Abstract
Ebola virus (EBOV) is a filamentous lipid-enveloped virus that causes hemorrhagic fever with a high fatality rate. Viral protein 40 (VP40) is the major EBOV matrix protein and regulates viral budding from the plasma membrane. VP40 is a transformer/morpheein that can structurally rearrange its native homodimer into either a hexameric filament that facilitates viral budding or an RNA-binding octameric ring that regulates viral transcription. VP40 associates with plasma-membrane lipids such as phosphatidylserine (PS), and this association is critical to budding from the host cell. However, it is poorly understood how different VP40 structures interact with PS, what essential residues are involved in this association, and whether VP40 has true selectivity for PS among different glycerophospholipid headgroups. In this study, we used lipid-binding assays, MD simulations, and cellular imaging to investigate the molecular basis of VP40–PS interactions and to determine whether different VP40 structures (i.e. monomer, dimer, and octamer) can interact with PS-containing membranes. Results from quantitative analysis indicated that VP40 associates with PS vesicles via a cationic patch in the C-terminal domain (Lys224, 225 and Lys274, 275). Substitutions of these residues with alanine reduced PS-vesicle binding by >40-fold and abrogated VP40 localization to the plasma membrane. Dimeric VP40 had 2-fold greater affinity for PS-containing membranes than the monomer, whereas binding of the VP40 octameric ring was reduced by nearly 10-fold. Taken together, these results suggest the different VP40 structures known to form in the viral life cycle harbor different affinities for PS-containing membranes.
Keywords: Ebola virus, lipid-binding protein, oligomerization, phosphatidylserine, plasma membrane, VP40, filovirus, viral budding
Introduction
The Filoviridae family includes Ebola virus (EBOV)2 and Marburg (MARV) virus, which use a negative sense RNA genome to replicate in the host cell (2). To date, no Food and Drug Administration-approved vaccines or therapeutics are available for EBOV or MARV, and with fatality rates as high as 90% (3, 4), these viruses are classified as category A pathogens by the National Institutes of Health. The EBOV genome encodes seven genes that can express eight proteins, including nucleoprotein, VP24, VP30, VP35, and L protein, which together constitute the nucleocapsid (5); the viral-surface glycoprotein (GP), a secreted glycoprotein GP; and the viral matrix protein VP40. VP40 is a peripheral membrane protein that regulates budding as well as virus structure and stability (6–8). VP40 is the most abundant protein contained in mature virions (9) and can associate with (10) and oligomerize on (8, 11–14) the plasma membrane (PM) inner leaflet, where it acquires the viral lipid envelope.
VP40, as the only EBOV protein expressed in mammalian cells, is sufficient to form virus-like particles (VLPs) that are nearly indistinguishable from authentic virions (10, 15). Thus, VP40 has served as a safe and viable model to study the binding, assembly, and egress processes of EBOV outside of BSL-4 laboratories. The absence of VP40 prevents proper trafficking of the nucleocapsid to the plasma membrane (16). Therefore, understanding how VP40 regulates assembly of VLPs both in vitro and in cells is crucial for understanding the viral life cycle and could have a significant impact in identifying potential therapeutic targets.
The first X-ray structure of VP40 (17) demonstrated that VP40 harbored an N-terminal domain that has been shown to mediate VP40 oligomerization (1, 7, 18) and a C-terminal domain that can interact with anionic membranes (1, 13, 19–22). Additionally, VP40 harbors an ∼43-amino acid-long N-terminal region that has not been resolved in crystal structures. This region contains the late domains (23–25) and the ALIX-binding motif (25), and it may also have latch-like properties with the C terminus (26).
Membrane interaction of the C-terminal domain has been hypothesized to induce structural rearrangement of VP40 to regulate oligomerization (1, 8, 13, 21, 22). Although the molecular basis of VP40 association with the anionic inner leaflet of the PM is unknown, it is clear that both electrostatic (19) and hydrophobic interactions (10, 13) play an important role. Hydrophobic residues in the C-terminal domain are critical for localization, membrane penetration, and budding (13), whereas interactions with phosphatidylserine (PS) have been shown to be important for association with lipid vesicles in vitro (19, 20, 22, 27) and budding from the plasma membrane of mammalian cells (27). Moreover, VP40 oligomers have been detected in VLPs and UV-inactivated virions (7, 8) and reside predominantly in filamentous structures emanating from the PM (12, 13, 28, 29). Larger oligomeric structures of VP40 may also play a role in viral assembly and egress (1, 12, 14).
Recently, VP40 was shown to be a dimer, which is mediated through an N-terminal domain α-helical interface mediated mostly by hydrophobic interactions (1). VP40 is a transformer/morpheein (30) as it can also form an octameric ring structure that binds RNA (6) as well as a hexameric filament through C-terminal domain (CTD) contacts (1). The dimer also revealed a cationic interface (Fig. 1) that lies adjacent to the hydrophobic residues that penetrate the PM (13). Mutation or deletion of these cationic residues, namely Lys224, Lys225, Lys274, and Lys275, greatly dampened budding and PM localization of VP40. To determine whether VP40 binds PS in a selective manner and whether these CTD cationic residues play a role in VP40 membrane binding and viral budding, we performed lipid binding and PM-localization analysis of WT VP40 and a panel of VP40 mutations. Together these studies reveal that VP40 associates selectively with PS compared with other glycerophospholipids, and electrostatic interactions between the C-terminal domain and the PM are critical to VP40 binding and PM association, and the VP40 dimer has higher affinity for PS-containing membranes than the VP40 monomer or VP40 octamer.
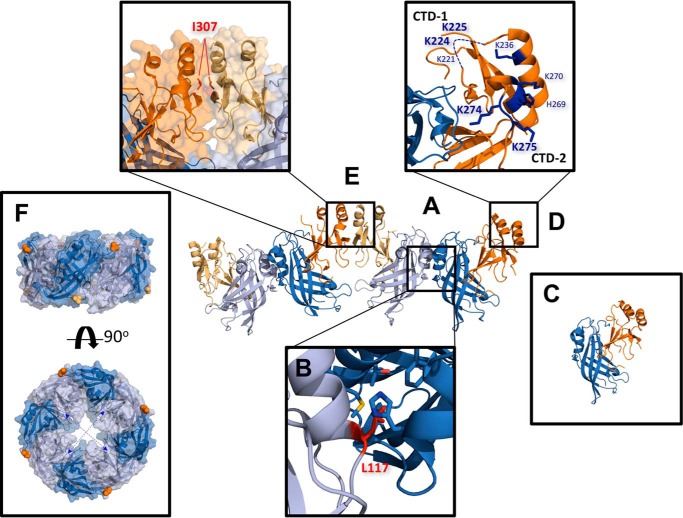
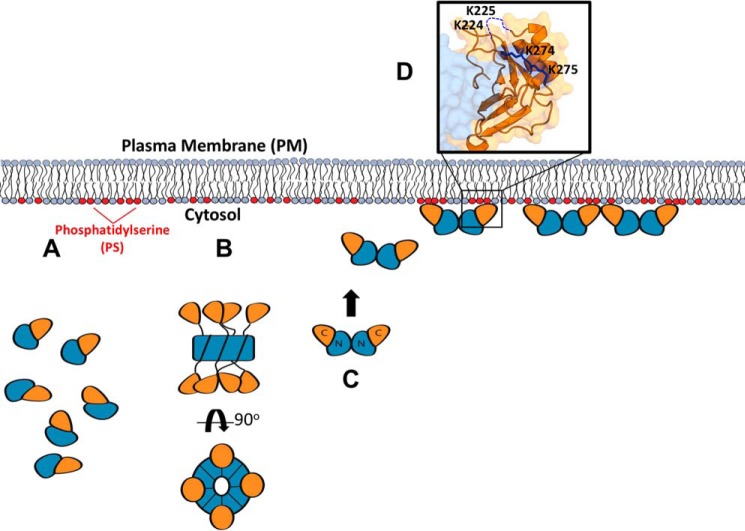
Figure 1.
Important structural features of VP40. A, two VP40 dimers are depicted (Protein Data Bank (PDB) code 4LDB). VP40 exists as a homodimer in solution consisting of an NTD (light/dark blue) and a CTD (light/dark orange). B, dimer is stabilized by an α-helical hydrophobic interface in which Leu117 (red stick) inserts into a hydrophobic pocket formed by Ala55, His61, Ala113, Thr112, and Met116 (blue sticks) of the other VP40 protomer. C, mutating Leu117 to arginine abrogates this interaction and forms predominantly monomeric VP40 in solution (1). D, CTD of VP40 contains a cationic patch enriched with basic residues (blue sticks) implicated in plasma membrane interactions (1). This study focuses on the highly conserved lysine residues, Lys224, Lys225, Lys274, and Lys275. As they occupy two distinct loops in the cationic patch, Lys224 and Lys225 are referred to as C-terminal motif 1 (CTD-1) and Lys274 and Lys275 as C-terminal motif 2 (CTD-2). Residues 221–230 form a highly flexible loop and are not resolved in the crystal structure (dashed blue line). E, additionally, dimer–dimer interactions, as well as hexamer–hexamer (not shown here), are mediated by a hydrophobic CTD interface. Embedded in this interface is residue Ile307 (red). Mutating Ile307 to arginine interferes with this interaction and induces a rearrangement of the VP40 subunits into an octameric ring structure (1). F, octameric ring (PDB code 4LDM) assembles via antiparallel NTD contacts, which requires the CTD of each VP40 subunit to disengage from its NTD via a 10-residue linker to expose the oligomerization interfaces. Because of the flexible nature of the linker, the detached CTDs of the octamer are not resolved in the crystal structure but are expected to alternatingly project above and below the ring as indicated by the first residue of each CTD linker (Ala188, light/dark orange spheres). The RNA-binding interface is located within the interior of the ring structure (dashed blue arrows) (PDB 1H2C).
Results
Generation of eVP40 conformational mutants
VP40 was recently established to be a dimeric protein in which dimerization is mediated by an N-terminal domain α-helical interface, a critical interaction necessary for viral budding (Fig. 1) (1). Recent and extensive structural analysis provided by Bornholdt et al. (1) indicates that VP40 exists in separate structural states for separate functions and that point mutations may affect one for more of these states or may change the equilibrium among them. Mutations of VP40, such as I307R, can lock VP40 in an octameric ring structure, for example.
A number of earlier studies have examined the role of PS in lipid vesicle binding and cellular assembly of VP40 demonstrating an important role for PS in membrane binding and membrane curvature changes (13, 19, 20, 31, 32) as well as a critical requirement of plasma membrane PS levels in VLP formation (27). However, the majority of this previous work predated knowledge on purification conditions and mutations that may influence VP40 dimeric, monomeric, hexameric, or octameric ring structure. In fact, previous work from the Stahelin laboratory using a GST-VP40 expression system with subsequent cleavage of the GST tag (13, 20) demonstrated purification of monomeric VP40 identified by chemical cross-linking.3
Thus, we sought to better understand the mode by which different VP40 structures interact with PS-containing membranes. We also wanted to examine the role of the 43 amino acids of unknown structure at the N terminus of VP40 in PS binding. We employed several different VP40 expression constructs for in vitro measurements, including a full-length WT His6-tag–expression system, a truncation (Δ43) of VP40, or point mutations of VP40 that lock VP40 into different structures.
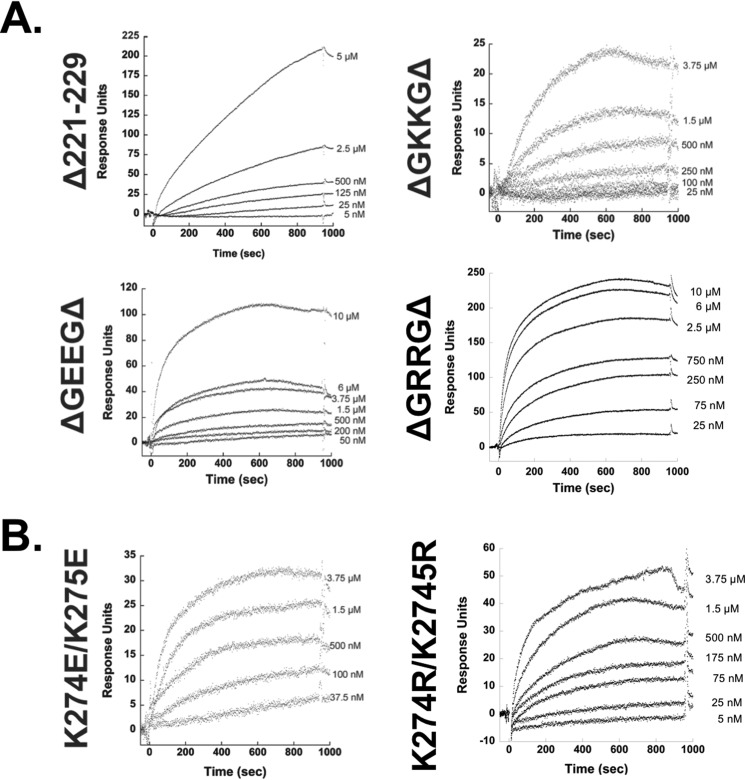
These point mutations included an L117R mutation that prevents the NTD α-helical dimerization and forms monomeric VP40 (Fig. 2) (1) and an I307R mutation that generates the VP40 octameric ring structure (Fig. 2) (1). Several mutations of cationic residues were also employed and included deletion of a CTD loop containing Lys224 and Lys225 (Δ221–229), deletion of residues adjacent to Lys224 and Lys225 and their respective charge reversal (ΔGEEGΔ, which deleted residues 221, 222, 227, 228, and 229), and partial cationic charge restoration (ΔGKKGΔ, which deleted residues 221, 222, 227, 228, and 229). We also prepared mutations of an adjacent cationic loop, which included Lys274 and Lys275 (K274E/K275E). These aforementioned Lys residues are conserved among the five types of ebolaviruses (Fig. 3). Finally, charge reversal mutations (ΔGRRGΔ and K274R/K275R) were also included to test the role of specific versus non-specific electrostatics in the hypothesized PS selectivity.
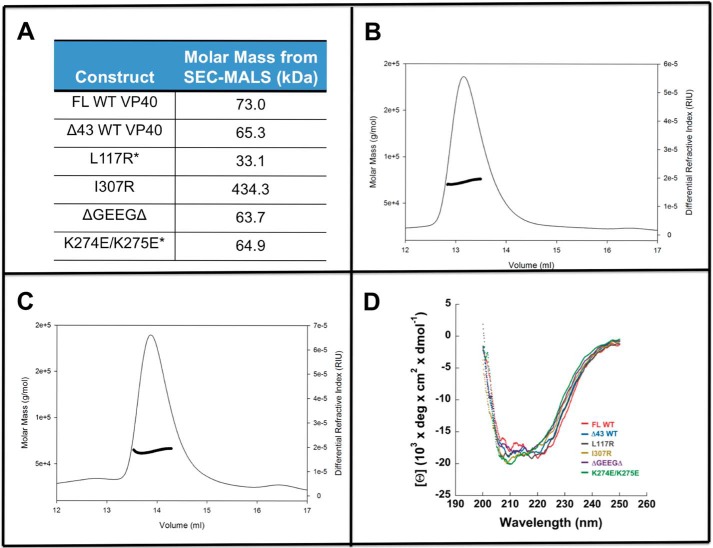
Figure 2.
SEC-MALS and CD spectroscopy analysis of different VP40 proteins employed in in vitro lipid-binding measurements. SEC-MALS analysis was used to confirm the stoichiometry of VP40 and mutants in solution. A, table of WT VP40 and mutants confirming the molecular weight of each species as calculated within the SEC-MALS trace. Asterisk indicates that a small secondary population of higher-order structure was observed in these samples. B, SEC-MALS trace of FL WT VP40. C, SEC-MALS trace of ΔGEEGΔ. D, CD spectroscopy analysis of WT VP40 and mutants confirming the agreement of α-helical and β-sheet content among all constructs.
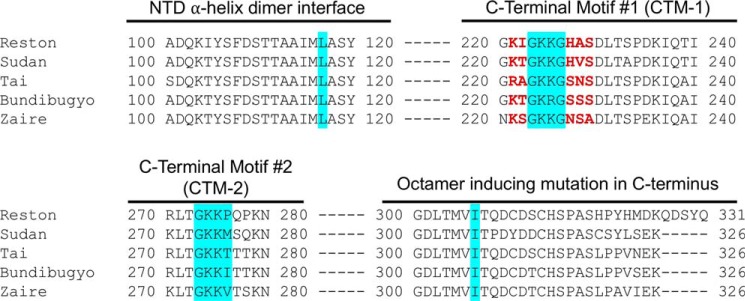
Figure 3.
Sequence alignment of the CTD found in the five different types of ebolaviruses. Mutations were made in the Zaire sequence of VP40, as highlighted in red.
To confirm that VP40 mutants to be used in in vitro assays were properly folded and in the anticipated oligomerization state, we performed CD spectroscopy and SEC-MALS, respectively, on each purified protein. As shown in Fig. 2D, CD spectroscopy studies demonstrated a similar and overlapping structural profile of full-length VP40, the Δ43 truncation, and point mutations consistent with α-helical and β-sheet content and previous reports of VP40 CD spectra (20, 29). SEC-MALS analysis (Fig. 2, A–C) demonstrated that VP40 and respective mutations were in the previously reported oligomeric state under the purification conditions used here. This included full-length and Δ43 VP40 being primarily dimeric, L117R being predominantly monomeric, with some formation of octameric ring as reported previously (1), I307R forming an octameric ring, and mutations or deletion of cationic residues in the CTD retaining the predominantly dimeric state. It is important to note that the K274E/K275E mutation primarily was found as a dimer, but a larger size aggregate was also detected by SEC-MALS (Fig. 2).
VP40 selectively binds lipid vesicles containing phosphatidylserine
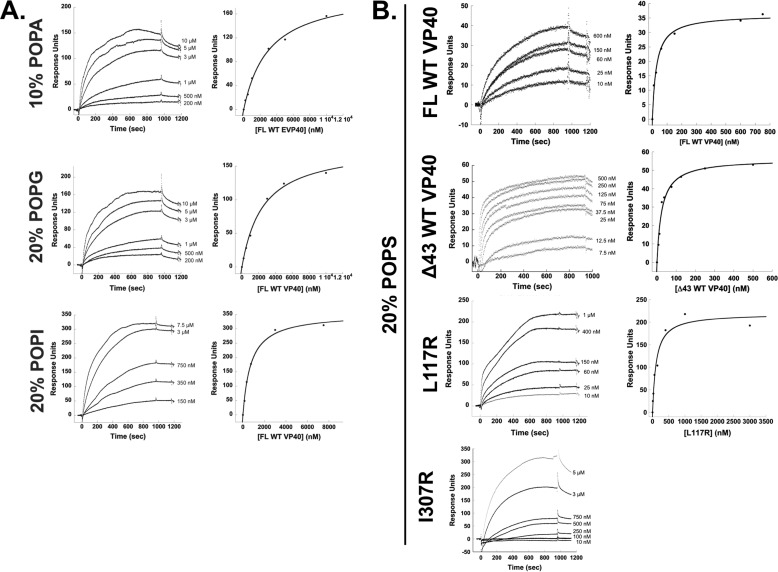
To examine the selectivity of VP40 for PS-containing vesicles, we monitored binding of WT VP40 using surface plasmon resonance (SPR) to lipid vesicles containing POPC/POPE/POPX (60:20:20) with a control flow cell of POPC/POPE (80:20) vesicles, where X is an anionic phospholipid. Along with PS, we tested WT VP40-binding affinity to lipid vesicles containing other anionic glycerophospholipids at an equimolar percentage of overall anionic charge. Vesicles containing either 20% phosphatidylinositol (POPI), 20% phosphatidylglycerol (POPG), or 10% phosphatidic acid (POPA) (note: net anionic charge of POPA is −2 as opposed to the other anionic glycerophospholipids' net charge of −1 at physiological pH) were compared with control vesicles. Table 1 and Fig. 4 summarize the findings that VP40 interacts selectively with phosphatidylserine-containing vesicles in comparison with vesicles containing PI, PG, or PA. VP40 dimer was found to associate with POPS-containing vesicles 20–60-fold more strongly than to vesicles containing POPI, POPA, or POPG (Fig. 4, A and B, and Table 1).
Table 1.
Apparent binding affinities (Kd) of full-length WT VP40 to lipid vesicles containing anionic glycerophospholipids
Apparent Kd values were measured for WT VP40 to liposomes containing 20% POPS, 20% POPI, 20% POPG, or 10% POPA. WT VP40 preferentially binds to lipids containing phosphatidylserine.
| Lipid composition | Apparent Kd |
|---|---|
| nm | |
| 70:20:10 POPC/POPE/POPA | 2500 ± 800 |
| 60:20:20 POPC/POPE/POPG | 2400 ± 200 |
| 60:20:20 POPC/POPE/POPI | 710 ± 40 |
| 60:20:20 POPC/POPE/POPS | 35 ± 15 |
Figure 4.
Binding measurements of VP40 with different anionic lipids. A, SPR of FL WT VP40-binding experiments with LUVs containing either 10% POPA, 20% POPG, or 20% POPI was tested to confirm WT VP40 selectivity to POPS-containing vesicles. n = 2 for all measurements. B, SPR sensorgrams and respective RU plots for FL WT VP40, Δ43 VP40, L117R, and I307R binding to vesicles containing 20% POPS. Concentrations of VP40 noted on sensorgrams account for stoichiometry of VP40 in solution. All experiments were measured in triplicate or greater.
Conformational selectivity of VP40-phosphatidylserine interactions
We next monitored binding using SPR of different VP40 protein constructs to lipid vesicles containing POPC/POPE/POPS (60:20:20) with a control flow cell of POPC/POPE (80:20) vesicles. We compared the binding of full-length (FL) VP40 to the N-terminally truncated (Δ43) construct. The apparent Kd values and SPR sensorgrams are shown in Table 2 and Fig. 4B, respectively. Both of these WT constructs bind with similar affinity (apparent Kd of 35 ± 15 and 35 ± 10 nm, respectively) to PS-containing vesicles, concluding that the N-terminal 43 amino acids do not appreciably contribute to the PS-vesicle binding in vitro.
Table 2.
Apparent binding affinities (Kd) of VP40 and mutants to lipid vesicles containing phosphatidylserine
Apparent Kd values were measured for WT VP40 and VP40 mutants to liposomes containing 20% POPS. WT VP40 and Δ43 VP40 both display similar binding affinity. Monomeric construct L117R also displays WT levels of affinity. Various mutations within the C-terminal cationic regions abrogate binding to varying degrees. n ≥ 3 for all measurements. (n = 3 for all measurements, except L117R n = 5 and FL WT VP40 n = 6). Error is reported as S.E. of the combined replicates.
| Construct | Predicted effect | Apparent Kd |
|---|---|---|
| FL WT VP40 | WT, full functionality | 35 ± 15 nm |
| Δ43 WT | WT, full functionality | 35 ± 10 nm |
| (Δ43) L117R | Monomer | 70 ± 30 nm |
| (Δ43) I307R | Octomeric ring | >3 μm |
| (Δ43) Δ221–229 | Loop deletion (still dimer) | >6 μm |
| (Δ43) ΔGKKGΔ | Partial loop restoration | >2 μm |
| (Δ43) ΔGEEGΔ | Partial loop restoration, charge reversed | >6 μm |
| (Δ43) ΔGRRGΔ | Partial loop restoration, charge maintained | 400 ± 70 nm |
| (Δ43) K274E/K275E | Charge reversal (still dimer) | >6 μm |
| (Δ43) K274R/K275R | Charge maintained (still dimer) | 600 ± 300 nm |
To investigate the structural basis of PS-vesicle binding, we first used a mutant of VP40, L117R, which exists primarily as a monomer (1), as this mutation disrupts the homodimer interface, and previous lipid-binding studies likely employed VP40 monomer in the PS-binding measurements (13, 19, 20, 31). The L117R monomeric mutant was shown to bind ∼2-fold less (apparent Kd of 60 ± 30 nm) than WT dimeric VP40. In contrast, another structural mutant, I307R, which has been shown to lock VP40 in the octameric RNA-binding ring structure (1), had >25-fold reduction in PS-vesicle binding (Table 2). The apparent Kd value for I307R was difficult to estimate as the response at 1 μm I307R was not sufficient to represent true saturation of binding, so this apparent Kd value should be considered an estimate. Nonetheless, PS binding is still evident for the octameric ring but significantly lower than that of VP40 monomers or dimers.
Ebola VP40 binds the plasma membrane through a cationic patch in the C-terminal domain
The interactions of VP40 with vesicles containing PS (19, 31) are thought to be mediated by cationic residues in the C-terminal domain (1). However, the molecular basis of this interaction is still poorly understood. To elucidate which highly conserved cationic regions of VP40 were important for PS binding, we looked at two primary regions: the basic patch from amino acids 221–229, which we have termed “C-terminal motif 1,” as well as the basic patch from amino acids 270–280, which we have termed “C-terminal motif 2.” Bornholdt et al. (1) first hypothesized these regions interacted with the anionic PM as mutation of these residues abrogated PM localization and VLP formation.
To determine whether the first motif was important for PS binding, we utilized a construct generated by Bornholdt et al. (1), where the loop of amino acids 221–229 (221KSGKKGNSA229) in the CTD was completely deleted. We also utilized other VP40 expression constructs generated by Bornholdt et al. (1), which partially restored this loop by re-incorporating only amino acids 223–226 (223GKKG226), forming the construct termed ΔGKKGΔ. We also wanted to determine whether the charges of these amino acids were important, so we mutated lysine 224 and 225 to form constructs ΔGEEGΔ (charge-reversal) and ΔGRRGΔ (charge-maintained) (1). In all of these cases, the amino acids flanking the 223–226 region have been deleted (yielding 223GXXG226). All mutants reduced binding by at least 15-fold (Table 2 and Fig. 5A), strongly suggesting the cationic residues in this loop region are an important determinant of association with vesicles containing PS. Notably, keeping Lys224 and Lys225 in the VP40 sequence but eliminating residues 221–222 and 227–229 also significantly reduced PS-dependent binding suggesting the position of this loop and/or other amino acids that are deleted are also important determinants of the PS-dependent association with the membrane interface.
Figure 5.
SPR of VP40 mutants within motifs of the CTD. A, sensorgrams and RU plots are shown for mutants of VP40 within amino acid region 221–229. Δ221–229 is a complete truncation of the C-terminal motif 1. ΔGKKGΔ has partial loop restoration of amino acids 223–226. ΔGEEGΔ has partial loop restoration of amino acids 223–226 but with amino acids 224 and 225 converted from lysine to glutamic acid. ΔGRRGΔ has partial loop restoration of amino acids 223–226 but with amino acids 224 and 225 converted from lysine to arginine. B, SPR sensorgrams for the C-terminal motif 2 mutants: K274E/K275E is a charge reversal from WT, and K274R/K275R is a charge-maintaining mutant to WT.
To determine the selectivity of Lys residues versus cationic charge in CTD-1, we replaced Lys224 and Lys225 with Arg. Notably, the Arg residues in the ΔGRRGΔ construct were sufficient to compensate for Lys. This suggests cationic charge is a major contributor of VP40 PS binding in CTD-1 but that Lys residues may be dispensable for sufficient PS interactions. Although the introduction of Arg was able to provide a higher capacity of binding to PS vesicles than that of Lys, the partial deletion construct harbored an ∼11-fold decrease in PS binding relative to WT, again underscoring the importance or positioning of loop residues (221, 222, or 227–229) in CTD-1.
We employed a similar strategy in examining motif 2 by measuring the association of K274E/K275E and with PS-containing vesicles. This mutation reduced PS-vesicle binding by >80-fold (Table 2 and Fig. 5B) demonstrating the important role of cationic residues at positions 274 and 275 in PS-vesicle binding. Furthermore, conservation of charge in CTD-2 K274R/K275R was sufficient to increase the binding affinity in comparison with the K274E/K275E mutation; however, when Arg was present at positions 274 and 275 instead of Lys, the mutation harbored a 17-fold reduction in PS-vesicle binding affinity. This strongly suggests the importance of these Lys residues in PS selectivity as Arg can compensate for Lys in non-specific electrostatic interactions between peripheral proteins and membranes. Thus, Lys may be indispensable in CTD-2 compared with those lysines found in CTD-1 for VP40 PS selectivity.
VP40 cellular localization analysis
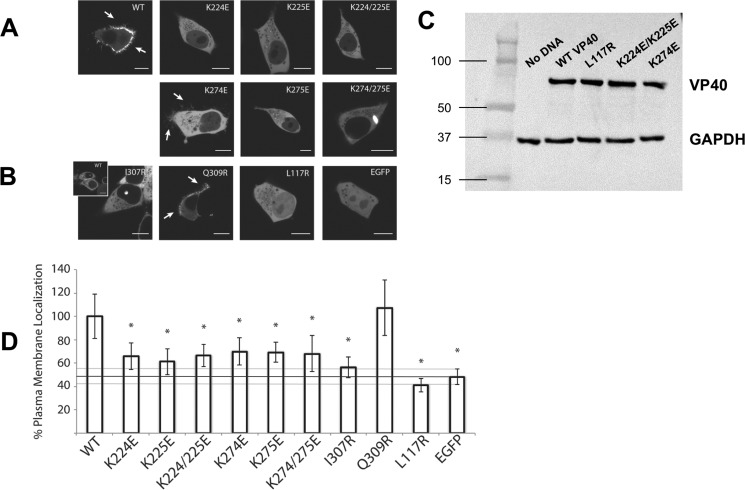
To assess the localization of the VP40 monomer and octamer in live cells, mutants L117R and I307R were made in a previously utilized EGFP-VP40 construct (12, 27). Constructs were transfected into HEK293 cells and imaged using confocal microscopy (Fig. 6). As demonstrated previously, EGFP-VP40 WT localized predominantly to the PM and formed extensions indicative of pre-assembled VLPs (33). In contrast, the monomeric mutant L117R displayed little to no detectable PM or internal membrane localization but was diffusely distributed between the cytoplasm and the nucleus, similar to free EGFP (Fig. 6B).
Figure 6.
Confocal live cell imaging of EGFP-VP40 and respective mutations. HEK293 cells were transfected with the indicated mutant constructs and imaged on a Zeiss LSM 710 confocal microscope 22–24 h post-transfection. A, WT EGFP-VP40 exhibited strong localization at the plasma membrane with extensive membrane protrusions (white arrows). CTD-1 mutants (K224E, K225E, and K224E/K225E) were mainly diffusely localized in the cytoplasm with no detectable VLP formation at the PM. Likewise, charge reversal mutations in the CTD-2 (K274E, K275E, K274E/K275E) resulted in little to no detectable PM localization. However, the mutant K274E demonstrated a partial ability to form VLPs as evidenced by the fractured protrusion at the PM (white arrows). Additionally, the double mutant K274/K275E formed bright intracellular globules. B, structural mutants (I307R and L117R) were also not detected at the PM but remained predominantly cytosolic with L117R diffuse in the nucleus as well. However, I307R occasionally exhibited a unique punctate nuclear localization similar to WT (inset). The control Q309R, which is near I307R but does not spontaneously induce octameric ring formation (1), displayed WT-like enrichment at the PM and VLP formation (white arrows). C, Western blot confirming equivalent heterologous expression of various VP40 mutants in cell culture. Western blot for GAPDH is shown as a loading control. D, quantification of %PM localization of WT and all mutants (%PM localization = PM intensity/(PM + cytosol intensity)·100%). Between 16 and 25 cells were analyzed for each mutant from three independent experiments. All bars represent the mean ± S.E. and are normalized to WT. A Student's t test was calculated for all mutants with respect to WT (*, p < 0.005). The %PM localization of EGFP is indicated with a black line as a marker for baseline PM localization. Scale bars, 10 μm.
The I307R octameric ring mutant also did not localize to the PM or internal membranes but was predominantly cytoplasmic with no detectable free diffusion into the nucleus, likely due to its larger size. Unexpectedly, I307R sometimes appeared to accumulate as a small globule within the nucleus in addition to its diffuse cytoplasmic distribution (Fig. 6B). WT VP40 was also found to localize in a similar subnuclear region in a fraction of the cells observed. Upon further examination, WT VP40 and I307R were observed to form this nuclear punctate in ∼5% of cells following a 24-h transfection period (data not shown). The control mutant Q309R, which was previously shown not to interfere with the CTD-CTD oligomerization interface (1), behaved like WT VP40 and was predominantly enriched on the PM (Fig. 6B).
To determine whether heterologous expression effected the outcomes of cellular localization analysis for WT VP40 and respective mutations, several Western blottings were performed to determine the cellular expression level of constructs versus that of a GAPDH background (Fig. 6C). WT VP40 and L117R harbored a similar level of expression demonstrating the reduced PM localization of L117R was not attributed to an overall lower level of expression. Similarly, several of the Lys mutations of VP40 employed expressed at a similar level to WT VP40. In contrast, we consistently detected a reduced expression level of I307R compared with other VP40 constructs. This lower level of I307R detection relative to WT VP40 has also been observed previously in a 293E cell line (1). Although the reasons for the lower level of I307R expression are unknown, we speculate it may be attributed to the distinct cellular role of the VP40 octamer and the nuclear localization observed (discussed more below).
Several mutations of the Lys residues described were made in the EGFP-VP40 construct to analyze the role of the highly conserved cationic patch (Fig. 1) residues in mediating PM localization and VLP formation in live cells. To assess the role of the motif 1 lysines, charge reversal mutants K224E/K225E, K224E, and K225E were generated. K224E/K225E failed to localize to the PM in HEK293 cells (Fig. 6). Additionally, single mutations of either Lys224 or Lys225 to glutamic acid abolished PM localization and evidence of VLP budding. In a like manner, we assessed mutation of the adjacent motif 2 lysines. Similar to the motif 1 double mutant, K274E/K275E failed to localize to the PM. However, instead of a completely diffuse cytosolic distribution, K274E/K275E also was found as bright clusters in a number of cells. Single mutations of either Lys274 or Lys275 greatly reduced detectable PM localization. Interestingly, neither single mutant produced intracellular accumulations like K274E/K275E, and K274E still formed some filamentous protrusions at the PM.
Molecular dynamics (MD) simulations implicate C-terminal cationic residues of VP40 for phosphatidylserine binding
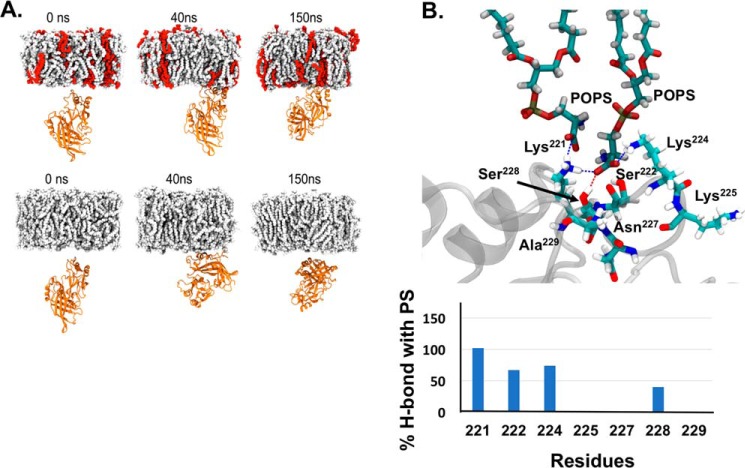
MD simulations of eVP40 protomer–membrane systems with and without POPS lipids in the membrane show that the presence of POPS lipids has a significant effect on eVP40 association with the PM. The CTD of the eVP40 protomer is readily associated with the PM consisting of POPS lipids, as shown in Fig. 7A, top panel. Several cationic lysine residues as well as some polar residues, such as Ser222, Lys224, Lys225, Ser233, Lys236, Lys270, Lys274, Lys275, Thr277, and Ser278, participate in hydrogen bonding with POPS. A typical hydrogen-bonded configuration is shown in Fig. 7, A, top panel, and B, and the dynamics of association is shown in Movie S1. Lys221, Ser222, Lys224, and Asn228 provided the most significant number of H-bonds with PS among the CTD-1 residues throughout the simulation time (Fig. 7B). The orientation of PS binding in CTD-1 shown in Fig. 7B supports the in vitro PS selectivity data where at least three points of contact with the PS headgroup are made. Moreover, residues in addition to Lys224 and Lys225 in CTD-1 are important for PS binding throughout the simulation, which is also supported by the extensive in vitro data detailing a role for residues 221, 222, and 227–229 when they were deleted from the VP40 construct or deleted in the presence of Arg224 and Arg225.
Figure 7.
MD simulation of VP40 to PS lipids. A, snapshots at different times during the MD simulations of eVP40 protomer association with the (top panel) membrane consisting of POPS lipids (POPS, red; other lipids, gray) and (bottom panel) membrane without POPS. B, residues Lys221, Ser222, Lys224, and Ser228 involved in the formation of hydrogen bonds with PS. For these illustrated residues, there is no PI binding except for Lys224 which shows ∼75% PS binding versus ∼8% PI binding (data not shown). In addition to the illustrated residues, Lys225 also makes H-bonds with PS. The bar graph shows the number of H-bonds made with PS by respective residues throughout one of the simulations times.
In contrast to when POPS was in the membrane, the eVP40 protomer had only weak and transient interactions with the membrane that did not contain POPS (Fig. 7A, bottom panel, and Movie S2). The weak CTD interactions allowed the eVP40 protomer to rotate and make lipid interactions through its NTD surface as shown in Fig. 7A, bottom panel, and Movie S2
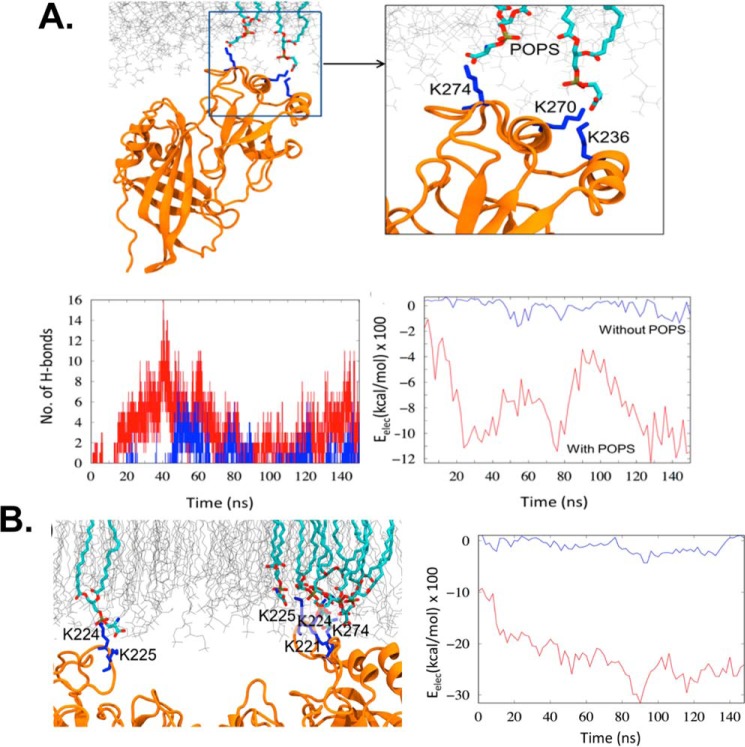
To compare the interaction of the eVP40 protomer and the PM with and without POPS, we calculated the hydrogen bonds and electrostatic interaction energy. Fig. 8A illustrates the time evolution of the number of hydrogen bonds. Fig. 8B highlights the C-terminal motif 1 residues that are involved in hydrogen bonding with the serine headgroup on POPS. In the absence of POPS in the membrane, the eVP40 protomer formed a few transient bonds with the POPC and POPE but to a significantly lesser extent (Fig. 7A). This is also displayed in Fig. 8A, which compares the attractive, electrostatic interaction energy between eVP40 and lipids in the presence and absence of POPS. As the eVP40 associates with the membrane-containing POPS, the electrostatic interaction energy decreases from −200 kcal/mol to −1200 kcal/mol, whereas in the absence of POPS it remains much higher.
Figure 8.
MD simulation of VP40 hydrogen-bond formation. A, eVP40 protomer–lipid interactions. Top, representative hydrogen-bonded configuration of eVP40 in which cationic lysine residues make hydrogen-bond interactions with the anionic headgroups of POPS. Bottom left, number of hydrogen bonds, and bottom right, electrostatic energy of interaction between protomer and the PM. Red curves, with POPS, and blue curves, without POPS. B, eVP40 dimer association with PM. Left, typical snapshot of the lysine residues in the CTD basic patch interacting with the POPS. Right, electrostatic energy of interaction between eVP40 dimer and the PM. Red, with POPS; blue, without POPS.
We compared the membrane association and lipid specificity of the eVP40 protomer with that of eVP40 dimer. As with the eVP40 protomer, the dimer strongly associated with the PM in the presence of POPS. Compared with only one CTD interaction in the eVP0 protomer, the CTD interactions on both ends of the dimer provide more stability for the protein at the membrane surface (Fig. 8B). The two CTDs anchored to the membrane also reduce the dimer flexibility by removing the rotational degree of freedom available for the protomer (34). As shown in Fig. 8B, the electrostatic interaction energy for the final eVP40 dimer–membrane complex showed a 2-fold decrease compared with the protomer–membrane complex indicating the enhanced stability of the dimer at the membrane interface. In the absence of POPS, some transient interactions with the membrane can occur but not enough to stabilize the complex.
Discussion
In this study, we have examined how different VP40 structures interact with PS-containing membranes. In vitro binding studies demonstrated that both full-length WT VP40 and a VP40 construct with an N-terminal truncation of 43 amino acids (Δ43 WT) both harbor similar binding affinity for PS-containing membranes. The N-terminal region of VP40 contains two established late domains (23) and a third recently elucidated interaction site for ALIX (25). The absence of these motifs or other basic amino acids (4 Arg residues in the N terminus) in the mutant VP40 constructs does not seem to affect binding to vesicles containing PS. Although all of the in vitro data (with the exception of FL VP40) was collected using a mutant library based off of the Δ43 WT parent sequence, we have confirmed that both FL WT and Δ43 WT bind with similar affinities to PS-containing vesicles, suggesting that the N-terminal 43 amino acids may not play a critical role in association with PS.
Additional in vitro studies with different anionic lipids demonstrated that VP40 binds selectively to PS-containing vesicles by greater than 1 order of magnitude compared with phosphatidylinositol, phosphatidylglycerol, or phosphatidic acid, and it is not merely acting as an anionic charge sensor as the MARV matrix protein (35). Although selectivity for PS among glycerophospholipids has been clarified in this study, selectivity for PS based upon acyl chain length and/or saturation is not known and warrants further investigation. Acyl chain saturation has been shown to be an important factor in membrane binding of HIV-1 Gag (36, 37), but it has been relatively unexplored in filovirus work.
Next, we compared the WT dimer construct to VP40 bearing an L117R mutation, which disrupts the N-terminal homodimeric interface yielding primarily monomeric VP40 (1). From the in vitro SPR analysis, we find that this monomeric construct binds only 2-fold less than WT (Table 1 and Fig. 4). This is unexpected, as other viral matrix proteins have shown lipid binding only when the matrix maintains assembling ability (38, 39). The similar affinity of the VP40 monomer and dimer could be explained in as much as the PS-binding site on VP40 may not be near the dimeric interface, leading both monomeric VP40 and dimeric (WT) VP40 to bind membranes via the C-terminal domain with similar affinity. This may be further supported by earlier studies that demonstrated association with PS-containing membranes using purification conditions that likely produced a recombinant VP40 monomer (17, 30, 31). Additionally, the CTD lipid-binding surfaces may not be equivalent in the dimer, and only one of the cationic patches may be predominantly accessible to interact with the anionic membrane interface. Indeed, MARV VP40, which also forms a dimer, seems to require dimerization for significant avidity and anionic lipid binding (35, 40). However, MARV VP40 and EBOV VP40 have only 16% sequence identity in their CTDs and also harbor significantly different CTD structures in terms of cationic charge and shape of the lipid-binding surface (40). Moreover, MARV VP40 acts as an anionic charge sensor (35, 40), whereas EBOV VP40 harbors PS selectivity as shown here and in previous studies (14, 27), which further supports PS-selective binding properties of the EBOV VP40 CTD.
Interestingly, L117R does not localize to the PM in cells or even internal membranes but remains diffusely distributed throughout the cell. Because L117R-VP40 is monomeric, its lipid-binding site may be unobstructed and therefore able to display binding in a simplified, highly controlled environment. In cells, however, VP40 may require interactions with various host factors for transport and membrane budding (41). Interaction with these factors may be dependent upon the VP40 oligomerization state. Cellular transport of VP40 may be mediated in a structure-dependent manner, and perhaps the dimer is necessary to interact with Sec24C (42) and/or other aspects of the cellular transport machinery.
Considering that the monomer L117R binds to PS with an apparent affinity tantamount to the PS biosensor Lact C2 (27, 43), it is remarkable that it fails to bind to the plasma membrane in cells. Perhaps multimerization enhances VP40 plasma membrane affinity, as has been observed for HIV-1 Gag (44), which would be prohibited in L117R (Fig. 9). Alternatively, VP40 is known to harbor important interactions with phosphoinositides at the plasma membrane, which have been shown to stabilize VP40 oligomers (14). Notably, VP40 oligomers are abundant at the plasma membrane interface (12, 13); perhaps efficient assembly of VP40 is necessary for phosphoinositide interactions (14), whereas assembly of L117R is not possible due to mutation of the NTD α-helical dimerization interface.
Figure 9.
Summary of VP40 structures in the life cycle of EBOV. As described throughout, VP40 forms a monomer, dimer, and RNA-binding octamer. A, monomer is the simplest unit of VP40 but may not possess biologically relevant lipid-binding properties. Despite harboring high affinity for PS-containing membranes, mutations of the NTD α-helical interface that mediate dimerization disrupt PM localization of VP40 as described herein as well as in Bornholdt et al. (1). VP40 trafficking to the PM may require the intact dimer to interact with host proteins such as Sec24C. B, octameric ring conformation renders VP40 less fit for PS binding, suggesting alternative cellular functions consistent with its proposed role in regulation of viral transcription (1). Presumably, the PS-binding surface in the VP40 CTD is not optimized for high-affinity PS binding in the octameric form. Additionally, the octameric VP40 may not be sufficient for proper trafficking to the PM. C, VP40 dimers harbor the requisite PS-binding properties for PM localization and budding. D, selective PS binding is mediated through CTD residues Lys224, Lys225, Lys274, and Lys275 in VP40 dimers.
WT VP40 is able to conformationally rearrange into the octameric ring structure that can bind RNA. However, although VP40 ring structures are found in infected cells, they were not found in mature virions (6, 7). This suggests that although WT VP40 can adopt a ring conformation that may provide some transcriptional regulatory function within the cell, it is unclear whether this ring form possesses PS-binding capabilities. Using the I307R mutant, which locks VP40 in the octameric structure (1), PS binding was detected but nearly an order of magnitude lower than that of the dimer (Table 2). The I307R mutation likely presents the C-terminal domain in a different manner than VP40 dimers or monomers that is not optimal for selective PS binding.
The I307R mutant did not localize to the plasma membrane or intracellular membranes in our live cell experiments but instead remained diffuse in the cytoplasm. This suggests that the free CTDs of the octamer may be serving an alternative function to plasma membrane binding in the cellular context. I307R occasionally accumulated in a subnuclear compartment in addition to its cytoplasmic distribution. Indeed, VP40 has been reported to localize in the nucleus during the early stages of infection (45), and WT VP40 was observed to accumulate in a similar subnuclear region in our experiments, but the mechanisms that regulate this event are still unknown. Our data suggest that VP40 may adopt the octamer conformation to localize in the nucleus. The domains of VP40 that mediate import into the nucleus are unclear, but nuclear localization signals typically consist of short sequences of positively charged lysines or arginines (46) and have been identified in other related viral matrix proteins (47, 48). As the octamer binds PS with weaker affinity than the dimer, it is an attractive hypothesis that this structural rearrangement orients VP40 to preferentially interact with the nuclear transport system instead of PM lipids by presenting the CTDs in a new manner. It is also possible that the octamer can bind non-PS lipids more specific to the nucleus that we have not yet screened in our assays.
The analysis of C-terminal domain residues involved in PS binding indicated the importance of a C-terminal domain loop (motif 1) consisting of residues 221–229, which confirmed the hypothesis by Bornholdt et al. (1). This region is a flexible loop where a GKKG motif is highly conserved among the five types of ebolavirus (Zaire, Sudan, Reston, Tai Forest, and Bundibugyo) (Fig. 3). The resulting mutations in this region all abrogated PS binding by at least 11-fold, even when the highly conserved GKKG motif was reintroduced without several adjacent residues. This suggested that the local structure and overall cationic patch charge are important in promoting the high-affinity PS binding of VP40.
The complex structure of this loop also seems to confer some degree of selectivity. Although the amino acids surrounding the GKKG motif are not as sequence-conserved among the five types of ebolavirus, they are functionally conserved, i.e. there are amino acids that can function as hydrogen-bond acceptors (Thr, Ser, or Asn) or as flexible linkers (Gly), for example. Perhaps not only the conserved, positively-charged amino acids are important, but the length and positioning of the loop itself may prove to be important in PS binding and/or electrostatic interactions. Although a conserved PS-binding domain or motif has yet to be discovered in nature, proteins that harbor PS selectivity have typically three points of contact for stereospecific binding to PS. PS-selective binding modules include C2 domains (49) and some pleckstrin homology domains (50–52). Interactions of these domains with PS are most often mediated by H-bonding or electrostatic interactions by Thr, Ser, Asn, Lys, or Arg residues. Indeed, binding studies performed herein suggest important roles for residues 221, 222, and 227–229 as their deletion greatly reduced PS binding, which was supported by significant H-bonding by Lys221, Ser222, and Asn228 throughout the MD simulation time with PS-containing membranes. Additionally, H-bonds were also be observed throughout the simulations between PS and Ser233, Lys236, Lys270, Thr277, and Ser278 in addition to Lys224, Lys225, Lys274, and Lys275.
Calcium can also be an important determinant of PS-dependent membrane binding as shown for some C2 domains (49) and annexin V (53). Here, calcium acts as a bridge between the phosphate on the PS headgroup and a calcium coordination site on the protein. Although the role of calcium in VP40 membrane binding has not been explored, it is important to note that calcium is an important factor in VLP formation and EBOV budding (54). In fact, VP40 was sufficient to increase cellular levels of calcium in mammalian cells (55), and its role in promoting some degree of VP40 lipid binding cannot be ruled out at this time.
Maintaining a certain threshold of positive charge in the CTD is necessary to achieve effective contacts with the plasma membrane; individual mutation of either Lys224 or Lys225 abolished plasma membrane localization and evidence of VLP budding. Interestingly, it has been demonstrated that VP40 requires a minimum of 10–20 mol % PS, representative of the plasma membrane, in order to interact with synthetic lipid vesicles in vitro as well as a certain threshold of PS in cells (27, 32). It appears that the initial interaction of VP40 with the plasma membrane is electrostatically driven. Therefore, it seems probable that these lysine residues help facilitate initial contacts with the plasma membrane, whereas these or adjacent amino acid residues (e.g. 221–229) may fine-tune interactions with PS.
In addition to the cationic residues in motif 1 of the VP40 CTD, there is a second highly conserved cationic patch (motif 2) adjacent to motif 1 and roughly on the same interface. It has been proposed that this region contains a “switch” to induce VP40 conformational changes following anionic lipid binding (19). As with motif 1, mutation of motif 2 (K274E/K275E) abrogated binding to PS vesicles (Table 2 and Fig. 5B) suggesting both regions play an important role in electrostatic interactions and/or PS-selective interactions. CTD-2 Lys residues (Lys274 and Lys275) seem to play an important role in PS selectivity as replacement with Arg residues was not sufficient to restore PS binding as K274R/K275R had greater than an order of magnitude loss in PS vesicle affinity.
Our EGFP-tagged K274E/K275E mutant failed to accumulate on the PM in live cells but instead had a diffuse cytosolic appearance and in some cells aggregated intracellularly (Fig. 6). It has been established that VP40 interacts with several key cellular proteins to facilitate trafficking and budding (11, 42, 56). As Lys274 and Lys275 have been hypothesized to promote the oligomerization of VP40 on the PM (1), it is possible that losing both lysine residues could shift the equilibrium of VP40 from interacting with the PM to host cellular proteins, stalling it at various points of protein transport, resulting in the observed VP40 accumulations in the cytosol. Indeed, this appearance has been observed in other VP40 CTD mutations (57) and attributed to enhanced association with host cellular factors.
Re-introducing either lysine may partially rescue this stalled trafficking phenotype as cytosolic accumulations were not observed in either single mutant, and even some degree of PM interaction was detected in the K274E mutant (Fig. 6). It may be that the re-introduced lysine restores the equilibrium of trafficking to the PM but is insufficient to mediate efficient accumulation of VP40 on the PM above the detection limit of our confocal system. Nevertheless, the final bridge to the PM seems to be abrogated in the K274E/K275E double mutant, indicating that these lysine residues are critical for PM targeting and supporting proper accumulation of VP40 on the PM inner leaflet, in agreement with previous work (1).
Experimental procedures
Materials
POPC, POPE, POPG, POPI, POPA, and POPS were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and used without further purification. A QuikChange site-directed XL mutagenesis kit was from Agilent Technologies, (Santa Clara, CA). Rosetta2 BL21DE3 pLysS cells were from Novagen (Madison, WI). Nickel-nitrilotriacetic acid resin was from Qiagen (Valencia, CA), and the L1 sensor chip from was GE Healthcare (Little Chalfont, UK).
Protein expression and purification
VP40 bacterial expression plasmids were a kind gift from the laboratory of Dr. Erica Ollmann Saphire (The Scripps Research Institute, La Jolla, CA). VP40 and respective mutations were expressed in Rosetta2 BL21DE3 pLysS cells (Novagen, Madison, WI) to an A600 nm of 0.4–0.6 at 37 °C, and subsequently induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside. Induced cells were allowed to express for 5 h at room temperature and were purified according to the QIAExpressionist batch method (Qiagen, Valencia, CA). The eluted proteins were dialyzed against 10 mm Tris, pH 8.0, containing 300 mm NaCl and stored at a concentration of 1–2 mg/ml at 4 °C and used within 2 weeks. Protein concentrations were determined with the BCA method (Pierce).
CD spectroscopy
VP40 protein (WT and mutants) was purified as described above. Protein samples were prepared at 0.25 mg/ml and imaged on a Jasco J-815 through a 1-mm quartz cuvette (Starna Cells Inc., Atascadero, CA), all as described in Greenfield et al. (58). Measurements reported are the average of five separate wave scans.
Surface plasmon resonance
POPC/POPE (80:20) large unilamellar vesicles (LUVs) and POPC/POPE/POPS (60:20:20) LUVs were prepared as described previously (27). SPR was performed using a Biacore X instrument and an L1 sensor chip (GE Healthcare). Data were collected at a flow rate of 5 μl/min using 80-μl injections of VP40 and respective mutations. Protein concentrations typically were in the range of 10-fold below or 15-fold above the respective apparent Kd value of each construct. Protein concentrations were calculated to account for the stoichiometry of dimerization or oligomerization (SEC-MALS) in lipid-binding analysis. The saturation response (Req) was determined at each protein concentration. Data were analyzed using Kaleidagraph (Synergy Software, Reading, PA), and trials were repeated in triplicate (note: L117R was repeated five times; I307R and FL WT-VP40 was repeated six times) to determine the mean and standard deviation of the apparent Kd. Req values (saturation RU values) were then plotted versus protein concentrations (C), and the Kd value was determined by a nonlinear least-squares analysis of the binding isotherm using the equation, Req = Rmax/(1 + Kd/C).
SEC-MALS
1 mg of purified proteins was loaded onto a Superdex 200 Increase 10/300 gel-filtration column (GE Healthcare), pre-equilibrated with buffer (10 mm Tris-HCl, pH 8.0, containing 300 mm NaCl). Following elution from the column, the FPLC system was connected in-line with a miniDAWN TREOS followed by an Optilab T-rEX refractometer (Wyatt Technologies). Data processing and absolute molar mass calculation were performed using ASTRA software (Wyatt Technologies).
Molecular biology
EGFP-VP40 mutants for live-cell imaging were generated in a pcDNA3.1 vector using a Quikchange site-directed XL mutagenesis kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The mutations were confirmed by automated DNA sequencing.
Cell culture and cell imaging
HEK293 cells were cultured and maintained at 37 °C and 5% CO2 supplemented with low-glucose Dulbecco's modified Eagle's medium (Life Technologies, Inc.), 10% fetal bovine serum, and 1% penicillin/streptomycin. Preceding transfection, cells were grown to 80–90% confluency in a T-25 flask, trypsinized for 4–5 min at 37 °C and 5% CO2, and then seeded into an 8-well Lab-Tek IITM chambered coverglass (Thermo Fisher Scientific) to a final confluency of 50–70%. Cells were then transfected the next day with 300 ng/well DNA using Lipofectamine® LTX and PLUSTM reagent (Invitrogen) according to the manufacturer's instructions. Cells were imaged 22–24 h post-transfection with a Zeiss LSM 710 confocal microscope using a Plan Apochromat 63× 1.4 NA oil objective. The 488-nm line of the argon laser was used to excite EGFP, and the laser attenuator transmission was held constant at 2% throughout the experiments with the emission collected from 493 to 797 nm. A total of 16–25 cells were imaged for each mutant from three independent transfections. The identity of the mutants was concealed during imaging to minimize bias. Heterologous expression levels were confirmed via Western blotting as described previously (59). Antibodies used include monoclonal rabbit anti-EBOV VP40 (IBT Bioservices 0301-010); monoclonal mouse anti-eGFP (Thermo Fisher Scientific F56–6A1.2.3); mouse anti-GAPDH antibody (Abcam ab8245); sheep anti-mouse IgG H&L (Abcam ab6808); and goat anti-rabbit IgG H&L (Abcam ab205718).
Image analysis
Cellular data were quantified using a MATLAB script (Mathworks, Natick, MA) provided by Johnson et al. (14). The script measures the average EGFP intensity of the pixels within the MATLAB script-identified plasma membrane and cytoplasm of the cell under analysis. The % PM localization was calculated by dividing the EGFP PM intensity by the total EGFP (PM + cytosol) intensity × 100%.
Molecular dynamics simulations on eVP40 membrane interactions
The protein-membrane systems were built with the Charmm-Gui Membrane Builder (60). Two different lipid compositions were used for each of the eVP40 protomer systems: a membrane with POPS, POPE, and POPC in the ratio of 20:20:60 and a membrane with POPC and POPE in the ratio of 20:80, with at least 100 lipids in each leaflet. Systems were neutralized using counter-ions. All simulations were performed using NAMD2.11 (61) with the CHARMM36 force field (62). The particle mesh Ewald (PME) method (63) was used for the long-range electrostatic interactions. The SHAKE (64) algorithm was used to constrain the covalent bonds involving hydrogen atoms.
Each system was minimized for 10,000 steps and equilibrated with a six-step Charmm-Gui protocol. A Nose-Hoover Langevin-piston method (65) was used with a piston period of 50 fs and a decay of 25 fs to control the pressure. The temperature was controlled using Langevin temperature coupling with a friction coefficient of 1 ps−1. The simulation time step was set to 2 fs for all productions runs. Production runs were performed for 150 ns for each system. For calculations of hydrogen bonds, the cutoff distance and cutoff angle were 3.5 Å and 30 °C, respectively. Visual molecular dynamics (66) was used to visualize and analyze the data. Visual molecular dynamics namdenergy plugin was used to calculate the interaction energies.
Author contributions
K. D., E. O. S., and R. V. S. designed and planned the experiments. K. D., C. T. F., and S. O. performed the experiments and analyzed the data. J. B. G., B. S. G., and P. P. C. designed the molecular dynamics simulations, and J. B. G. performed the molecular dynamics simulations. K. D. and R. V. S. wrote the manuscript with input, review, and approval of the final version of the text from all authors.
Supplementary Material
Acknowledgments
The imaging studies were supported by the Indiana University School of Medicine-South Bend Imaging and Flow Cytometry Core. We thank Kristen A. Johnson for generating the MATLAB script and providing helpful advice in analyzing cellular localization.
This work was supported by National Institutes of Health Grants AI081077 and AI121841 (to R. V. S.) and National Institutes of Health CBBI Predoctoral Training Fellowship T32GM075762 (to K. D. and C. T. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Movies S1 and S2.
E. Adu-Gyamfi and R.V. Stahelin, unpublished data.
- EBOV
- Ebola virus
- MARV
- Marburg virus
- RU
- response unit
- SPR
- surface plasmon resonance
- MD
- molecular dynamics
- PS
- phosphatidylserine
- PM
- plasma membrane
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- POPI
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol
- POPA
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine
- VLP
- virus-like particle
- EGFP
- enhanced GFP
- GP
- glycoprotein
- LUV
- large unilamellar vesicle
- SEC-MALS
- size-exclusion chromatography–multiangle light scattering
- FL
- full length.
References
- 1. Bornholdt Z. A., Noda T., Abelson D. M., Halfmann P., Wood M. R., Kawaoka Y., and Saphire E. O. (2013) Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154, 763–774 10.1016/j.cell.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ortin J., and Martin-Benito J. (2015) The RNA synthesis machinery of negative-stranded RNA viruses. Virology 479–480, 532–544 [DOI] [PubMed] [Google Scholar]
- 3. Smetana J., Chlíbek R., and Vacková M. (2006) Outbreak of Marburg hemorrhagic fever in Angola. Epidemiol. Mikrobiol. Imunol. 55, 63–67 [PubMed] [Google Scholar]
- 4. Bwaka M. A., Bonnet M. J., Calain P., Colebunders R., De Roo A., Guimard Y., Katwiki K. R., Kibadi K., Kipasa M. A., Kuvula K. J., Mapanda B. B., Massamba M., Mupapa K. D., Muyembe-Tamfum J. J., Ndaberey E., et al. (1999) Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: clinical observations in 103 patients. J. Infect. Dis. 179, Suppl. 1, 1–7 [DOI] [PubMed] [Google Scholar]
- 5. Choi J., and Croyle M. (2013) Emerging targets and novel approaches to Ebola virus prophylaxis and treatment. BioDrugs 27, 563–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomis-Rüth F. X., Dessen A., Timmins J., Bracher A., Kolesnikowa L., Becker S., Klenk H. D., and Weissenhorn W. (2003) The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure 11, 423–433 10.1016/S0969-2126(03)00050-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoenen T., Biedenkopf N., Zielecki F., Jung S., Groseth A., Feldmann H., and Becker S. (2010) Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J. Virol. 84, 7053–7063 10.1128/JVI.00737-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Panchal R. G., Ruthel G., Kenny T. A., Kallstrom G. H., Lane D., Badie S. S., Li L., Bavari S., and Aman M. J. (2003) In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. U.S.A. 100, 15936–15941 10.1073/pnas.2533915100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han Z., Boshra H., Sunyer J. O., Zwiers S. H., Paragas J., and Harty R. N. (2003) Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77, 1793–1800 10.1128/JVI.77.3.1793-1800.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jasenosky L. D., Neumann G., Lukashevich I., and Kawaoka Y. (2001) Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75, 5205–5214 10.1128/JVI.75.11.5205-5214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Timmins J., Schoehn G., Kohlhaas C., Klenk H.-D., Ruigrok R. W., and Weissenhorn W. (2003) Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312, 359–368 10.1016/S0042-6822(03)00260-5 [DOI] [PubMed] [Google Scholar]
- 12. Adu-Gyamfi E., Digman M. A., Gratton E., and Stahelin R. V. (2012) Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys. J. 102, 2517–2525 10.1016/j.bpj.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adu-Gyamfi E., Soni S. P., Xue Y., Digman M. A., Gratton E., and Stahelin R. V. (2013) The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 288, 5779–5789 10.1074/jbc.M112.443960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson K. A., Taghon G. J., Scott J. L., and Stahelin R. V. (2016) The Ebola virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 6, 19125 10.1038/srep19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noda T., Sagara H., Suzuki E., Takada A., Kida H., and Kawaoka Y. (2002) Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76, 4855–4865 10.1128/JVI.76.10.4855-4865.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noda T., Ebihara H., Muramoto Y., Fujii K., Takada A., Sagara H., Kim J. H., Kida H., Feldmann H., and Kawaoka Y. (2006) Assembly and budding of Ebolavirus. PLoS Pathog. 2, e99 10.1371/journal.ppat.0020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dessen A., Volchkov V., Dolnik O., Klenk H. D., and Weissenhorn W. (2000) Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19, 4228–4236 10.1093/emboj/19.16.4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harty R. (2009) No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res. 81, 189–197 10.1016/j.antiviral.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruigrok R. W., Schoehn G., Dessen A., Forest E., Volchkov V., Dolnik O., Klenk H. D., and Weissenhorn W. (2000) Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300, 103–112 10.1006/jmbi.2000.3822 [DOI] [PubMed] [Google Scholar]
- 20. Soni S. P., Adu-Gyamfi E., Yong S. S., Jee C. S., and Stahelin R. V. (2013) The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys. J. 104, 1940–1949 10.1016/j.bpj.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Radzimanowski J., Effantin G., and Weissenhorn W. (2014) Conformational plasticity of the Ebola virus matrix protein. Protein Sci. 23, 1519–1527 10.1002/pro.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stahelin R. V. (2014) Membrane binding and bending in Ebola VP40 assembly and egress. Front. Microbiol. 5, 300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Licata J. M., Simpson-Holley M., Wright N. T., Han Z., Paragas J., and Harty R. N. (2003) Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77, 1812–1819 10.1128/JVI.77.3.1812-1819.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freed E. O. (2002) Viral late domains. J. Virol. 76, 4679–4687 10.1128/JVI.76.10.4679-4687.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han Z., Madara J. J., Liu Y., Liu W., Ruthel G., Freedman B. D., and Harty R. N. (2015) ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of Ebola VP40: a role for ALIX in Ebola virus egress. J. Infect. Dis. 212, Suppl. 2, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silva L. P., Vanzile M., Bavari S., Aman J. M., and Schriemer D. C. (2012) Assembly of Ebola virus matrix protein VP40 is regulated by latch-like properties of N- and C- terminal tails. PLoS One 7, e39978 10.1371/journal.pone.0039978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adu-Gyamfi E., Johnson K. A., Fraser M. E., Scott J. L., Soni S. P., Jones K. R., Digman M. A., Gratton E., Tessier C. R., and Stahelin R. V. (2015) Host cell plasma membrane phosphatidylserine regulates the assembly and budding of Ebola virus. J. Virol. 89, 9440–9453 10.1128/JVI.01087-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adu-Gyamfi E., Digman M.A, Gratton E., Stahelin R V. (2012) Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys. J. 103, L41–L43 10.1016/j.bpj.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adu-Gyamfi E., Soni S. P., Jee C. S., Digman M. A., Gratton E., and Stahelin R. V. (2014) A loop region in the N-terminal domain of Ebola virus VP40 is important in viral assembly, budding, and egress. Viruses 6, 3837–3854 10.3390/v6103837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaffe E. K. (2005) Morpheeins–a new structural paradigm for allosteric regulation. Trends Biochem. Sci. 30, 490–497 10.1016/j.tibs.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 31. Scianimanico S., Schoehn G., Timmins J., Ruigrok R. H., Klenk H. D., and Weissenhorn W. (2000) Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19, 6732–6741 10.1093/emboj/19.24.6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soni S. P., and Stahelin R. V. (2014) The Ebola virus matrix protein VP40 selectively induces vesiculation from phosphatidylserine-enriched membranes. J. Biol. Chem. 289, 33590–33597 10.1074/jbc.M114.586396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., and Kawaoka Y. (2010) Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6, e1001121 10.1371/journal.ppat.1001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gc J. B., Gerstman B. S., and Chapagain P. P. (2017) Membrane association and localization dynamics of the Ebola virus matrix protein VP40. Biochim. Biophys. Acta 1859, 2012–2020 10.1016/j.bbamem.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 35. Wijesinghe K. J., and Stahelin R. V. (2015) Investigation of the lipid binding properties of the Marburg virus matrix protein VP40. J. Virol. 90, 3074–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dick R. A., Goh S. L., Feigenson G. W., and Vogt V. M. (2012) HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. U.S.A. 109, 18761–18766 10.1073/pnas.1209408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vlach J., and Saad J. (2013) Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc. Natl. Acad. Sci. U.S.A. 110, 3525–3530 10.1073/pnas.1216655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson B. A., Reed J. C., Geary C. D., Swain J. V., and Lingappa J. R. (2014) A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J. Virol. 88, 5718–5741 10.1128/JVI.03609-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghanam R. H., Samal A. B., Fernandez T. F., and Saad J. S. (2012) Role of the HIV-1 matrix protein in Gag intracellular trafficking and targeting to the plasma membrane for virus assembly. Front. Microbiol. 3, 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oda S., Noda T., Wijesinghe K. J., Halfmann P., Bornholdt Z. A., Abelson D. M., Armbrust T., Stahelin R. V., Kawaoka Y., and Saphire E. O. (2015) Crystal structure of Marburg virus VP40 reveals a broad, basic patch for matrix assembly and a requirement of the N-terminal domain for immunosuppression. J. Virol. 90, 1839–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R. W., and Weissenhorn W. (2003) Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326, 493–502 10.1016/S0022-2836(02)01406-7 [DOI] [PubMed] [Google Scholar]
- 42. Yamayoshi S., Noda T., Ebihara H., Goto H., Morikawa Y., Lukashevich I. S., Neumann G., Feldmann H., and Kawaoka Y. (2008) Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 3, 168–177 10.1016/j.chom.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kay J. G., Koivusalo M., Ma X., Wohland T., and Grinstein S. (2012) Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23, 2198–2212 10.1091/mbc.E11-11-0936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dick R. A., and Vogt V. M. (2014) Membrane interaction of retroviral Gag proteins. Front. Microbiol. 5, 187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nanbo A., Watanabe S., Halfmann P., and Kawaoka Y. (2013) The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 3, 1206 10.1038/srep01206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freitas N., and Cunha C. (2009) Mechanisms and signals for the nuclear import of proteins. Curr. Genomics 10, 550–557 10.2174/138920209789503941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ghildyal R., Ho A., Wagstaff K. M., Dias M. M., Barton C. L., Jans P., Bardin P., and Jans D. A. (2005) Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin β1 independent of importin α. Biochemistry 44, 12887–12895 10.1021/bi050701e [DOI] [PubMed] [Google Scholar]
- 48. Wang Y. E., Park A., Lake M., Pentecost M., Torres B., Yun T. E., Wolf M. C., Holbrook M. R., Freiberg A. N., and Lee B. (2010) Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 6, e1001186 10.1371/journal.ppat.1001186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cho W., and Stahelin R. V. (2006) Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761, 838–849 10.1016/j.bbalip.2006.06.014 [DOI] [PubMed] [Google Scholar]
- 50. Uchida Y., Hasegawa J., Chinnapen D., Inoue T., Okazaki S., Kato R., Wakatsuki S., Misaki R., Koike M., Uchiyama Y., Iemura S., Natsume T., Kuwahara R., Nakagawa T., Nishikawa K., et al. (2011) Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. U.S.A. 108, 15846–15851 10.1073/pnas.1109101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang B. X., Akbar M., Kevala K., and Kim H. Y. (2011) Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192, 979–992 10.1083/jcb.201005100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lucas N., and Cho W. (2011) Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 286, 41265–41272 10.1074/jbc.M111.300806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. van Genderen H. O., Kenis H., Hofstra L., Narula J., and Reutelingsperger C. P. (2008) Extracellular annexin A5: functions of phosphatidylserine-binding and two-dimensional crystallization. Biochim. Biophys. Acta 1783, 953–963 10.1016/j.bbamcr.2008.01.030 [DOI] [PubMed] [Google Scholar]
- 54. Han Z., and Harty R. N. (2005) Packaging of actin into Ebola virus VLPs. Virol. J. 2, 92 10.1186/1743-422X-2-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han Z., Madara J. J., Herbert A., Prugar L. I., Ruthel G., Lu J., Liu Y., Liu W., Liu X., Wrobel J. E., Reitz A. B., Dye J. M., Harty R. N., and Freedman B. D. (2015) Calcium regulation of hemorrhagic fever virus budding: mechanistic implications for host-oriented therapeutic intervention. PLoS Pathog. 11, e1005220 10.1371/journal.ppat.1005220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruthel G., Demmin G. L., Kallstrom G., Javid M. P., Badie S. S., Will A. B., Nelle T., Schokman R., Nguyen T. L., Carra J. H., Bavari S., and Aman M. J. (2005) Association of ebola virus matrix protein VP40 with microtubules. J. Virol. 79, 4709–4719 10.1128/JVI.79.8.4709-4719.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reynard O., Nemirov K., Page A., Mateo M., Raoul H., Weissenhorn W., and Volchkov V. (2011) Conserved proline-rich region of Ebola virus matrix protein VP40 is essential for plasma membrane targeting and virus-like particle release. J. Infect. Dis. 204, Suppl. 3, 884–891 [DOI] [PubMed] [Google Scholar]
- 58. Greenfield N. J. (2006) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gc J. B., Johnson K. A., Husby M. L., Frick C. T., Gerstman B. S., Stahelin R. V., and Chapagain P. P. (2016) Interdomain salt-bridges in the Ebola virus protein VP40 and their role in domain association and plasma membrane localization. Protein Sci. 25, 1648–1658 10.1002/pro.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qi Y., Ingólfsson H. I., Cheng X., Lee J., Marrink S. J., and Im W. (2015) CHARMM-GUI martini maker for coarse-grained simulations with the martini force field. J. Chem. Theory Comput. 11, 4486–4494 10.1021/acs.jctc.5b00513 [DOI] [PubMed] [Google Scholar]
- 61. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mackerell A. D., Jr. (2004) Empirical force fields for biological macromolecules: overview and issues. J. Comput. Chem. 25, 1584–1604 10.1002/jcc.20082 [DOI] [PubMed] [Google Scholar]
- 63. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., and Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Physics 103, 8577–8593 10.1063/1.470117 [DOI] [Google Scholar]
- 64. Ryckaert J.-P., Ciccotti G., and Berendsen H. J. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Physics 23, 327–341 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- 65. Brooks M. M., Hallstrom A., and Peckova M. (1995) A simulation study used to design the sequential monitoring plan for a clinical trial. Stat. Med. 14, 2227–2237 10.1002/sim.4780142006 [DOI] [PubMed] [Google Scholar]
- 66. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.