.rbmo.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Ren Y, Zhi X, Zhu X, Huang J, Lian Y, Li R, Jin H, Zhang Y, Zhang W, Nie Y, et al. Clinical applications of MARSALA for preimplantation genetic diagnosis of spinal muscular atrophy. J Genet Genomics. 2016;43:541–547. doi: 10.1016/j.jgg.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Lukaszuk K, Pukszta S, Ochman K, Cybulska C, Liss J, Pastuszek E, Zabielska J, Woclawek-Potocka I. Healthy baby born to a robertsonian translocation carrier following next-generation sequencing-based preimplantation genetic diagnosis: A case report. AJP Rep. 2015;5:e172–e175. doi: 10.1055/s-0035-1558402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Diao Z, Xu Z, Zhou J, Wang W, Li J, Yan G, Sun H. The clinical application of preimplantation genetic diagnosis for the patient affected by congenital contractural arachnodactyly and spinal and bulbar muscular atrophy. J Assist Reprod Genet. 2016;33:1459–1466. doi: 10.1007/s10815-016-0760-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liss J, Chromik I, Szczyglińska J, Jagiełło M, Łukaszuk A, Łukaszuk K. Current methods for preimplantation genetic diagnosis. Ginekol Pol. 2016;87:522–526. doi: 10.5603/GP.2016.0037. [DOI] [PubMed] [Google Scholar]
- 24.Roy S, LaFramboise WA, Nikiforov YE, Nikiforova MN, Routbort MJ, Pfeifer J, Nagarajan R, Carter AB, Pantanowitz L. Next-generation sequencing informatics: Challenges and strategies for implementation in a clinical environment. Arch Pathol Lab Med. 2016;140:958–975. doi: 10.5858/arpa.2015-0507-RA. [DOI] [PubMed] [Google Scholar]
- 25.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Tenedini E, Artuso L, Bernardis I, Artusi V, Percesepe A, De Rosa L, Contin R, Manfredini R, Pellacani G, Giannetti A, et al. Amplicon-based next-generation sequencing: An effective approach for the molecular diagnosis of epidermolysis bullosa. Br J Dermatol. 2015;173:731–738. doi: 10.1111/bjd.13858. [DOI] [PubMed] [Google Scholar]
- 27.Drakopoulos P, Blockeel C, Stoop D, Camus M, de Vos M, Tournaye H, Polyzos NP. Conventional ovarian stimulation and single embryo transfer for IVF/ICSI. How many oocytes do we need to maximize cumulative live birth rates after utilization of all fresh and frozen embryos? Hum Reprod. 2016;31:370–376. doi: 10.1093/humrep/dev316. [DOI] [PubMed] [Google Scholar]
- 28.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: Towards a single blastocyst transfer. Fertil Steril. 2000;73:1155–1158. doi: 10.1016/S0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 29.Yin A, Liu C, Zhang Y, Wu J, Mai M, Ding H, Yang J, Zhang X. The carrier rate and mutation spectrum of genes associated with hearing loss in South China hearing female population of childbearing age. BMC Med Genet. 2013;14:57. doi: 10.1186/1471-2350-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang QJ, Zhao YL, Rao SQ, Guo YF, He Y, Lan L, Yang WY, Zheng QY, Ruben RJ, Han DY, Shen Y. Newborn hearing concurrent gene screening can improve care for hearing loss: A study on 14,913 Chinese newborns. Int J Pediatr Otorhinolaryngol. 2011;75:535–542. doi: 10.1016/j.ijporl.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 32.Postelmans JT, Cleffken B, Stokroos RJ. Post-operative complications of cochlear implantation in adults and children: Five years' experience in Maastricht. J Laryngol Otol. 2007;121:318–323. doi: 10.1017/S0022215106003471. [DOI] [PubMed] [Google Scholar]
- 33.Liss J, Mirecka A, Kitowska K, Lukaszuk K. Preimplantaion genetic diagnosis of hearing loss with 35delG mutation in GJB2 gene-preliminary report. Otolaryngol Pol. 2011;65:443–446. doi: 10.1016/S0030-6657(11)70738-7. (In Polish) [DOI] [PubMed] [Google Scholar]
- 34.Wu CC, Lin SY, Su YN, Fang MY, Chen SU, Hsu CJ. Preimplantation genetic diagnosis (embryo screening) for enlarged vestibular aqueduct due to SLC26A4 mutation. Audiol Neurootol. 2010;15:311–317. doi: 10.1159/000284349. [DOI] [PubMed] [Google Scholar]
- 35.Xiong W, Wang D, Gao Y, Gao Y, Wang H, Guan J, Lan L, Yan J, Zong L, Yuan Y, et al. Reproductive management through integration of PGD and MPS-based noninvasive prenatal screening/diagnosis for a family with GJB2-associated hearing impairment. Sci China Life Sci. 2015;58:829–838. doi: 10.1007/s11427-015-4936-y. [DOI] [PubMed] [Google Scholar]
- 36.Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT., Jr Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99:1377–1384.e6. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Idan N, Brownstein Z, Shivatzki S, Avraham KB. Advances in genetic diagnostics for hereditary hearing loss. J Basic Clin Physiol Pharmacol. 2013;24:165–170. doi: 10.1515/jbcpp-2013-0063. [DOI] [PubMed] [Google Scholar]
- 38.Watson CT, Marques-Bonet T, Sharp AJ, Mefford HC. The genetics of microdeletion and microduplication syndromes: An update. Annu Rev Genomics Hum Genet. 2014;15:215–244. doi: 10.1146/annurev-genom-091212-153408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo CD, Di Giacomo G, Cignini P, Padula F, Mangiafico L, Mesoraca A, D'Emidio L, McCluskey MR, Paganelli A, Giorlandino C. Comparative study of aCGH and Next Generation Sequencing (NGS) for chromosomal microdeletion and microduplication screening. J Prenat Med. 2014;8:57–69. [PMC free article] [PubMed] [Google Scholar]