Abstract
The bacterial cell wall is an important and highly complex structure that is essential for bacterial growth because it protects bacteria from cell lysis and environmental insults. A typical Gram-positive bacterial cell wall is composed of peptidoglycan and the secondary cell wall polymers, wall teichoic acid (WTA) and lipoteichoic acid (LTA). In many Gram-positive bacteria, LTA is a polyglycerol-phosphate chain that is decorated with d-alanine and sugar residues. However, the function of and proteins responsible for the glycosylation of LTA are either unknown or not well-characterized. Here, using bioinformatics, genetic, and NMR spectroscopy approaches, we found that the Bacillus subtilis csbB and yfhO genes are essential for LTA glycosylation. Interestingly, the Listeria monocytogenes gene lmo1079, which encodes a YfhO homolog, was not required for LTA glycosylation, but instead was essential for WTA glycosylation. LTA is polymerized on the outside of the cell and hence can only be glycosylated extracellularly. Based on the similarity of the genes coding for YfhO homologs that are required in B. subtilis for LTA glycosylation or in L. monocytogenes for WTA glycosylation, we hypothesize that WTA glycosylation might also occur extracellularly in Listeria species. Finally, we discovered that in L. monocytogenes, lmo0626 (gtlB) was required for LTA glycosylation, indicating that the encoded protein has a function similar to that of YfhO, although the proteins are not homologous. Together, our results enable us to propose an updated model for LTA glycosylation and also indicate that glycosylation of WTA might occur through two different mechanisms in Gram-positive bacteria.
Keywords: Gram-positive bacteria, Bacillus, Staphylococcus aureus (S. aureus), cell wall, glycosyltransferase, bacterial glycobiology, lipoteichoic acid, wall teichoic acid, teichoic acid, glycosylation
Introduction
The bacterial cell wall is a highly complex and very important structure; it maintains the cell shape and protects bacteria from cell lysis and environmental insults. The main cell wall components present in Gram-positive bacteria, such as Bacillus subtilis, Listeria monocytogenes, and Staphylococcus aureus, are peptidoglycan and teichoic acids. Teichoic acids are anionic carbohydrate-containing polymers that are present in two forms: wall teichoic acid (WTA),4 which is covalently linked to the N-acetylmuramic acid residues of the peptidoglycan polymer, and lipoteichoic acid (LTA), which is embedded in the cytoplasmic membrane via a lipid anchor (1–3). Recent studies have shown that WTA is crucial for the virulence and β-lactam resistance of S. aureus and L. monocytogenes (4–6), whereas LTA is important for cell viability and cell division in these human pathogens (7, 8). In the soil bacterium B. subtilis, the absence of LTA affects divalent cation homeostasis and leads to increased sensitivity to diverse antibiotics and lysozyme, and the absence of WTA leads to drastic morphological alterations (9). It has also been shown that the deficiency in LTA synthesis leads to smaller colony sizes due to a failure in the execution of the colony developmental program in B. subtilis (10). Additionally, B. subtilis cells lacking both WTA and LTA are not viable, reflecting the importance of these cell polymers (9). Due to the impact of WTA and LTA on cell viability and virulence, the enzymes required for their synthesis are considered suitable targets for the development of new antimicrobial compounds (11–13).
The biosynthesis of LTA has been extensively studied in B. subtilis, S. aureus, and L. monocytogenes (3, 8, 14–17). B. subtilis produces type I LTA, which is composed of an unbranched 1–3-linked polyglycerol-phosphate (GroP) backbone chain that is attached to the outer layer of the bacterial membrane via a glycolipid anchor (14, 16). Each GroP subunit can be modified with d-alanine or GlcNAc residues (18–21). L. monocytogenes also produces a type I LTA; however, in this organism, the GroP subunits are substituted with d-alanine and galactose residues (22, 23). The enzymes required for the d-alanylation of LTA are encoded by the dltABCD operon and have been characterized in a number of studies (2, 24, 25). In contrast to the d-alanylation process, little is known about the enzymes responsible for LTA glycosylation. Fischer and others proposed a model for the addition of sugar residues to LTA based on biochemical studies performed three decades ago (21, 26–28). According to this model, a cytoplasmic glycosyltransferase (GT) uses a nucleotide-activated sugar to form a C55-P sugar intermediate, which is subsequently transported across the membrane by a flippase enzyme. The sugar molecule is subsequently transferred onto LTA by the action of a second GT (27, 28). As the polyglycerol-phosphate backbone of LTA is polymerized on the outside of the cell, this final step needs to be catalyzed by a GT with an extracellular active site. Recently, GtlA (locus tag Lmo0933 in strain EGD-e) has been identified as the putative cytoplasmic GT involved in the glycosylation process of LTA in the L. monocytogenes strain 10403S (29), although biochemical evidence for such an activity is still lacking. This protein is anchored by two C-terminal transmembrane helices to the membrane and contains a large N-terminal cytoplasmic enzymatic domain. NMR analysis of LTA produced by a gtlA mutant strain confirmed the absence of galactose modifications. Additionally, cell extracts obtained from the gtlA mutant strain showed a stronger LTA signal on western blots using a polyglycerol phosphate–specific antibody as compared with a WT strain, suggesting that the LTA backbone structure is better recognized by the antibody in the absence of sugar modifications (29). GtlA belongs to the GT2 family of glycosyltransferases and is characterized by a GT-A fold. GT-A fold glycosyltransferases assume a Rossmann fold with seven or more β-sheets, which is typical for proteins that bind nucleotides (30, 31); in L. monocytogenes, the likely substrate of GtlA is UDP-galactose. A second feature of enzymes with a GT-A fold is the presence of a conserved DXD motif, which interacts with the phosphate group of the nucleotide-activated sugar. This interaction requires a divalent cation, which in many cases is a Mn2+ ion (31–33). However, the remaining proteins required for the glycosylation of LTA in L. monocytogenes are still unknown. Also, none of the enzymes involved in the LTA glycosylation process of B. subtilis, including the enzyme required for the production of the C55-P sugar intermediate, the enzyme required for the transfer of this intermediate from the inner to the outer leaflet of the membrane, or the enzyme that transfers the sugar residue to the polyglycerol backbone, have been identified. Here, we show that deletion of the csbB and yfhO genes in B. subtilis led to a loss of sugar modifications on LTA. Interestingly, the L. monocytogenes YfhO homolog, Lmo1079, is not involved in LTA but WTA glycosylation. Instead, we found that the absence of Lmo0626 (here renamed GtlB) has an impact on LTA glycosylation, and we hypothesize that this protein performs the extracellular LTA glycosylation step in L. monocytogenes. With this, not only did we discover additional genes required for LTA glycosylation, but the work also allowed us to propose an alternative, extracellular, glycosylation mechanism for wall teichoic acid.
Results
CsbB is required for LTA glycosylation in B. subtilis
We have recently reported that the annotated glycosyltransferase GtlA probably catalyzes the first step of the LTA glycosylation process in L. monocytogenes (29). However, the enzymes involved in this process in other bacteria, including B. subtilis, remain unknown. To identify proteins required for LTA glycosylation in B. subtilis, the L. monocytogenes GtlA protein sequence was used as a query sequence in a BLASTP search against the B. subtilis 168 genome. This identified three homologs, YkcC (e-value: 1e−158), CsbB (e-value: 5e−68), and YkoT (e-value: 1e−60), all of which are encoded in a two-gene operon (Fig. 1A). Analysis using the Pfam database (http://pfam.xfam.org/)5 indicated that YkcC, CsbB, and YkoT encode GT-A fold family 2 glycosyltransferases, which are known to transfer sugar moieties from nucleotide-activated sugars, such as UDP-glucose, UDP-GlcNAc, or UDP-galactose to a variety of substrates, including the lipid carrier C55-P (34–36). To assess whether YkcC, CsbB, and YkoT are involved in the LTA glycosylation process in B. subtilis, ykcC, csbB, and ykoT deletion strains were constructed by replacing the respective gene in B. subtilis 168 with an antibiotic resistance marker. To determine whether deletion of one of these genes impacts LTA synthesis, anti-LTA western blot analysis was performed on cell extracts derived from the WT and mutant B. subtilis strains. The LTA isolated from strain 168ΔcsbB yielded a stronger signal as compared with the WT strain (Fig. 1B), indicating that the structure or the amount of the LTA polymer is changed in the absence of CsbB. In contrast, the anti-LTA signal for strains with a ykcC or ykoT deletion was indistinguishable from that seen for the WT (Fig. 1B). To determine the chemical structure of LTA in the WT and mutant strains, the polymer was isolated and analyzed by 1D 1H NMR. LTA purified from WT B. subtilis 168 showed the expected spectrum; peaks derived from the CH2 groups of the GroP backbone (colored green in Fig. 2A), the CH2 and CH3 groups of the fatty acid chain (colored orange in Fig. 2A), the non-exchangeable protons from the d-Ala substitutions (colored blue in Fig. 2A), and the GlcNAc modification (colored yellow in Fig. 2A) could be assigned, as described previously (29, 37–39). The spectra, including the peaks for the nonexchangeable protons derived from the GlcNAc residues, were essentially identical for the LTA isolated from the ykcC and ykoT deletion strains to that of the WT strain, indicating that the proteins encoded by these two genes are not involved in the LTA glycosylation process in B. subtilis under the conditions used (Fig. S1). In contrast, the GlcNAc-specific peaks were absent in the NMR spectra obtained from the LTA isolated from the csbB mutant (Fig. 2B). To confirm that the phenotype is due to inactivation of csbB, a copy of csbB expressed from its native promoter was introduced into the amyE locus of the csbB mutant. The LTA western blot signal (Fig. 1B) and NMR peaks corresponding to GlcNAc (Fig. 2C) were restored to WT levels in the complementation strain. These results highlight that CsbB is required for the decoration of LTA with GlcNAc residues in B. subtilis during vegetative growth. Although biochemical evidence is still lacking, we presume that CsbB functions as the cytoplasmic LTA glycosyltransferase.
Figure 1.
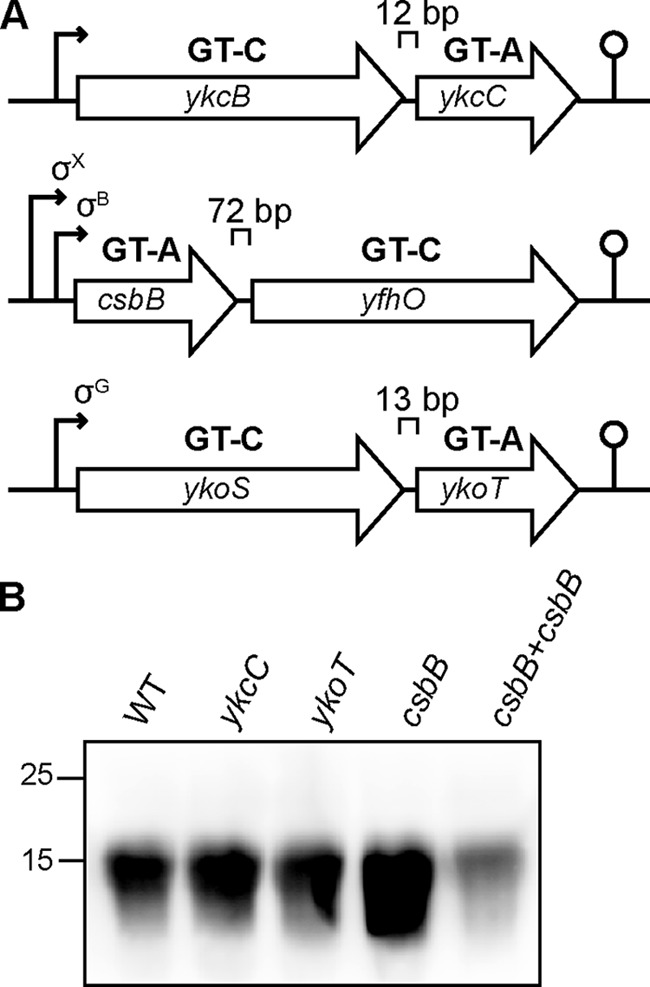
B. subtilis operons coding for L. monocytogenes GtlA homologs. A, B. subtilis operons coding for predicted GT-A-fold glycosyltransferases YkcC, CsbB, and YkoT with homology to the L. monocytogenes GtlA enzyme and the predicted GT-C-fold glycosyltransferases YkcB, YfhO, and YkoS, respectively. The distance between adjacent genes is given in base pairs (bp), and the gene orientation is indicated by the arrowheads. Promoters are indicated by black arrows, and transcription terminators are indicated by a loop structure. The ykcBC operon is part of the YclJ and YrkP regulon (82, 83). Earlier studies indicated that the csbB-yfhO operon is expressed from σX- and σB-dependent promoters and that the ykoST operon is expressed from a σG-dependent promoter (40, 48, 71, 84). B, LTA analysis by western blotting. Cell extracts of B. subtilis strains 168 (WT), ykcC, ykoT, csbB mutants, and the complementation strain csbB+csbB were prepared and separated on a 15% SDS-polyacrylamide gel. LTA was detected by western blotting using a humanized monoclonal LTA-specific antibody and an HRP-linked anti-human antibody. A representative blot from four independent experiments is shown.
Figure 2.
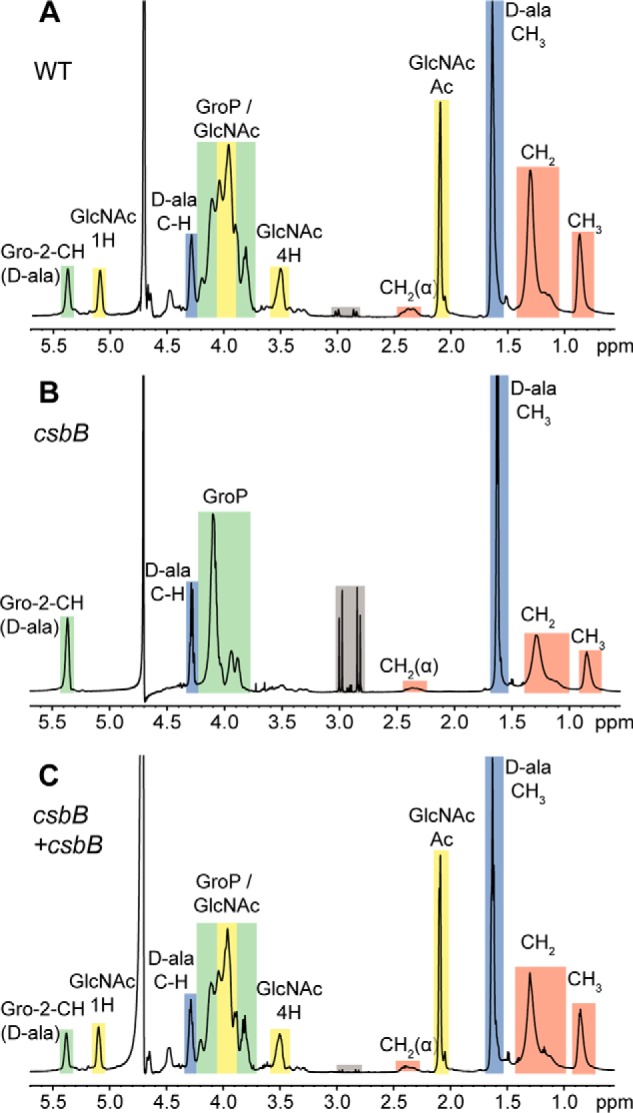
NMR analysis of LTA isolated from WT B. subtilis 168, csbB mutant, and complementation strains. Shown are NMR spectra of LTA derived from B. subtilis strains 168 (WT) (A), csbB mutant (B), and the csbB+csbB complementation strain (C). Colored boxes and labels indicate nonexchangeable protons derived from the different LTA components. Peaks were assigned as described previously (17, 37–39). The different peaks for the protons and acetyl group of GlcNAc are labeled with 1H, 4H, and Ac, respectively. Gray boxes highlight peaks resulting from residual citrate, a buffer component used during the LTA purification procedure. The spectra are representative of three independent experiments.
The B. subtilis YfhO protein is involved in the LTA glycosylation process in B. subtilis
Analysis of the genomic ykcC, csbB, and ykoT regions highlighted that all three genes are encoded in an operon with a second gene, namely ykcB, yfhO, and ykoS, respectively (Fig. 1A). YkcB, YfhO, and YkoS show similarities to members of the GT-C family glycosyltransferases, which are characterized by 8–13 transmembrane helices and a DXD or modified (DXE, EXD, DDX, or DEX) motif in an extracellular loop (34). GT-C enzymes are good candidate enzymes for transferring the glycosyl group from the lipid intermediate to the LTA backbone on the outside of the cell. Because CsbB is required for LTA glycosylation in B. subtilis, we hypothesized that YfhO, the predicted GT-C enzyme encoded in the same operon, could also be involved in the LTA glycosylation process. To test this, a yfhO mutant was constructed, and the LTA isolated from this strain was analyzed by western blotting. Similar to the csbB mutant, the LTA signal for the yfhO deletion strain was stronger than that of the WT strain (Fig. 3). This phenotype could be complemented by expressing the complete csbB-yfhO operon from the native promoter from the amyE locus in the yfhO mutant strain (yfhO+csbB-yfhO; Fig. 3). To ensure that the complementation does not result from the overexpression of csbB, the csbBD97A-yfhO operon, which produces a non-functional CsbB variant, was constructed and introduced into the yfhO mutant and the csbB mutant. Previous work had shown that the CsbBD97A variant, in which the second Asp residue of the conserved DXD motif predicted to be required for binding of a divalent cation is mutated, is inactive (40). An increased LTA signal was observed on western blots for cell extracts obtained from the B. subtilis csbB+csbBD97A-yfhO strain, and no GlcNAc-specific peaks were observed by NMR analysis of purified LTA, confirming the inactivity of CsbBD97A (Fig. 3 and Fig. S2). On the other hand, introduction of the csbBD97A-yfhO operon into the yfhO background strain led to partial complementation of the phenotype (Fig. 3). These data indicate that YfhO is involved in the LTA glycosylation process in B. subtilis. To investigate this further, LTA was isolated from the yfhO mutant and the yfhO+csbBD97A-yfhO complementation strain, as well as the WT control strain, and analyzed by NMR. GlcNAc-specific peaks were absent in the sample derived from the yfhO deletion strain, but again present in the complementation strain (Fig. 4). We also constructed B. subtilis ykcB and ykoS mutants, with deletions in the other two genes coding for predicted GT-C–like glycosyltransferases. However, LTA glycosylation was not abolished in these strains under the conditions tested (Fig. S3). Taken together, these data show that YfhO is involved in the LTA glycosylation process in B. subtilis. Although biochemical evidence is still lacking, we hypothesize that the B. subtilis YfhO protein probably acts as extracellular GT-C-type glycosyltransferase and moves the GlcNAc sugar moieties from a C55-P intermediate onto the LTA backbone on the outside of the cell.
Figure 3.
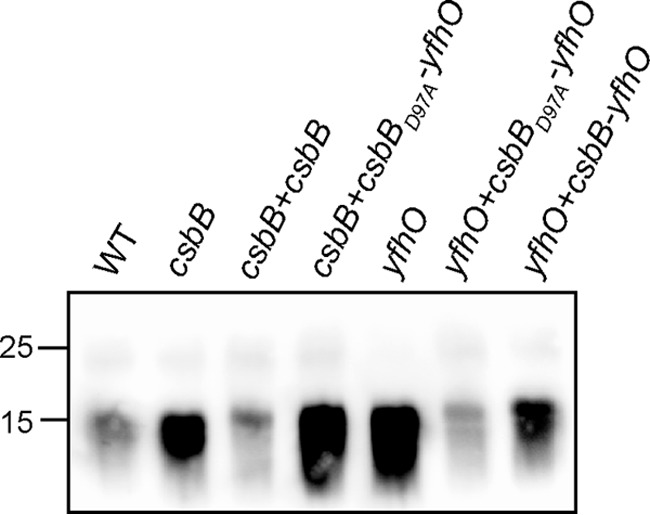
LTA production in WT B. subtilis 168, csbB, and yfhO deletion and complementation strains. Cell extracts were prepared from overnight cultures of B. subtilis 168 (WT), csbB, csbB+csbB, csbB+csbBD97A-yfhO, yfhO, yfhO+csbBD97A-yfhO, and yfhO+csbB-yfhO mutant and complementation strains and separated on a 15% SDS-polyacrylamide gel. The LTA was detected by western blotting using a humanized monoclonal LTA-specific antibody and an HRP-linked anti-human antibody. A representative blot from five independent experiments is shown.
Figure 4.
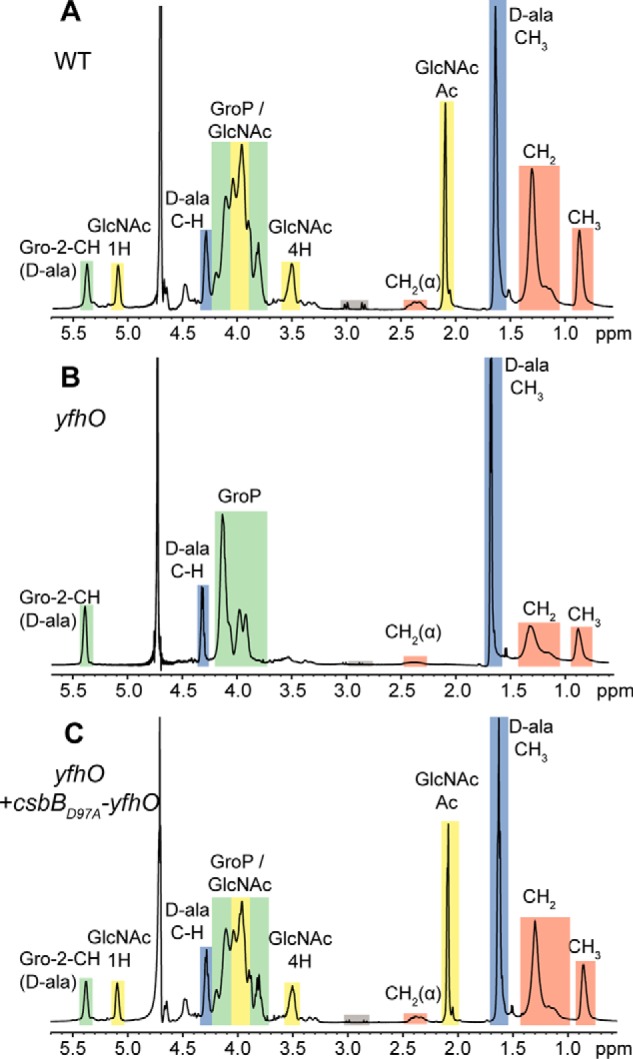
NMR analysis of LTA isolated from B. subtilis wildtype 168, yfhO mutant, and complementation strains. A–C, NMR spectra of LTA obtained from B. subtilis 168 (WT) (A), the yfhO mutant (B), and the yfhO+csbBD97A-yfhO complementation strain (C). Colored boxes and labels indicate nonexchangeable protons derived from the different LTA components. Peaks were assigned as described previously (17, 37–39). The different peaks for the protons and acetyl group of GlcNAc are labeled with 1H, 4H, and Ac, respectively. Gray boxes highlight peaks resulting from residual citrate, a buffer component used during the LTA purification procedure. The spectra are representative of three independent experiments. The spectrum for the B. subtilis 168 (WT) LTA is the same as shown in Fig. 2A.
LTA glycosylation has no impact on chain length and percentage of d-alanine substitutions in B. subtilis
The polymerization of the LTA backbone takes place on the outside of the cell, and hence the transfer of d-alanine and sugar residues must also take place on the outside of the cell. Furthermore, individual glycerol-phosphate subunits are modified with either a d-alanine or sugar residue but not with both. It is conceivable that the LTA polymerization and modification processes are coordinated and that the absence of sugar modifications could impact the amount of d-alanine substitutions or even the chain length of LTA, in case sugar residues are introduced while the LTA backbone is still polymerized. To determine whether the chain length and/or the percentage of d-alanine modifications are changed in the absence of sugar modifications, the peaks of the NMR spectra were integrated, and the LTA chain length and percentage modification were calculated using a previously published method and as described under “Experimental procedures” (29). For each strain, three NMR spectra obtained from three independent LTA extractions were analyzed. The LTA isolated from WT B. subtilis 168 had a calculated chain length of 17.74 ± 1.04 GroP units, and 62.07 ± 11.32 and 27.05 ± 1.79% of the GroP residues were modified with d-alanine and GlcNAc residues, respectively (Fig. 5). No significant difference in chain length (21.62 ± 2.46 and 20.16 ± 2.19 GroP units; Fig. 5A) or percentage of d-alanine substitutions (63.89 ± 12.92 and 67.95 ± 13.15%; Fig. 5B) was observed for the csbB or yfhO mutant strain. The percentage of GlcNAc residues in the csbB+csbB and yfhO+csbBD97A-yfhO complementation strains were 33.74 ± 3.21 and 34.33 ± 1.22% and therefore comparable with the percentage of sugar substitutions present in the LTA isolated from WT B. subtilis strain 168 (Fig. 5B). Taken together, these data suggest that the LTA chain length and the amount of d-alanine substitutions are not affected by the presence or absence of sugar modifications in B. subtilis 168 when grown under standard laboratory conditions.
Figure 5.
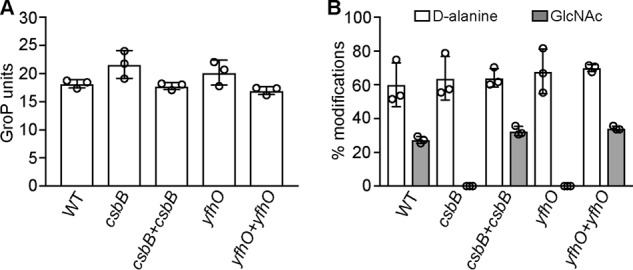
Determination of LTA chain length and percentages of d-alanine and GlcNAc substitutions. The peaks in the NMR spectra obtained from LTA isolated from B. subtilis strains 168 (WT), csbB, csbB+csbB, yfhO, and yfhO+csbBD97A-yfhO (yfhO+yfhO) were integrated, and the chain length (A) and percentages of d-alanine and GlcNAc substitutions (B) were determined. The average values and S.D. (error bars) from three independent experiments were determined and plotted. Two-tailed unpaired t tests identified only significant differences for the percentage of GlcNAc substitution between the WT B. subtilis 168 and csbB or yfhO mutant strains with p values < 0.05.
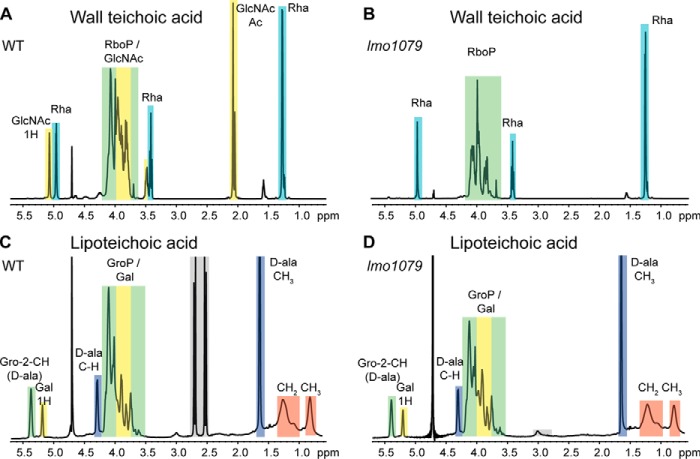
The L. monocytogenes YfhO homolog Lmo1079 is involved in WTA but not LTA glycosylation
So far, the only protein known to be involved in the LTA glycosylation process in L. monocytogenes is GtlA, an annotated GT with a predicted cytoplasmic catalytic domain (29). To identify additional L. monocytogenes proteins involved in the LTA glycosylation process, we performed a homology search using the B. subtilis YfhO protein as a query sequence. This yielded Lmo1079 as the only close homolog with an e-value of 1e−39. In previous work, it has been shown that deletion of lmo1079 prevents binding of the phage-derived protein CBDP35, which recognizes terminal GlcNAc residues on the bacterial surface, to L. monocytogenes EGD-e cells (41). LTA in L. monocytogenes is modified with galactose residues, whereas terminal GlcNAc moieties are found on WTA in L. monocytogenes strains EGD-e and 10403S. Therefore, these previous findings indicate that Lmo1079 is probably required for the glycosylation of WTA with GlcNAc residues (41), but the structure of WTA produced by an lmo1079 deletion strain has not yet been analyzed. To specifically determine the role of Lmo1079 in WTA glycosylation, and potentially in the glycosylation of LTA, a strain lacking lmo1079 (10403SΔlmo1079) was constructed. Next, the WTA and LTA polymers were isolated from WT 10403S and the lmo1079 mutant and analyzed by NMR. As expected, GlcNAc moieties were present on the WTA polymer isolated from the WT strain but absent from the lmo1079 mutant (Fig. 6, A and B). In contrast, no differences between the NMR spectra for LTA isolated from the WT and lmo1079 mutant strains were observed, and galactose residues were present on the LTA isolated from both strains (Fig. 6, C and D). Taken together, these data show that Lmo1079 is involved in a different process than YfhO, its closest homolog in B. subtilis; Lmo1079 is needed for the glycosylation of WTA in L. monocytogenes, whereas YfhO is essential for LTA glycosylation in B. subtilis. The wider implications that closely related proteins are involved in WTA glycosylation in one bacterial species but LTA glycosylation in another bacterial species will be addressed in detail under “Discussion.”
Figure 6.
Detection of sugar modifications on WTA and LTA using 1H NMR. A and B, NMR spectra of WTA derived from L. monocytogenes 10403S (WT) (A) and the lmo1079 mutant (B). Peaks of nonexchangeable protons were assigned to the different WTA components according to previously published spectra (45, 74, 78). The different peaks for the protons and acetyl group of GlcNAc are labeled with 1H, 4H, and Ac, respectively. One representative spectrum from three independent experiments is shown. C and D, NMR spectra of LTA derived from L. monocytogenes 10403S (WT) (C) and the lmo1079 mutant strain (D). Peaks of nonexchangeable protons were assigned to the different LTA components using previously published spectra and are highlighted in colored boxes (29, 37–39). Gray boxes indicate residual citrate, a component of the buffer used for LTA purification. The spectra for lmo1079 are representative of three independent experiments. LTA of strain 10403S was isolated once as a control.
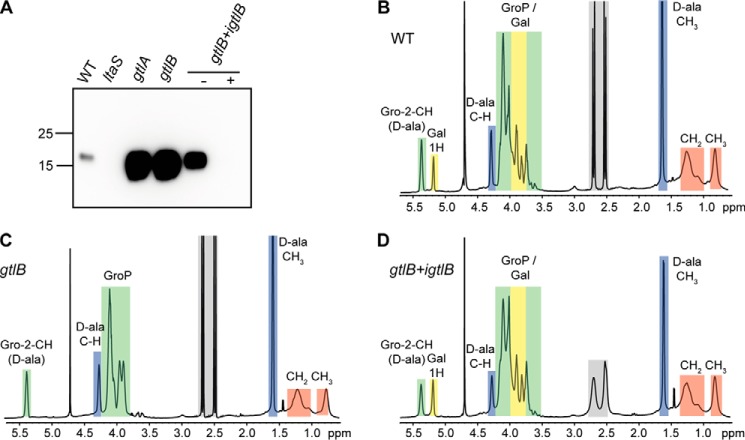
The L. monocytogenes GtlB protein is required for LTA glycosylation
Lmo1079 is the only L. monocytogenes protein that shows clear homology to the B. subtilis YfhO protein. However, as shown above, the protein could be excluded as the potential extracellular LTA GT in L. monocytogenes. Therefore, we used a bioinformatics approach to try to identify candidate proteins that could potentially act as extracellular GT involved in the LTA glycosylation process in L. monocytogenes. We reasoned that such a protein probably assumes a GT-C fold. As enzymes with such a fold have 8–13 transmembrane helices and a big extracellular loop containing the catalytic site of the protein (34), we performed a bioinformatics analysis to identify membrane proteins of unknown function in L. monocytogenes containing eight or more transmembrane helices and at least one extracellular loop of at least 50 amino acids. Genes coding for possible candidates were deleted in L. monocytogenes 10403S, and the LTA in these mutant strains was analyzed by western blotting. A strain with a deletion of lmo0626, which encodes a membrane protein with eight transmembrane helices and an extracellular loop of 61 amino acids, showed a similar increase in the anti-LTA signal as seen for the gtlA mutant (Fig. 7A). The L. monocytogenes protein Lmo0626 was renamed GtlB for glycosyltransferase LTA B. To confirm that the increased LTA signal is due to the deletion of gtlB, a complementation strain was constructed by introducing a functional copy of gtlB placed under IPTG-inducible expression control into the gtlB mutant strain, yielding strain 10403SΔgtlB pIMK3-gtlB (gtlB+igtlB). Anti-LTA western blot analysis using cell extracts isolated from this strain grown in the absence of IPTG showed the expected increased LTA signal (Fig. 7A). However, the signal was not as strong as for extracts prepared from the original gtlB mutant, indicating low-level gtlB expression even in the absence of IPTG. When the gtlB complementation strain was grown in the presence of IPTG, the LTA signal was reduced, indicating successful complementation (Fig. 7A). Indeed, no signal was detected in the complementation strain, probably due to overexpression of GtlB upon IPTG induction (Fig. 7A). Next, LTA was isolated from WT 10403S, the gtlB mutant, and the complementation strain (gtlB+igtlB) grown in the presence of IPTG and subsequently analyzed by 1H NMR. The NMR spectra for the LTA isolated from the WT 10403S strain showed the expected peaks, which were assigned to the different LTA components according to published spectra (29, 37–39) (Fig. 7B). On the other hand, the peaks derived from the galactose residues were absent in the gtlB deletion strain but again present in the complementation strain (Fig. 7, C and D). To determine whether the absence of GtlB affects the LTA chain length and percentage of d-alanine substitutions, these parameters were calculated from the integral values of the different peaks. LTA isolated from WT 10403S had a chain length of 13.11 ± 0.95 GroP units, and 56.38 ± 4.05 and 29.32 ± 2.65% were modified with d-alanine and galactose, respectively (Fig. 8). Similar values were calculated for the LTA isolated from the gtlB complementation strain (13.62 ± 0.41 GroP units, 52.56 ± 1.37% d-alanine and 35.84 ± 1.08% galactose substitutions). Interestingly, deletion of gtlB did not only result in an absence of galactose residues, but the LTA polymer was also significantly longer with a calculated length of 18.11 ± 1.39 GroP units (Fig. 8A). On the other hand, the absence of gtlB did not influence the percentage of d-alanine modifications, which remained at 53.3 ± 1.99% (Fig. 8B). Altogether, these results show that GtlB is necessary for the glycosylation of LTA in L. monocytogenes, and we hypothesize that this protein functions as extracellular glycosyltransferase, similar to the B. subtilis YfhO protein, and transfers galactose residues onto the LTA backbone.
Figure 7.
LTA production in WT L. monocytogenes 10403S, the gtlB mutant, and the complementation strain. A, LTA detection by western blotting. Cell extracts were prepared from the L. monocytogenes strains 10403S (WT), the ltaS and gtlA mutant control strains, and the gtlB mutant and complementation strain gtlB+igtlB grown in the absence (−) or presence (+) of IPTG. The extracts were separated on a 15% SDS-polyacrylamide gel, and the LTA was detected by western blotting using a polyglycerol phosphate–specific monoclonal antibody. Strains ltaS and gtlA were included as negative and positive control, respectively. B–D, NMR spectra of LTA isolated from strains 10403S (WT) (B), gtlB (C), and gtlB+igtlB (D) (grown in the presence of IPTG). Colored boxes and labels indicate nonexchangeable protons derived from the different LTA components. Peaks were assigned as described previously (29, 37–39). Gray boxes indicate residues of citrate, a buffer component used for the LTA purification. The spectra are representative of three independent experiments.
Figure 8.
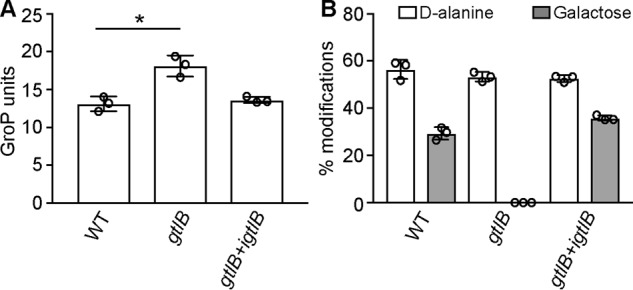
Determination of chain length, percentage of d-alanine, and galactose modifications of LTA from WT L. monocytogenes, the gtlB mutant, and the complementation strain. The different peaks in the NMR spectra obtained from LTA isolated from L. monocytogenes strains 10403S (WT), gtlB, and gtlB+igtlB were integrated, and the chain length (A) and percentages of d-alanine and galactose substitutions (B) were determined. The average values and S.D. (error bars) from three independent experiments were calculated and plotted. Two-tailed unpaired t tests identified significant differences for the number of GroP units of LTA isolated from the WT versus gtlB deletion strain with a p value < 0.05 (indicated by an asterisk).
Global analysis of GtlB proteins and identification of conserved aspartic acid residues required for function
Although the L. monocytogenes GtlB and B. subtilis YfhO proteins show only limited sequence identity, our results suggest that both proteins are involved in the LTA glycosylation process. The B. subtilis YfhO protein forms part of the YfhO-domain Pfam protein family PF09586. Currently, more than 1800 sequences in over 1000 species are listed in this Pfam family with a large number of proteins found in Firmicutes, but also some in Actinobacteria and Bacteroidetes. Consistent with our hypothesis that YfhO acts as a glycosyltransferase, some members of this family are annotated GT-C glycosyltransferases, which are characterized by 8–13 transmembrane helices and a DXD or modified DXE, EXD, DDX, or DEX motif, followed by a short stretch of hydrophobic residues (34, 42). B. subtilis YfhO has 13 predicted transmembrane helices and a large extracellular loop of 389 amino acids connecting the last two transmembrane helices (Fig. 9A). Two possible DXD motifs, D450LD and D480DD, are present in this loop of the B. subtilis YfhO protein. However, a Jalview alignment of 870 B. subtilis YfhO homologs, which share at least 30% sequence identity, indicated that neither of these DXD motifs is conserved, making the prediction of residues potentially involved in enzyme catalysis difficult. No Pfam family is currently available for proteins that are homologous to the L. monocytogenes GtlB protein. Using the L. monocytogenes GtlB protein sequence as a query sequence in a BLASTP search, just over 1000 proteins with >20% identity could be identified. These proteins are mainly present in Listeria and Bacillus sp. (Table S3). The L. monocytogenes GtlB protein has only eight predicted transmembrane helices and a much smaller extracellular loop, which connects the first and second transmembrane helices (Fig. 9B). Although no DXD or modified DXD motif could be identified, GtlB-like proteins contain a conserved DXXD (D35NLD38 in GtlB) motif in this loop (Fig. 9C), and these aspartic acid residues could potentially form part of an active site. In addition, several highly conserved hydrophobic residues are found downstream of the DXXD motif, which is similar to what has been reported for the active sites of other GT-C-fold glycosyltransferases (Fig. 9C). To test whether the conserved aspartic acid residues are essential for the function of GtlB, an alanine mutagenesis experiment was performed. gtlB alleles with the appropriate base substitutions to change amino acids Asp35, Asp38, and as control Ser39 to an alanine were placed under IPTG-inducible expression control and integrated into the chromosome of the L. monocytogenes gtlB mutant strain, allowing for IPTG-dependent expression of the different GtlB variants. Cell extracts of strains expressing gtlB, gtlBD35A, gtlBD38A, and gtlBS39A were prepared, and the LTA was detected by western blot (Fig. 9D). As described earlier, an increased signal was observed for a gtlB mutant containing the empty plasmid vector, and the signal was again reduced upon expression of a functional GtlB protein (Fig. 9D). The expression of the GtlBS39A variant complemented the gtlB mutant phenotype, as indicated by the weak anti-LTA signal, suggesting that residue Ser39 is dispensable for GtlB function (Fig. 9D). However, no complementation was observed in strains expressing the GtlBD35A and GtlBD38A variants (Fig. 9D). These data highlight that the aspartate residues at positions 35 and 38 in GtlB, with a predicted location in the first extracellular loop, are required for protein function and hence could form part of an extracellular active site. The L. monocytogenes strain used in this study is a 1/2a serovar strain; however, based on flagella and cell wall (somatic) antigens, 13 different L. monocytogenes serovars have been described (43, 44). Differences in WTA structure and glycosylation pattern provide the bases for differences in the somatic antigen (45), and for instance GlcNAc modifications are not present on WTA in all L. monocytogenes serovars (46, 47). Consistent with this, close homologs to Lmo1079, which is required in Listeria for the addition of GlcNAc residues onto WTA are only present in some but not all L. monocytogenes serovars (Table S3). On the other hand, a close homolog to GtlB, which is required for LTA glycosylation, is present in all 13 Listeria serovars as well as Listeria innocua, Listeria ivanovii, and Listeria seeligeri (Table S3), indicating that the LTA is probably modified with galactose residues in these strains.
Figure 9.
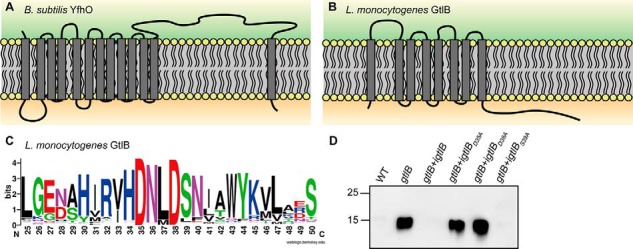
Topology of YfhO and GtlB-like proteins and identification of conserved residues required for GtlB function. A and B, topology models of B. subtilis YfhO (A) and L. monocytogenes GtlB (B) as predicted using the TMHMM version 2.0 server (85). C, WebLogo showing the conserved DXXD motif in the first extracellular loop of GtlB. Protein sequences of GtlB homologs with ≥20% identity were aligned in Jalview (80), and the alignment was used to create a WebLogo motif (81). The WebLogo motif for amino acid region 25–50 of GtlB is shown, with amino acids 35 and 38 being the conserved aspartic acid residues. D, LTA production in strains expressing different GtlB variants. L. monocytogenes strains 10403S (WT), gtlB, gtlB+igtlB, gtlB+igtlBD35A, gtlB+igtlBD38A, and gtlB+igtlBS39A were grown overnight in BHI medium at 37 °C in the presence of 1 mm IPTG. Cell extracts were prepared and separated on a 15% SDS-PAGE, and the LTA was detected using a polyglycerol phosphate–specific monoclonal LTA antibody. A representative image of three independent experiments is shown.
Discussion
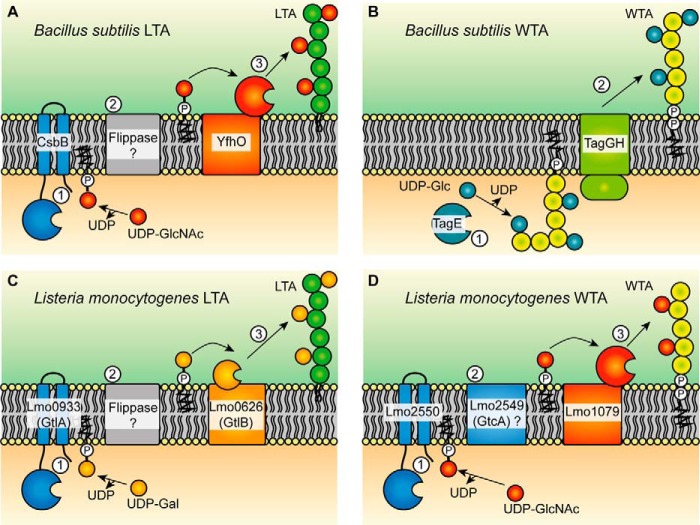
As part of this work, we identified several new proteins required for the glycosylation of LTA in B. subtilis and L. monocytogenes and, as described below, propose a new model for the WTA glycosylation process in L. monocytogenes. In previous work, GtlA (Lmo0933) was shown to be involved in the glycosylation of LTA in L. monocytogenes (29). The three close B. subtilis homologs, YkcC, YkoT, and CsbB, are predicted glycosyltransferases with N-terminal cytoplasmic catalytic domains that assume a GT-A fold and are linked by two C-terminal transmembrane helices to the membrane. Here, we show that a B. subtilis csbB mutant, but not mutants with deletions in the other two genes, lacks GlcNAc modifications on LTA (Figs. 1B and 2B and Fig. S1). Previously, it was speculated that CsbB functions in a cell wall synthesis–related process (48, 49), and our study shows that this process is the glycosylation of the cell wall polymer LTA.
Fischer and others presented a model for the LTA glycosylation process in which a cytoplasmic GT initially transfers a sugar moiety onto a C55-P lipid carrier (21, 26–28). After the carrier is “flipped” from the inner leaflet to the outer leaflet of the membrane by a flippase enzyme, a second extracellular GT was suggested to transfer the sugar moiety onto the GroP units of LTA (21, 26–28). Due to the similarity of CsbB to the C55-P glycosyltransferase GtrB in Gram-negative bacteria, Inoue et al. (40) suggested that CsbB can glycosylate the lipid carrier C55-P. This is in good agreement with our hypothesis that CsbB produces a C55-P–GlcNAc intermediate (Fig. 10A). csbB is co-transcribed with yfhO (40), and the data presented in our study show that its absence leads to the lack of sugar modifications on LTA, suggesting that yfhO might encode a GT-C-fold glycosyltransferase responsible for the transfer of the GlcNAc residues onto the LTA polymer on the outside of the cell (Figs. 4B and 10A). YfhO is a membrane protein with 13 predicted transmembrane helices and a large extracellular loop between the last two transmembrane helices (Fig. 9A), which could aid in the transfer of the GlcNAc residues onto the LTA chain (Figs. 4B and 10A). The identity of the protein that facilitates the flipping of the C55-P–GlcNAc intermediate across the membrane remains unknown; potentially, YfhO could be responsible for the flipping of the C55-P–sugar intermediate in addition to the transfer of the sugar onto LTA, which will be interesting to experimentally test in future biochemical studies.
Figure 10.
Models for the LTA and WTA glycosylation processes in B. subtilis and L. monocytogenes. A and B, proposed model for the glycosylation process of LTA (A) and WTA (B) in B. subtilis. A, based on the genetic data presented in this study, we suggest that LTA in B. subtilis is glycosylated with the aid of the cytoplasmic GT CsbB, which we predict transfers GlcNAc residues to the C55-P lipid carrier (step 1). The C55-P-GlcNAc intermediate is then transported across the membrane by an unknown flippase enzyme (step 2). We further hypothesize that the GlcNAc residues are then transferred onto LTA by the GT candidate enzyme YfhO (step 3). B, WTA of B. subtilis is modified with glucose residues in a process that takes place in the cytoplasm of the cell and that is catalyzed as previously reported by the GT TagE, which uses UDP-glucose as substrate (step 1) (58). The glycosylated WTA is subsequently transported across the membrane via TagGH (step 2) (57). C and D, proposed model for the LTA (C) and WTA (D) glycosylation process in L. monocytogenes. C, LTA in L. monocytogenes is modified with galactose residues. In the first step, GtlA, which is thought to act as cytoplasmic GT, transfers galactose molecules onto the C55-P lipid carrier (29). The sugar-lipid intermediate is then transported across the membrane by an unknown flippase enzyme (step 2), and the galactose residues are subsequently transferred onto LTA on the outside of the cell, and we suggest that this step is catalyzed by GtlB (step 3). D, for WTA glycosylation in L. monocytogenes, the cytoplasmic GT Lmo2550 probably produces a C55-P-GlcNAc lipid intermediate (step 1) that is subsequently transported across the membrane by a flippase, a function that might be provided by Lmo2549 (also referred to as GtcA) (step 2). As final step, we suggest that Lmo1079, a B. subtilis YfhO homolog, transfers the GlcNAc molecule onto WTA on the outside of the cell (step 3).
Not only LTA, but also WTA, is glycosylated in Gram-positive bacteria. This process is best characterized in S. aureus, where two cytoplasmic GTs, TarM and TarS, transfer α- and β-GlcNAc residues, respectively, onto the growing WTA chain (4, 50–52). Once glycosylated, the WTA polymer is transported across the bacterial membrane by the TarGH ABC transporter and subsequently linked to peptidoglycan by Lcp enzymes (53–56). WTA of B. subtilis is modified with glucose residues by the glycosyltransferase TagE in the cytoplasm of the cell and subsequently moved across the membrane by the TagGH transporter (Fig. 10B) (57, 58). Glycosylation of WTA can take place in the cytoplasm of the cell as WTA is polymerized within the cell. This is in contrast to polyglycerol-phosphate LTA, which is polymerized on the outside of the cell and hence can only by glycosylated on the extracellular side of the membrane. However, based on the homology of the L. monocytogenes WTA glycosylation enzymes with the enzymes involved in the B. subtilis LTA glycosylation process uncovered as part of this work, we propose that glycosylation of WTA might not universally take place in the cytoplasm of the cell. Instead, we hypothesize that the glycosylation of WTA in L. monocytogenes 10403S and EGD-e, 1/2a serovar strains, and probably a number of other serovar strains (Table S3) takes place on the outside of the cell and proceeds via the production of a C55-P–sugar intermediate that is transported across the membrane and subsequently utilized for the glycosylation of WTA on the outside of the cell (Fig. 10D). WTA in L. monocytogenes 10403S is decorated with GlcNAc residues, and several L. monocytogenes proteins required for the glycosylation of WTA have been identified in previous studies (41, 59–61). One of these is Lmo2550, a GT-A fold glycosyltransferase with a predicted cytoplasmic catalytic domain that is linked via two C-terminal transmembrane helices to the membrane. Lmo2550 shares a high degree of similarity to the B. subtilis CsbB protein (e-value: 4e−91) (59, 60, 62) (Table S3). Hence, it is likely that both proteins, L. monocytogenes Lmo2550 and B. subtilis CsbB, catalyze the same reaction, which we predict is the formation of a C55-P–GlcNAc lipid intermediate in the cytoplasm of the cell (Fig. 10). Another protein required for the WTA glycosylation process in L. monocytogenes is Lmo2549 (59, 60). Lmo2549, also named GtcA, is a membrane protein with four predicted transmembrane helices and a GtrA domain. The exact function of GtcA is not known; however, based on homology to small multidrug resistance transporters, GtcA and other members of this family could be involved in the flipping of lipid-linked intermediates across the membrane, as suggested previously (Fig. 10D) (63–65). In the case of L. monocytogenes 10403S, we hypothesize that this would be the C55-P–GlcNAc lipid intermediate. However, it should be noted that biochemical evidence for such flippase activity is still lacking. Finally, the Lmo1079 protein has been implicated in the WTA glycosylation pathway in L. monocytogenes (41, 61). Lmo1079 shows a high degree of similarity to the B. subtilis transmembrane protein YfhO (e-value: 1e−39), which, we suggest, is a GT-C-fold glycosyltransferase responsible for the transfer of the GlcNAc residues to the B. subtilis LTA polymer on the outside of the cell (Fig. 10A). As part of this work, we purified WTA and LTA from an L. monocytogenes lmo1079 mutant strain and show that this protein is indeed required for WTA glycosylation but not required for the glycosylation of LTA (Fig. 6). It is tempting to speculate that both proteins, L. monocytogenes Lmo1079 and B. subtilis YfhO, act as extracellular glycosyltransferases and move GlcNAc residues from C55-P lipid intermediates onto the WTA or LTA polymers, respectively, on the outside of the cell. Taken together, this suggests that WTA in Gram-positive bacteria cannot only be glycosylated on the inside of the cell as described until now but also on the outside of the membrane (Fig. 10).
LTA in L. monocytogenes strain 10403S is modified with galactose residues (22, 23, 29). Because inactivation of Lmo1079, the only clear homolog to the B. subtilis YfhO protein in L. monocytogenes, did not abolish the glycosylation of LTA (Fig. 6D), a different protein must have been responsible for the transfer of galactose residues onto the LTA polymer. Our work indicates that the transmembrane protein GtlB (Lmo0626) is a good candidate protein to catalyze this reaction, as the sugar modifications on LTA were absent in a gtlB mutant (Figs. 7C and 10C). The B. subtilis YfhO and L. monocytogenes GtlB proteins have different topologies and share little similarity on an amino acid level (Fig. 9, A and B). Nevertheless, our results show that both proteins are involved in the LTA glycosylation process, and we propose that both proteins transfer C55-P–linked sugar residues onto cell wall polymers on the outside of the cell. GtlB is characterized by eight transmembrane helices and a relatively large extracellular loop of 61 amino acids between the first and second predicted TM helix that may be involved in the sugar transfer (Fig. 9B). Furthermore, the GtlB protein in L. monocytogenes strain EGD-e, which is 100% identical to the L. monocytogenes 10403S protein analyzed in this study, is annotated to contain a PMT-2 domain (GenBankTM number CAC98704.1). PMT domains are found in proteins of the dolichyl-phosphate-mannose protein mannosyltransferase family, which catalyze O-linked glycosylation of proteins. Members of the PMT family include the l-Ara4-N lipid A transferase ArnT of Escherichia coli and Salmonella typhimurium, which is responsible for the glycosylation of lipid A, and the Pmt (Cg1014) protein of Corynebacterium glutamicum, which is necessary for the glycosylation of secreted proteins (66, 67). Further bioinformatics analysis revealed that the L. monocytogenes GtlB protein shows some similarity to the Pseudomonas aeruginosa arabinosyltransferase TfpW, which has been described as a GT-C-fold glycosyltransferase associated with the glycosylation of type IV pili (42). These findings strengthen our hypothesis that the L. monocytogenes GtlB protein might act as the second glycosyltransferase responsible for the transfer of galactose residues on the outside of the cell to the LTA polymer (Fig. 10C), although biochemical evidence is still lacking. GtlB possesses a modified DXXD motif in the first predicted extracellular loop that is a characteristic of GT-C glycosyltransferases. Our alanine substitution analysis indicated that both aspartic acid residues, Asp35 and Asp38, are crucial for GtlB function and for the glycosylation of LTA. A conserved DXXD motif has also been identified in other GT-C-type glycosyltransferases, including the WecA protein from E. coli, the α-(1–6)-mannopyranosyltransferases Rv1459c and Rv2174 of M. tuberculosis, and MptB of C. glutamicum (68–70), further highlighting that the enzymatic site of GtlB is probably on the extracellular site of the membrane.
Glycosyltransferases are widely distributed among bacteria. In particular, GT-A-type glycosyltransferases, such as the B. subtilis CsbB and L. monocytogenes GtlA proteins characterized as part of this and a previous study (29), are found in a large number of bacteria. The distribution of such glycosyltransferases, which are probably responsible for the production of C55-P–sugar intermediates, is presented in Table S3 for the different L. monocytogenes serovars as well as a selected number of other well-studied Gram-positive bacteria. YfhO and GtlB homologs were not present in all bacteria listed in Table S3. However, we could still identify around 5150 and 1005 proteins that share >20% identity to YfhO of B. subtilis and GtlB of L. monocytogenes, respectively. YfhO-like proteins are not only found in Bacillus and Listeria sp., but are also present in a range of other Gram-positive bacteria, including different Staphylococcus sp. and Streptococcus pneumoniae (Table S3). We would expect that these GT-C-fold enzymes are also involved in the glycosylation of an extracellular cell wall component in these species. However, as highlighted by the findings presented in this study, the exact nature of the cell wall polymer that is modified cannot easily be predicted solely based on homology and will await future experimental studies. GtlB-like proteins are less widely distributed and primarily found in Listeria and Bacillus sp. Interestingly, the L. monocytogenes GtlB protein shares 49% identity with the B. subtilis YkoS protein (Table S3). ykoS is co-transcribed with ykoT, which codes for a GT-A-fold glycosyltransferase that we analyzed as part of this study (Fig. 1A and Fig. S1). The ykoST operon of B. subtilis is expressed during sporulation and regulated by SigG and SpoVT (71) and has also been shown to be under the expression control of the YkoH-YkoG two-component system (72). Neither of these two proteins is required for LTA glycosylation (Figs. S1 and S3) under standard growth conditions, and hence the target for the predicted glycosyltransferases YkoS and YkoT is unknown. However, it is tempting to speculate that the proteins are required for the glycosylation of an extracellular cell wall polymer or perhaps even protein during the sporulation process.
Taken together, we identified several new enzymes required for the glycosylation of LTA in B. subtilis and L. monocytogenes and speculate that these enzymes function as cytoplasmic or extracellular glycosyltransferases, although biochemical evidence is still lacking. GT-C-fold glycosyltransferases are notoriously difficult to predict bioinformatically, and based on the genetic evidence presented in this work, we suggest that YfhO- and GtlB-like proteins are novel classes of GT-C-fold glycosyltransferases. Taken together, our findings will help us to determine the function of sugar modifications on LTA in Gram-positive bacteria, which has remained elusive up to date. But perhaps most importantly, another implication of our work is that WTA in Gram-positive bacteria might be glycosylated via two different mechanisms, one taking place inside the cell and a second novel mechanism taking place on the outside of the cell.
Experimental procedures
Bacterial strains and growth conditions
All strains and plasmids used in this study are listed in Table S1. E. coli and B. subtilis strains were grown in Lysogeny Broth (LB) medium and L. monocytogenes strains in brain heart infusion (BHI) medium at 37 °C unless otherwise stated. If necessary, antibiotics and supplements were added to the medium at the following concentrations: for E. coli cultures, ampicillin at 100 μg/ml, chloramphenicol (Cam) at 20 μg/ml, erythromycin (Erm) at 10 μg/ml, and kanamycin (Kan) at 30 μg/ml; for B. subtilis cultures, Cam at 5 μg/ml, Erm at 5 μg/ml, and Kan at 10 μg/ml; and for L. monocytogenes cultures, Cam at 10 μg/ml, Kan at 30 μg/ml, and IPTG at 1 mm. In this study, we used L. monocytogenes strain 10403S and derivatives thereof. However, L. monocytogenes EGD-e gene and locus tag numbers were used, as this was the first fully sequenced L. monocytogenes strain, and the corresponding 10403S locus tag numbers are given in parenthesis.
Strain and plasmid construction
All primers used in this study are listed in Table S2. For the construction of B. subtilis strains with gene deletions, 1-kb DNA fragments upstream and downstream of the respective gene were amplified, and the products were digested with ApaI and XhoI and ligated to an antibiotic resistance cassette. The antibiotic resistance cassettes were obtained by digesting plasmids pCN34 (Kan), pCN38 (Cam), and pCN49 (Erm) with restriction enzymes ApaI and XhoI. Up- and downstream ykcC fragments were generated by PCR using primers ANG1534/1535 and ANG1536/1537, and ykoS fragments were generated using primers ANG1564/1565 and ANG1566/1567. The fragments were digested with ApaI and XhoI and ligated with the Cam cassette. Up- and downstream fragments for ykcB were generated by PCR using primer pairs ANG1540/1541 and ANG1542/1543, and csbB fragments were generated with primer pairs ANG1546/1547 and ANG1548/1549, and these fragments were digested and ligated with a Kan cassette. The up- and downstream regions of yfhO and ykoT were amplified using primer pairs ANG1552/1553 and ANG1554/1555 and primer pairs ANG1558/1559 and ANG1560/1561, respectively, digested, and fused to an Erm cassette. Subsequently, the purified ligation products were transformed into B. subtilis 168, and transformants were selected on LB agar plates containing the appropriate antibiotics. The replacement of the target gene with the antibiotic marker was verified by PCR, resulting in the construction of the B. subtilis strains 168ΔykcC::cam (ANG2747), 168ΔykcB::kan (ANG2748), 168ΔcsbB::kan (ANG2749), 168ΔyfhO::erm (ANG2750), 168ΔykoT::erm (ANG2751), and 168ΔykoS::cam (ANG2752). For complementation analysis of the csbB mutant, plasmid pDG1662-csbB was constructed. To this end, the csbB gene, including the upstream promotor region, was amplified from B. subtilis 168 chromosomal DNA using primers ANG1624/1625, and the product was digested with BamHI and HindIII and inserted into plasmid pDG1662, which had been cut with the same enzymes. The resulting plasmid pDG1662-csbB was recovered in E. coli XL1-Blue, yielding strain ANG2905. The yfhO mutation in strain 168ΔyfhO::erm was complemented by introducing the complete csbB-yfhO operon, including the native promotor preceding csbB. The csbB-yfhO operon was amplified with primers ANG1624/1628, and the fragment was cut with BamHI and HindIII and inserted into plasmid pD1662, yielding plasmid pDG1662-csbB-yfhO. In addition, plasmid pDG1662-csbBD97A-yfhO was produced for the expression of an inactive CsbB variation with an D97A amino acid substitution. For this purpose, PCR products were generated with primer pairs ANG1624/1731 and ANG1628/1732 introducing the required base change in csbB as part of the primer sequences. The products were fused by a second PCR using primers ANG1624 and ANG1628. The fragment was digested with BamHI and HindIII and ligated with pDG1662, yielding plasmid pDG1662-csbBD97A-yfhO. Plasmids pDG1662-csbB-yfhO and pDG1662-csbBD97A-yfhO were recovered in E. coli XL1-Blue, yielding strains ANG3508 and ANG3303, respectively. All three complementation plasmids were linearized with XhoI and transformed into B. subtilis 168 mutant strains, where the plasmids integrate by double crossover recombination into the amyE locus. This resulted in the generation of the B. subtilis strains 168ΔcsbB::kan amyE::csbB (ANG3017), 168ΔcsbB:: kan amyE::csbBD97A-yfhO (ANG3305), 168ΔyfhO:: erm amyE::csbB-yfhO (ANG3510), and 168ΔyfhO:: erm amyE::csbBD97A-yfhO (ANG3304).
For the markerless in-frame deletion of the L. monocytogenes genes lmo1079 (lmrg_00541) and lmo0626 (lmrg_00309, gtlB), 1-kb DNA fragments up- and downstream of the respective gene were amplified by PCR using primer pairs ANG1527/1528 and ANG1529/1530 (lmo1079) or ANG2516/2517 and ANG2518/2519 (gtlB). The resulting PCR products were fused by PCR using the outside primer ANG1527/ANG1530 (lmo1079) or ANG2516/ANG2519 (gtlB). The products were cut with BamHI and KpnI and ligated with plasmid pKSV7 that had been digested with the same enzymes. This resulted in the generation of plasmids pKSV7-Δlmo1079 and pKSV7-ΔgtlB, which were recovered in E. coli XL1-Blue, yielding strains ANG2793 and ANG4232, respectively. The plasmids were subsequently electroporated into L. monocytogenes 10403S, and the genes were deleted by allelic exchange using a procedure described previously (73). The deletion of the respective gene was verified by PCR, resulting in the construction of L. monocytogenes strains 10403SΔlmo1079 (ANG2794) and 10403SΔgtlB (ANG4264). For complementation analysis of the gtlB mutant, plasmid pIMK3-gtlB was constructed. Plasmid pIMK3 carries the Phelp promoter, which allows for IPTG-dependent gene expression. The gtlB gene was amplified using primers ANG2708 and ANG2709, and the product was digested with NcoI and SalI and ligated with pIMK3. Plasmid pIMK3-gtlB was recovered in E. coli XL1-Blue, yielding strain ANG4401. Additionally, point mutations were introduced into the gtlB gene for the expression of GtlB variants with D35A, D38A, and S39A amino acid substitutions. For this purpose, primer pairs ANG2708/2791 (D35A), ANG2708/2902 (D38A), and ANG2708/2793 (S39A) were used to amplify the 5′-end of gtlB introducing the respective base changes as part of the primer sequence. The 3′-end of gtlB was amplified using primers ANG2709/2790 (D35A), ANG2709/2901 (D38A), and ANG2709/2792 (S39A). The appropriate fragments were fused in a second PCR using primers ANG2708 and ANG2709. The PCR products were cut with NcoI and SalI and ligated with pIMK3 that had been cut with the same enzymes. The resulting plasmids were recovered in E. coli XL1-Blue, yielding strains XL1-Blue pIMK3-gtlBD35A (ANG4630), XL1-Blue pIMK3-gtlBD38A (ANG4769), and XL1-Blue pIMK3-gtlBS39A (ANG4631). The pIMK3 derivatives as well as the empty pIMK3 plasmid were transformed into L. monocytogenes strain 10403SΔgtlB by electroporation, resulting in the construction of strains 10403SΔgtlB pIMK3 (ANG4637), 10403SΔgtlB pIMK3-gtlB (ANG4443), 10403SΔgtlB pIMK3-gtlBD35A (ANG4648), 10403SΔgtlB pIMK3-gtlBD38A (ANG4775), and 10403SΔgtlB pIMK3-gtlBS39A (ANG4638).
Preparation of cell extract and western blot analysis
B. subtilis 168 (WT) and the indicted mutant strains were grown for 20–22 h in 5 ml of LB medium at 30 °C. Bacteria from 4 ml of culture were collected by centrifugation for 30 min at 17,000 × g, and bacterial pellets were suspended in 2× SDS-PAGE sample buffer to an A600 = 3. Samples were boiled for 45 min and centrifuged for 5 min, and 10 μl were loaded onto a 15% SDS-polyacrylamide gel. For LTA detection, the humanized monoclonal LTA antibody (gift from Biosynexus Inc., Gaithersburg, MD) and the HRP-conjugated polyclonal rabbit anti-human IgA, IgG, κ, λ antibody (DakoCytomation) were used at 1:5000 and 1:10,000 dilutions, respectively. L. monocytogenes 10403S (WT) and the different mutant strains were grown overnight in 5 ml of BHI medium at 37 °C. The expression of gtlB (and the different variants) from the Phelp promoter was induced by the addition of 1 mm IPTG to the medium. Cell extracts for LTA detection were prepared as described previously (8). LTA was detected using a polyglycerol phosphate–specific antibody (Clone 55 from Hycult Biotechnology) and an HRP-conjugated anti-mouse IgG (Cell Signaling Technologies) at 1:4000 and 1:10,000 dilutions, respectively. western blots were developed by the enhanced chemiluminescence method, and the signal was detected using a ChemiDoc Touch Imager (Bio-Rad). All experiments were performed at least three times, and representative images are shown.
LTA and WTA isolation
For the isolation of LTA from B. subtilis, strains were grown overnight in 3 liters of LB medium, and cells were collected by centrifugation. For the isolation of LTA from L. monocytogenes, the strains were grown overnight in 2–3 liters of BHI medium, and when required, the medium was supplemented with 1 mm IPTG, and the bacteria were harvested by centrifugation. LTA was purified and analyzed using 1D 1H NMR, as described previously (7, 29). Briefly, LTA was extracted with butanol and purified by hydrophobic interaction chromatography using a 24 × 1.6-cm octyl-Sepharose column. Fractions containing LTA were identified by western blotting using the humanized monoclonal LTA or polyglycerol phosphate–specific antibody. The LTA-containing fractions were pooled and lyophilized. WTA was purified and analyzed by NMR, as described previously (74). Briefly, bacteria from 5-liter cultures of L. monocytogenes 10403S and 10403SΔlmo1079 grown to mid-log phage (A600 of 0.6–1) in BHI medium at 37 °C were collected by centrifugation and subsequently washed with 1 m NaCl of pH 5.8. The cells were disrupted using a bead beater, and the cell debris was collected by centrifugation. After washing the cell wall material with 1 m NaCl, pH 5.8, 0.5% SDS, pH 5.8, and water, pH 5.8, the material was suspended in water and incubated for 30 min at 60 °C. The cell wall material was recovered by centrifugation, washed with water, and suspended in 0.15 mm Tris/HCl, pH 7.0, containing 0.2 mg/ml trypsin and incubated at 37 °C for 18 h. The next day, the material was collected by centrifugation; washed with 1 m Tris, pH 7.0, 1 m Tris, pH 7.0, containing 1 m NaCl, 1 m Tris, pH 7.0; and washed three times with water. The cell wall material was then incubated for 18 h at 4 °C with 10% TCA to hydrolyze the WTA from the peptidoglycan. The peptidoglycan was removed by centrifugation, and the WTA was precipitated with 0.1 volume of 3 m sodium acetate, pH 5.2, and 3 volumes of ice-cold 95% ethanol and left overnight at −80 °C. The WTA was recovered by centrifugation, washed with ethanol, and air-dried. As final step, the WTA was suspended in 1 ml of water and lyophilized.
NMR analysis of cell wall polymers
To analyze the LTA and WTA polymers by 1H NMR, 2 mg of LTA or 6 mg of WTA were suspended and lyophilized twice in 500 μl of D2O of 99.96% purity. In the final step, the LTA or WTA preparations were suspended in 500 μl of D2O of 99.99% purity, and NMR spectra were recorded on a 600-MHz Bruker Advance III spectrometer equipped with a TCl cryoprobe. To ensure accurate integration of the signals, NMR spectra were recorded at 303 K with a total recycling time of 5 s and a 1H flip angle of ∼30°. Three independent LTA and WTA extractions were performed for each strain shown here and once for the data presented in the supporting material. The data were analyzed using Topspin version 3.5 software (Bruker Biospin, Ltd.), and the spectra were annotated according to previously published NMR spectra (29, 37–39, 45, 74–78). To determine the LTA chain length and percentage of d-alanine and sugar modifications, the different peaks were integrated, and chain length and percentage modification calculations were performed for the B. subtilis and L. monocytogenes LTA as described previously (17, 29). Briefly, the area under the peak at 4.3 ppm corresponding to the CH group of d-alanine was used as a reference and set to 1. The integration values for the peaks of the different LTA components (namely GroP, d-alanine, GlcNAc, or Gal and fatty acids) were determined and adjusted by the number of nonexchangeable protons; for the LTA samples isolated from B. subtilis, these are 58 protons for the lipid anchor, 5 for GroP, 4 for d-alanine, and 10 for GlcNAc. For the LTA samples isolated from L. monocytogenes, the calculations are based on 58 protons for the lipid anchor, 5 for GroP, 4 for d-alanine, and 7 for Gal modifications. Dividing the proton-adjusted value for GroP by the proton-adjusted value for the lipid anchor gives the chain length of the LTA. To determine the percentage of substitution, the proton-adjusted values for d-alanine and GlcNAc (B. subtilis) or d-alanine and Gal (L. monocytogenes) were divided by the proton-adjusted value for GroP and then multiplied by 100. The average values and S.D. from three biological replicates were determined and plotted.
Sequence analysis and protein alignments
For the identification of potential homologs of the L. monocytogenes GtlA and GtlB proteins and of the B. subtilis YfhO protein, the corresponding protein sequences were used in BLASTP searches against the NCBI non-redundant protein sequence database (79). Sequences of proteins with a minimum query coverage of 60% and a minimum sequence identity of 20% to GtlB were downloaded and used for further analysis in Jalview (80). A multiple-sequence alignment of GtlB and its homologs was performed with Clustal Omega, setting the L. monocytogenes GtlB protein as a reference. The alignment was subsequently used to generate a sequence logo motif using the web-based application WebLogo (81).
Author contributions
J.R., M.G.P., and A.G. conceptualization; J.R., M.G.P., and A.G. resources; J.R. and M.G.P. data curation; J.R., M.G.P., and A.G. formal analysis; J.R. and A.G. funding acquisition; J.R., M.G.P., and A.G. investigation; J.R. and A.G. writing-original draft; M.G.P. and A.G. methodology; M.G.P. writing-review and editing; A.G. supervision; A.G. project administration.
Supplementary Material
This work was supported by European Research Council Grant 260371 and Wellcome Trust Grant 100289 (to A. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- WTA
- wall teichoic acid
- LTA
- lipoteichoic acid
- GT
- glycosyltransferase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- LB
- Lysogeny Broth
- BHI
- brain heart infusion
- Cam
- chloramphenicol
- Erm
- erythromycin
- Kan
- kanamycin
- HRP
- horseradish peroxidase.
References
- 1. Araki Y., and Ito E. (1989) Linkage units in cell walls of Gram-positive bacteria. Crit. Rev. Microbiol. 17, 121–135 10.3109/10408418909105745 [DOI] [PubMed] [Google Scholar]
- 2. Neuhaus F. C., and Baddiley J. (2003) A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 686–723 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Percy M. G., and Gründling A. (2014) Lipoteichoic acid synthesis and function in Gram-positive bacteria. Annu. Rev. Microbiol. 68, 81–100 10.1146/annurev-micro-091213-112949 [DOI] [PubMed] [Google Scholar]
- 4. Brown S., Xia G., Luhachack L. G., Campbell J., Meredith T. C., Chen C., Winstel V., Gekeler C., Irazoqui J. E., Peschel A., and Walker S. (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914 10.1073/pnas.1209126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krawczyk-Balska A., and Lipiak M. (2013) Critical role of a ferritin-like protein in the control of Listeria monocytogenes cell envelope structure and stability under β-lactam pressure. PLoS One 8, e77808 10.1371/journal.pone.0077808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spears P. A., Havell E. A., Hamrick T. S., Goforth J. B., Levine A. L., Thomas Abraham S. T., Heiss C., Azadi P., and Orndorff P. E. (2016) Listeria monocytogenes wall teichoic acid decoration in virulence and cell-to-cell spread. Mol. Microbiol. 101, 714–730 10.1111/mmi.13353 [DOI] [PubMed] [Google Scholar]
- 7. Gründling A., and Schneewind O. (2007) Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8478–8483 10.1073/pnas.0701821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Webb A. J., Karatsa-Dodgson M., and Gründling A. (2009) Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol. 74, 299–314 10.1111/j.1365-2958.2009.06829.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schirner K., Marles-Wright J., Lewis R. J., and Errington J. (2009) Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28, 830–842 10.1038/emboj.2009.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mamou G., Fiyaksel O., Sinai L., and Ben-Yehuda S. (2017) Deficiency in lipoteichoic acid synthesis causes a failure in executing the colony developmental program in Bacillus subtilis. Front. Microbiol. 8, 1991 10.3389/fmicb.2017.01991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xia G., and Peschel A. (2008) Toward the pathway of S. aureus WTA biosynthesis. Chem. Biol. 15, 95–96 10.1016/j.chembiol.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 12. Richter S. G., Elli D., Kim H. K., Hendrickx A. P., Sorg J. A., Schneewind O., and Missiakas D. (2013) Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for Gram-positive bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 3531–3536 10.1073/pnas.1217337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasquina L. W., Santa Maria J. P., and Walker S. (2013) Teichoic acid biosynthesis as an antibiotic target. Curr. Opin. Microbiol. 16, 531–537 10.1016/j.mib.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer W. (1990) Bacterial phosphoglycolipids and lipoteichoic acids. in Glycolipids, Phosphoglycolipids, and Sulfoglycolipids (Kates M., ed) pp. 123–234, Springer US, Boston, MA [Google Scholar]
- 15. Kiriukhin M. Y., Debabov D. V., Shinabarger D. L., and Neuhaus F. C. (2001) Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J. Bacteriol. 183, 3506–3514 10.1128/JB.183.11.3506-3514.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reichmann N. T., and Gründling A. (2011) Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319, 97–105 10.1111/j.1574-6968.2011.02260.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wörmann M. E., Corrigan R. M., Simpson P. J., Matthews S. J., and Gründling A. (2011) Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol. Microbiol. 79, 566–583 10.1111/j.1365-2958.2010.07472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fischer W., and Rösel P. (1980) The alanine ester substitution of lipoteichoic acid (LTA) in Staphylococcus aureus. FEBS Lett. 119, 224–226 10.1016/0014-5793(80)80257-2 [DOI] [PubMed] [Google Scholar]
- 19. Iwasaki H., Shimada A., and Ito E. (1986) Comparative studies of lipoteichoic acids from several Bacillus strains. J. Bacteriol. 167, 508–516 10.1128/jb.167.2.508-516.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer W. (1988) Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29, 233–302 10.1016/S0065-2911(08)60349-5 [DOI] [PubMed] [Google Scholar]
- 21. Iwasaki H., Shimada A., Yokoyama K., and Ito E. (1989) Structure and glycosylation of lipoteichoic acids in Bacillus strains. J. Bacteriol. 171, 424–429 10.1128/jb.171.1.424-429.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hether N. W., and Jackson L. L. (1983) Lipoteichoic acid from Listeria monocytogenes. J. Bacteriol. 156, 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uchikawa K., Sekikawa I., and Azuma I. (1986) Structural studies on lipoteichoic acids from four Listeria strains. J. Bacteriol. 168, 115–122 10.1128/jb.168.1.115-122.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heaton M. P., and Neuhaus F. C. (1992) Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J. Bacteriol. 174, 4707–4717 10.1128/jb.174.14.4707-4717.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abachin E., Poyart C., Pellegrini E., Milohanic E., Fiedler F., Berche P., and Trieu-Cuot P. (2002) Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43, 1–14 10.1046/j.1365-2958.2002.02723.x [DOI] [PubMed] [Google Scholar]
- 26. Mancuso D. J., and Chiu T. H. (1982) Biosynthesis of glucosyl monophosphoryl undecaprenol and its role in lipoteichoic acid biosynthesis. J. Bacteriol. 152, 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yokoyama K., Araki Y., and Ito E. (1988) The function of galactosyl phosphorylpolyprenol in biosynthesis of lipoteichoic acid in Bacillus coagulans. Eur. J. Biochem. 173, 453–458 10.1111/j.1432-1033.1988.tb14020.x [DOI] [PubMed] [Google Scholar]
- 28. Fischer W. (1994) Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med. Microbiol. Immunol. 183, 61–76 10.1007/BF00277157 [DOI] [PubMed] [Google Scholar]
- 29. Percy M. G., Karinou E., Webb A. J., and Gründling A. (2016) Identification of a lipoteichoic acid glycosyltransferase enzyme reveals that GW-domain-containing proteins can be retained in the cell wall of Listeria monocytogenes in the absence of lipoteichoic acid or its modifications. J. Bacteriol. 198, 2029–2042 10.1128/JB.00116-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rao S. T., and Rossmann M. G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 10.1016/0022-2836(73)90388-4 [DOI] [PubMed] [Google Scholar]
- 31. Breton C., Snajdrová L., Jeanneau C., Koca J., and Imberty A. (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16, 29R–37R 10.1093/glycob/cwj016 [DOI] [PubMed] [Google Scholar]
- 32. Breton C., Bettler E., Joziasse D. H., Geremia R. A., and Imberty A. (1998) Sequence-function relationships of prokaryotic and eukaryotic galactosyltransferases. J. Biochem. 123, 1000–1009 10.1093/oxfordjournals.jbchem.a022035 [DOI] [PubMed] [Google Scholar]
- 33. Breton C., and Imberty A. (1999) Structure/function studies of glycosyltransferases. Curr. Opin. Struct. Biol. 9, 563–571 10.1016/S0959-440X(99)00006-8 [DOI] [PubMed] [Google Scholar]
- 34. Liu J., and Mushegian A. (2003) Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 10.1110/ps.0302103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breazeale S. D., Ribeiro A. A., McClerren A. L., and Raetz C. R. (2005) A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose: identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280, 14154–14167 10.1074/jbc.M414265200 [DOI] [PubMed] [Google Scholar]
- 36. Lairson L. L., Henrissat B., Davies G. J., and Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 10.1146/annurev.biochem.76.061005.092322 [DOI] [PubMed] [Google Scholar]
- 37. Morath S., Geyer A., and Hartung T. (2001) Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193, 393–397 10.1084/jem.193.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morath S., Geyer A., Spreitzer I., Hermann C., and Hartung T. (2002) Structural decomposition and heterogeneity of commercial lipoteichoic acid preparations. Infect. Immun. 70, 938–944 10.1128/IAI.70.2.938-944.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morath S., Stadelmaier A., Geyer A., Schmidt R. R., and Hartung T. (2002) Synthetic lipoteichoic acid from Staphylococcus aureus is a potent stimulus of cytokine release. J. Exp. Med. 195, 1635–1640 10.1084/jem.20020322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inoue H., Suzuki D., and Asai K. (2013) A putative bactoprenol glycosyltransferase, CsbB, in Bacillus subtilis activates SigM in the absence of co-transcribed YfhO. Biochem. Biophys. Res. Commun. 436, 6–11 10.1016/j.bbrc.2013.04.064 [DOI] [PubMed] [Google Scholar]
- 41. Eugster M. R., Morax L. S., Hüls V. J., Huwiler S. G., Leclercq A., Lecuit M., and Loessner M. J. (2015) Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 97, 33–46 [DOI] [PubMed] [Google Scholar]
- 42. Kus J. V., Kelly J., Tessier L., Harvey H., Cvitkovitch D. G., and Burrows L. L. (2008) Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with d-Araf by a novel GT-C family arabinosyltransferase, TfpW. J. Bacteriol. 190, 7464–7478 10.1128/JB.01075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Graves L. M., Swaminathan B., and Hunter S. B. (1999) Subtyping Listeria monocytogenes. In Listeria, listeriosis and food safety (Ryser E. T., and Marth E. H., eds) pp. 251–297, Marcel Dekker Inc., New York [Google Scholar]
- 44. Seeliger H. P. R., and Höhne K. (1979) Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13, 31–49 10.1016/S0580-9517(08)70372-6 [DOI] [Google Scholar]
- 45. Kamisango K., Fujii H., Okumura H., Saiki I., Araki Y., Yamamura Y., and Azuma I. (1983) Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J. Biochem. 93, 1401–1409 10.1093/oxfordjournals.jbchem.a134275 [DOI] [PubMed] [Google Scholar]
- 46. Fiedler F. (1988) Biochemistry of the cell surface of Listeria strains: a locating general view. Infection 16, S92–S97 10.1007/BF01639729 [DOI] [PubMed] [Google Scholar]
- 47. Shen Y., Boulos S., Sumrall E., Gerber B., Julian-Rodero A., Eugster M. R., Fieseler L., Nyström L., Ebert M. O., and Loessner M. J. (2017) Structural and functional diversity in Listeria cell wall teichoic acids. J. Biol. Chem. 292, 17832–17844 10.1074/jbc.M117.813964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Akbar S., and Price C. W. (1996) Isolation and characterization of csbB, a gene controlled by Bacillus subtilis general stress transcription factor σ B. Gene 177, 123–128 10.1016/0378-1119(96)00287-9 [DOI] [PubMed] [Google Scholar]
- 49. Huang X., and Helmann J. D. (1998) Identification of target promoters for the Bacillus subtilis σ X factor using a consensus-directed search. J. Mol. Biol. 279, 165–173 10.1006/jmbi.1998.1765 [DOI] [PubMed] [Google Scholar]
- 50. Xia G., Maier L., Sanchez-Carballo P., Li M., Otto M., Holst O., and Peschel A. (2010) Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415 10.1074/jbc.M109.096172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winstel V., Xia G., and Peschel A. (2014) Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 304, 215–221 10.1016/j.ijmm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 52. Sobhanifar S., Worrall L. J., Gruninger R. J., Wasney G. A., Blaukopf M., Baumann L., Lameignere E., Solomonson M., Brown E. D., Withers S. G., and Strynadka N. C. (2015) Structure and mechanism of Staphylococcus aureus TarM, the wall teichoic acid α-glycosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 112, E576–E585 10.1073/pnas.1418084112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schirner K., Stone L. K., and Walker S. (2011) ABC transporters required for export of wall teichoic acids do not discriminate between different main chain polymers. ACS Chem. Biol. 6, 407–412 10.1021/cb100390w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N., Ogasawara N., Lewis R. J., Vollmer W., Daniel R. A., and Errington J. (2011) A widespread family of bacterial cell wall assembly proteins. EMBO J. 30, 4931–4941 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dengler V., Meier P. S., Heusser R., Kupferschmied P., Fazekas J., Friebe S., Staufer S. B., Majcherczyk P. A., Moreillon P., Berger-Bächi B., and McCallum N. (2012) Deletion of hypothetical wall teichoic acid ligases in Staphylococcus aureus activates the cell wall stress response. FEMS Microbiol. Lett. 333, 109–120 10.1111/j.1574-6968.2012.02603.x [DOI] [PubMed] [Google Scholar]
- 56. Chan Y. G., Frankel M. B., Dengler V., Schneewind O., and Missiakas D. (2013) Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J. Bacteriol. 195, 4650–4659 10.1128/JB.00544-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lazarevic V., and Karamata D. (1995) The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol. Microbiol. 16, 345–355 10.1111/j.1365-2958.1995.tb02306.x [DOI] [PubMed] [Google Scholar]
- 58. Allison S. E., D'Elia M. A., Arar S., Monteiro M. A., and Brown E. D. (2011) Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J. Biol. Chem. 286, 23708–23716 10.1074/jbc.M111.241265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Promadej N., Fiedler F., Cossart P., Dramsi S., and Kathariou S. (1999) Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J. Bacteriol. 181, 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eugster M. R., Haug M. C., Huwiler S. G., and Loessner M. J. (2011) The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol. Microbiol. 81, 1419–1432 10.1111/j.1365-2958.2011.07774.x [DOI] [PubMed] [Google Scholar]
- 61. Denes T., den Bakker H. C., Tokman J. I., Guldimann C., and Wiedmann M. (2015) Selection and characterization of phage-resistant mutant strains of Listeria monocytogenes reveal host genes linked to phage adsorption. Appl. Environ. Microbiol. 81, 4295–4305 10.1128/AEM.00087-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Autret N., Dubail I., Trieu-Cuot P., Berche P., and Charbit A. (2001) Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69, 2054–2065 10.1128/IAI.69.4.2054-2065.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kolly G. S., Mukherjee R., Kilacsková E., Abriata L. A., Raccaud M., Blaško J., Sala C., Dal Peraro M., Mikušová K., and Cole S. T. (2015) GtrA protein Rv3789 is required for arabinosylation of arabinogalactan in Mycobacterium tuberculosis. J. Bacteriol. 197, 3686–3697 10.1128/JB.00628-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Larrouy-Maumus G., Škovierová H., Dhouib R., Angala S. K., Zuberogoitia S., Pham H., Villela A. D., Mikušová K., Noguera A., Gilleron M., Valentínová L., Korduláková J., Brennan P. J., Puzo G., Nigou J., and Jackson M. (2012) A small multidrug resistance-like transporter involved in the arabinosylation of arabinogalactan and lipoarabinomannan in mycobacteria. J. Biol. Chem. 287, 39933–39941 10.1074/jbc.M112.400986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yerushalmi H., Lebendiker M., and Schuldiner S. (1995) EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270, 6856–6863 10.1074/jbc.270.12.6856 [DOI] [PubMed] [Google Scholar]
- 66. Trent M. S., Ribeiro A. A., Lin S., Cotter R. J., and Raetz C. R. (2001) An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276, 43122–43131 10.1074/jbc.M106961200 [DOI] [PubMed] [Google Scholar]
- 67. Mahne M., Tauch A., Pühler A., and Kalinowski J. (2006) The Corynebacterium glutamicum gene pmt encoding a glycosyltransferase related to eukaryotic protein-O-mannosyltransferases is essential for glycosylation of the resuscitation promoting factor (Rpf2) and other secreted proteins. FEMS Microbiol. Lett. 259, 226–233 10.1111/j.1574-6968.2006.00269.x [DOI] [PubMed] [Google Scholar]
- 68. Amer A. O., and Valvano M. A. (2002) Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148, 571–582 [DOI] [PubMed] [Google Scholar]
- 69. Berg S., Kaur D., Jackson M., and Brennan P. J. (2007) The glycosyltransferases of Mycobacterium tuberculosis: roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17, 35R–56R [DOI] [PubMed] [Google Scholar]
- 70. Mishra A. K., Alderwick L. J., Rittmann D., Wang C., Bhatt A., Jacobs W. R. Jr., Takayama K., Eggeling L., and Besra G. S. (2008) Identification of a novel α(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol. Microbiol. 68, 1595–1613 10.1111/j.1365-2958.2008.06265.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arrieta-Ortiz M. L., Hafemeister C., Bate A. R., Chu T., Greenfield A., Shuster B., Barry S. N., Gallitto M., Liu B., Kacmarczyk T., Santoriello F., Chen J., Rodrigues C. D., Sato T., Rudner D. Z., Driks A., Bonneau R., and Eichenberger P. (2015) An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol. Syst. Biol. 11, 839 10.15252/msb.20156236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kobayashi K., Ogura M., Yamaguchi H., Yoshida K., Ogasawara N., Tanaka T., and Fujita Y. (2001) Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183, 7365–7370 10.1128/JB.183.24.7365-7370.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Camilli A., Tilney L. G., and Portnoy D. A. (1993) Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8, 143–157 10.1111/j.1365-2958.1993.tb01211.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Reichmann N. T., Cassona C. P., and Gründling A. (2013) Revised mechanism of d-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 159, 1868–1877 10.1099/mic.0.069898-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kopp U., Roos M., Wecke J., and Labischinski H. (1996) Staphylococcal peptidoglycan interpeptide bridge biosynthesis: a novel antistaphylococcal target? Microb. Drug Resist. 2, 29–41 10.1089/mdr.1996.2.29 [DOI] [PubMed] [Google Scholar]
- 76. Strandén A. M., Ehlert K., Labischinski H., and Berger-Bächi B. (1997) Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179, 9–16 10.1128/jb.179.1.9-16.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bernal P., Zloh M., and Taylor P. W. (2009) Disruption of d-alanyl esterification of Staphylococcus aureus cell wall teichoic acid by the β-lactam resistance modifier (−)-epicatechin gallate. J. Antimicrob. Chemother. 63, 1156–1162 10.1093/jac/dkp094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Brauge T., Sadovskaya I., Faille C., Benezech T., Maes E., Guerardel Y., and Midelet-Bourdin G. (2016) Teichoic acid is the major polysaccharide present in the Listeria monocytogenes biofilm matrix. FEMS Microbiol. Lett. 363, fnv229 10.1093/femsle/fnv229 [DOI] [PubMed] [Google Scholar]
- 79. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 80. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ogura M., Ohsawa T., and Tanaka T. (2008) Identification of the sequences recognized by the Bacillus subtilis response regulator YrkP. Biosci. Biotechnol. Biochem. 72, 186–196 10.1271/bbb.70548 [DOI] [PubMed] [Google Scholar]
- 83. Ogura M., Tsukahara K., and Tanaka T. (2010) Identification of the sequences recognized by the Bacillus subtilis response regulator YclJ. Arch. Microbiol. 192, 569–580 10.1007/s00203-010-0586-4 [DOI] [PubMed] [Google Scholar]
- 84. Cao M., and Helmann J. D. (2004) The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186, 1136–1146 10.1128/JB.186.4.1136-1146.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sonnhammer E. L., von Heijne G., and Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.