Abstract
Landmark papers, even if they seem outdated, can still stimulate contemporary research and experiments to address new questions. Here is the tale of a classical experiment that inspired research over three decades.

Subject Categories: Membrane & Intracellular Transport, S&S: History & Philosophy of Science
Any PhD student or postdoc, who scrambles to write and submit a publication before getting scooped, can testify that science is a fast‐moving endeavor. Given our limited time and the ever‐increasing pace with which scientific studies are published, few students and postdocs—and PIs as a matter of fact—have time to keep up with the literature. “Reading” a manuscript often means just skimming through the abstract, having a quick glimpse at the figures and searching the PDF file for keywords of immediate interest. Notwithstanding these constraints, I would argue that the scientific literature is a treasure trove of information and ideas that go beyond contemporary papers to include classical publications. However, it can be difficult to motivate students and postdocs to read old landmark publications that reported groundbreaking discoveries and opened up new avenues of research. Many think that these papers are of merely historic interest and have little to contribute to scientists expected to use cutting‐edge methods to produce high‐impact publications.
Reading a manuscript often means just skimming through the abstract, having a quick glimpse at the figures and searching the PDF file for keywords of immediate interest.
However, classic papers still have a lot to offer. For once, they withstood the test of the time, as the reported results have shown to be correct and reproducible. This is unfortunately not the case for many high‐impact publications today, which, owing to the hype of selling it to the highest impact factor journal, just tell a cool story that in reality may be much less clear‐cut than reported. Furthermore, classic scientific publications often impress by a conceptual clarity and in many cases simplicity and thereby offer valuable lessons of how best science should be practiced. These are qualities I miss in many of today's papers that contain exceedingly large data sets from high‐throughput analyses and a dozen or more supplementary figures that no reader (or reviewer) is able to digest.
…classic scientific publications often impress by a conceptual clarity and in many cases simplicity…
To illustrate my point, I will discuss a 31‐year‐old paper that I consider to be such a classic study. It reported a groundbreaking discovery in the field of protein translocation, namely that that posttranslational import of mitochondrial proteins requires unfolding of the substrate 1. Furthermore, it has inspired various follow‐up experiments that further shed light on protein translocation.
My history of protein translocation
The reason I chose this specific paper is personal: 31 years ago, I was a PhD student working on the cytoskeleton of the parasitic protozoan Trypanosoma brucei in the laboratory of Thomas Seebeck at the University of Bern. My project was going well, I knew I wanted to stay in academia and was thinking about postdoc positions. This was when I read the paper “Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria” by Eilers and Schatz 1 for the first time. I was so fascinated that I knew protein translocation across membranes would be the scientific field I wanted to work on. Shortly afterward, I applied for a postdoc position in the Schatz laboratory at the Biocenter in Basel. This was quite naive as I was not aware at that time how famous Jeff Schatz was. Very surprisingly, since I wasn't too well prepared for the interview, I was offered a position and joined the Schatz laboratory in 1987 to start working on mitochondrial protein import in yeast. To my disappointment, the famous Eilers and Schatz experiment was not of great relevance for my postdoc project. Little did I know that, 31 years later, a variation of the experiment would help me to solve an important scientific question in my own laboratory.
In the following, I will discuss the long‐lasting impact of the Eilers and Schatz paper and present four of the many publications, which used variations of the experiment to answer important biological questions. As a disclaimer, the choice of these papers might not be representative as it was guided by my personal interests.
To unfold, or not to unfold, that is the question
In vitro systems allowing energy‐dependent import of substrate proteins into isolated mitochondria were already well established in 1986. However, not much was known about the mechanism of the process. At that time, the general rule was that proteins are cotranslationally transported across membranes, although there were a growing number of examples, including essentially all proteins imported into mitochondria, where protein folding occurred prior to membrane translocation. Thus, a key question was whether mitochondrial protein import requires unfolding of the substrate. How could this be addressed experimentally?
These experiments demonstrated for the first time that glycosomes are able to import folded proteins.
The basic concept behind the study of Eilers and Schatz is elegant and simple, and the consequences were far‐reaching (Fig 1A). The authors had the ingenious idea that high‐affinity binding of a ligand to the active center of an enzyme may stabilize its structure and prevent it from being unfolded. The enzyme they used was cytosolic dihydrofolate reductase (DHFR) from the mouse and the ligand the folate analogue methotrexate (MTX), a drug used to treat cancer and as an immunosuppressant. Eilers and Schatz expressed a chimaeric protein consisting of a N‐terminal presequence of the mitochondrial cytochrome oxidase subunit 4 (COX4) fused to mouse DFHR in Escherichia coli and purified it. Addition of this fusion protein (COX4‐DHFR) to isolated yeast mitochondria resulted in efficient import of the chimaeric protein and concomitant processing of the presequence. This was expected, since it had already been shown that a N‐terminal presequence is sufficient to transport non‐mitochondrial proteins into the matrix of mitochondria.
Figure 1. Graphical representation of the key experiments discussed here.
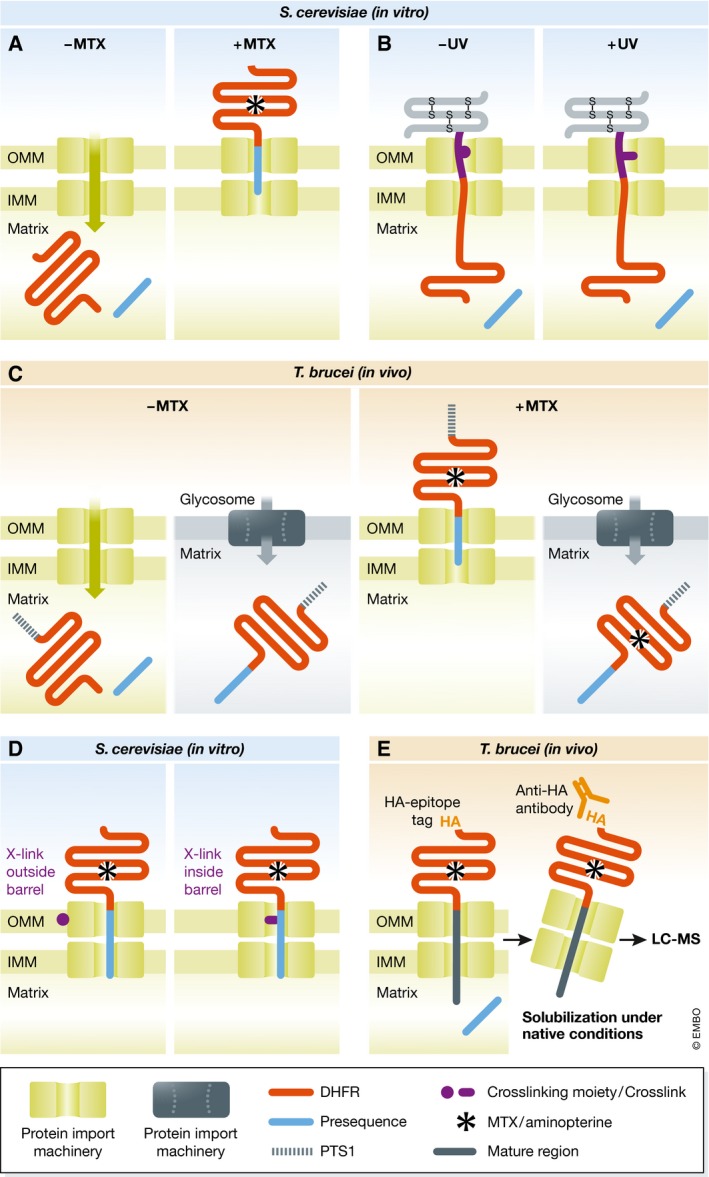
(A) Mitochondrial import of DHFR fused to a mitochondrial presequence was analyzed in the absence and presence of MTX which prevents its unfolding 1. (B) An import intermediate based on a chimaeric import substrate, consisting of a N‐terminal mitochondrial presequence fused to DHFR and a C‐terminal disulfide‐linked small protein with a trifunctional cross‐linker, allows the identification of the import machinery by photocross‐linking 2. (C) Import of a DHFR variant containing an N‐terminal mitochondrial presequence and a C‐terminal peroxisomal targeting signal (PTS1) into mitochondria and glycosomes, respectively, was analyzed in the absence and presence of the MTX‐analogue aminopterin 3. (D) Using MTX‐arrested DHFR fused to a mitochondrial presequence in mitochondria containing variants of Tom40 with strategically placed photocross‐linkable amino acids allows to retrace the path of the presequence in Tom40 at near atomic resolution 4. (E) Using an aminopterin‐arrested C‐terminally hemagglutinine epitope (HA)‐tagged DHFR fused to a mitochondrial presequence followed by a spacer consisting of the mature part of an imported protein enabled to identify the subunits of the protein import machineries by immunoprecipitation and subsequent differential mass spectrometric analysis 5.
When the same experiment was carried out in the presence of MTX, import was abolished. Moreover, DHFR became partially protease‐resistant, even though it was not imported into mitochondria and therefore, in principle, accessible to the protease (Fig 1A). Careful control experiments showed that MTX stabilized the enzyme in its 3D conformation, and created an import intermediate that spanned both mitochondrial membranes. The compelling conclusion of this elegant experiment was that translocation of proteins across the mitochondrial membranes indeed requires unfolding of the cargo proteins. The study also indicated that the mitochondrial protein import system must include an unfolding “enzyme”, which later was shown to be mitochondrial heat‐shock protein 70 that is peripherally associated with the inside of the inner membrane. Moreover, the paper had impact well beyond protein translocation: Active unfolding of proteins may also be required for other cellular processes such as proteolytic clearing of cytosolic proteins.
Catch me if you can
The Eilers and Schatz experiment provided the basis for another influential study in 1989 that identified the protein import channel in the mitochondrial outer membrane 2. Vestweber and Schatz constructed a tripartite fusion protein consisting of a C‐terminal small protein with internal disulfide bonds, a synthetic cross‐linker with a photoactivatable group, and DHFR with an N‐terminal presequence. The DHFR and the cross‐linker moieties of the fusion protein were imported into isolated mitochondria, whereas the small‐protein part could not be imported owing to the disulfide bonds (Fig 1B). The import substrate formed an import intermediate that, upon illumination, cross‐linked to the import channel, which allowed the identification of the first component of the membrane‐bound mitochondrial protein import machinery. The protein, initially termed import site protein 42, was later renamed Tom40 and shown to be a β‐barrel protein that forms the pore through which proteins are translocated across the outer membrane. Its discovery opened the way to the characterization of the entire translocase complex of the mitochondrial outer membrane (TOM complex), which consists of seven subunits and mediates import of essentially all mitochondrial proteins.
To unfold, or not to unfold, that is the question—part 2
Another variation of the experiment helped to characterize the protein import into peroxisomes, membrane‐bound organelles involved in oxidative processes. In contrast to mitochondria, it was thought these are able to import fully folded and even multimeric proteins. Häusler et al 3 used DHFR and aminopterin, a membrane permeable analogue of MTX, to test whether this also applies for glycosomes, a specialized peroxisome found in T. brucei and its relatives. In an elegant in vivo study, they showed that a DHFR fusion protein containing a N‐terminal mitochondrial and a C‐terminal peroxisomal targeting signal is imported into both mitochondria and glycosomes. Addition of aminopterin, which stabilizes the 3D conformation of the DHFR, greatly reduced mitochondrial import of the same substrate but did not affect its import into glycosomes (Fig 1C). These experiments demonstrated for the first time that glycosomes are able to import folded proteins.
Catch me if you can—part 2
The preliminary culmination of the Eilers and Schatz experiment was reached with the publication by Shiota et al in 2015, who resolved the path the presequence takes when it crosses Tom40 at near atomic resolution 4. Whereas Vestweber et al 2 used a cross‐linker attached to the substrate, Shiota et al placed an unnatural photoactivatable amino acid at 108 different positions of yeast Tom40.
The study by Shiota et al represents an amazing tour de force that provided unprecedented molecular details on the protein import mechanism…
Subsequent MTX‐induced stalling of DHFR‐containing import substrates in the β‐barrel allowed them to identify which of the 108 labeled residues of Tom40 interact with the substrate. Integration of these results with a computer model of the import channel finally showed that the positively charged presequence follows a path of aligned negatively charged patches inside the Tom40 pore, whereas a mitochondrial carrier protein takes a different route by interacting mainly with hydrophobic patches.
… methodological advances may offer, in some cases decades later, new ways of adapting the experiment.
The study also provided insight into the general architecture of the TOM complex since, depending on the position of the photoactivatable amino acid, Tom40 was not only cross‐linked to the arrested substrate but also to other TOM complex subunits. In the resulting model, three Tom40 molecules are linked to each other by three Tom22 subunits, each of which binds two Tom40 molecules. Moreover, the small Tom subunits Tom5, Tom6, and Tom7 were shown to bind at the periphery of the pore molecules. The study by Shiota et al represents an amazing tour de force that provided unprecedented molecular details on the protein import mechanism and on the TOM complex architecture at near atomic resolution.
And now for something completely different
A couple of years ago my laboratory decided to characterize the mitochondrial protein import system of T. brucei as previous bioinformatic analyses had indicated that it must be quite unique. This was surprising and highly interesting, but it also complicated the characterization of the system.
That was when we realized that our own variant of the Eilers and Schatz experiment, the one that prompted me to study mitochondrial biogenesis in the first place 31 years ago, could help us to identify the unique subunits of the trypanosomal protein import machineries. Using an epitope‐tagged DHFR‐based import substrate that was arrested in the import channel by aminopterin, followed by immunoprecipitation and proteomic analysis, allowed us to identify the seven known subunits of outer membrane protein translocase along with six previously unknown subunits of the translocase of the inner membrane (TIM complex) 5. Moreover, using a carrier protein that was arrested in the import pathway, we showed that T. brucei, in contrast to other eukaryotes, has only a single TIM complex that mediates import of presequence‐containing as well as of carrier proteins. Thus, the Eilers and Schatz experiment, in combination with cutting‐edge proteomic methods, can still be used to characterize a protein import system without prior knowledge of any of its subunits. All one needs to know is at least one substrate that is transported by the system and that unfolding is required for transport.
Conclusions
I hope that I could convince at least a few readers that it might be worthwhile to read old landmark papers. The Eilers and Schatz publication, together with many other studies derived from it, nicely illustrates that variations of classical experiments may be applicable to novel biological questions. Moreover, methodological advances may offer, in some cases decades later, new ways of adapting the experiment. I am convinced that we are not at the end yet and that future generations of scientists will find other ways of how to use DHFR and its ligand to probe protein translocation and to investigate new problems we do not even think about now.
Conflict of interest
The author declares that he has no conflict of interest.
References
- 1. Eilers M, Schatz G (1986) Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322: 228–232 [DOI] [PubMed] [Google Scholar]
- 2. Vestweber D, Brunner J, Baker A, Schatz G (1989) A 42K outer‐membrane protein is a component of the yeast mitochondrial protein import site. Nature 341: 205–209 [DOI] [PubMed] [Google Scholar]
- 3. Häusler T, Stierhof Y‐D, Wirz E, Clayton C (1996) Import of a DHFR hybrid protein into glycosomes in vivo is not inhibited by the folate‐analogue aminopterin. J Cell Biol 132: 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M et al (2015) Molecular architecture of the active mitochondrial protein gate. Science 349: 1544–1548 [DOI] [PubMed] [Google Scholar]
- 5. Harsman A, Oeljeklaus S, Wenger C, Huot JL, Warscheid B, Schneider A (2016) The non‐canonical mitochondrial inner membrane presequence translocase of trypanosomatids contains two essential rhomboid‐like proteins. Nat Commun 19: 13707 [DOI] [PMC free article] [PubMed] [Google Scholar]


