Abstract
The type I interferons (IFNs) are a family of cytokines with diverse biological activities, including antiviral, antiproliferative, and immunoregulatory functions. The discovery of the hormonally regulated, constitutively expressed IFNϵ has suggested a function for IFNs in reproductive tract homeostasis and protection from infections, but its intrinsic activities are untested. We report here the expression, purification, and functional characterization of murine IFNϵ (mIFNϵ). Recombinant mIFNϵ (rmIFNϵ) exhibited an α-helical fold characteristic of type I IFNs and bound to IFNα/β receptor 1 (IFNAR1) and IFNAR2, but, unusually, it had a preference for IFNAR1. Nevertheless, rmIFNϵ induced typical type I IFN signaling activity, including STAT1 phosphorylation and activation of canonical type I IFN signaling reporters, demonstrating that it uses the JAK–STAT signaling pathway. We also found that rmIFNϵ induces the activation of T, B, and NK cells and exhibits antiviral, antiproliferative, and antibacterial activities typical of type I IFNs, albeit with 100–1000-fold reduced potency compared with rmIFNα1 and rmIFNβ. Surprisingly, although the type I IFNs generally do not display cross-species activities, rmIFNϵ exhibited high antiviral activity on human cells, suppressing HIV replication and inducing the expression of known HIV restriction factors in human lymphocytes. Our findings define the intrinsic properties of murine IFNϵ, indicating that it distinctly interacts with IFNAR and elicits pathogen-suppressing activity with a potency enabling host defense but with limited toxicity, appropriate for a protein expressed constitutively in a sensitive mucosal site, such as the reproductive tract.
Keywords: immunology, innate immunity, interferon, protein expression, signal transduction
Introduction
The type I interferons (IFNs) are a family of cytokines comprising ∼20 members, including 14 α subtypes and one of each β, κ, ω, ϵ, τ, σ, and ς (1), that are critical in regulating innate and adaptive responses to infection and tumorigenesis. They induce this protection by a myriad of effects on cells, including the activation of antiviral and antibacterial states and regulation of cell proliferation, migration, and survival. In addition, the well-characterized “conventional” type I IFNs, such as IFNα subtypes and IFNβ, can regulate the development and activation of virtually every effector cell of the innate and adaptive immune response (2). Members of the type I IFN family of cytokines can promote survival of activated T and B cells (3, 4), activate natural killer (NK)5 cells (5), induce MHC-I up-regulation (6), and provide signals for dendritic cell maturation (7, 8). Their importance in host defense is underscored by the conservation of a multicomponent, species-specific type I IFN family found throughout vertebrates.
We identified the gene encoding the newest member of the type I IFN family of cytokines, IFNϵ, in the IFN locus on human chromosome 9 and the syntenic mouse chromosome 4 (9). We also showed it was unique in being constitutively expressed in the female reproductive tract and regulated by hormones but not by pathogens (10). Using IFNϵ−/− mice, we demonstrated that this new IFN was important in protection from herpes simplex virus 2 and Chlamydia infections of the reproductive tract (10). However, the mechanism of action was unclear in these studies because the intrinsic properties of IFNϵ protein had not been elucidated. Although some studies have proposed antiviral protection by IFNϵ constructs in mucosal immune responses, no protein product was characterized (10–12).
Therefore, to complement in vivo studies and to facilitate further work in murine models to understand the functions of this distinct protein, we undertook to define the intrinsic properties of murine IFNϵ. Here, we report the identification and characterization of the mature form of a mammalian IFNϵ, specifically the production and purification of recombinant murine (rm) IFNϵ, and have profiled its physicochemical and biological properties. rmIFNϵ showed the same broad range of biological activities (antiviral, antiproliferative, and immunoregulatory) as conventional IFNs α and β, but its potency was significantly lower. Consistent with this, we found that rmIFNϵ had a low affinity for binding IFNAR components relative to conventional type I IFNs. Another clear difference between rmIFNϵ and conventional type I IFNs was its high activity on human cells, which confirms its distinct interaction with the IFNAR receptor, a property that will make it suitable for study in humanized mouse models of disease. Indeed, we demonstrate here that rmIFNϵ induces HIV restriction factors and inhibits HIV replication in human T cells. Thus, we present new and critical data on the range and potency of a novel cytokine, murine IFNϵ, with unique characteristics fit for purpose as it functions to regulate mucosal immunity in the female reproductive tract.
Results
Expression and physicochemical characterization of rmIFNϵ
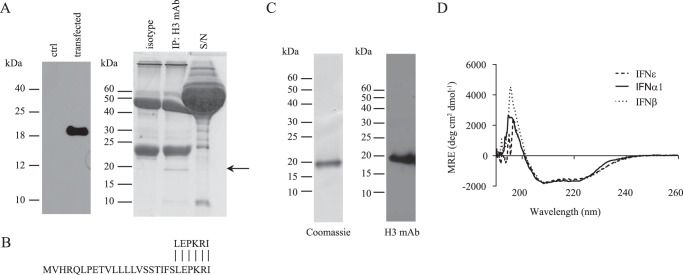
As a first step in characterizing the physicochemical and biological properties of murine IFNϵ, it was essential to elucidate where the signal peptide of this protein was cleaved to generate the mature, secreted protein as is the case with previously characterized type I IFNs. The Ifne1 gene was expressed under the control of a CMV promoter and transiently transfected into HEK293 cells. Supernatants from these cells were found to contain a protein of ∼20 kDa detected by SDS-PAGE and immunoblotting with an anti-IFNϵ monoclonal antibody (Fig. 1A, left panel). Immunoprecipitation of IFNϵ from these supernatants led to the visualization of a band at ∼20 kDa on Coomassie-stained SDS-PAGE that was not seen when immunoprecipitation was carried out with an isotype control antibody (Fig. 1A, right panel). Amino-terminal sequencing of this 20-kDa protein identified six amino acid residues, LEPKRI, representing residues 22–27 of the rmIFNϵ protein (RefSeq accession number NP_796322) (Fig. 1B). This result indicated that the mature IFNϵ polypeptide began at leucine 22 of the published sequence for rmIFNϵ (RefSeq accession number NP_796322) and, therefore, that the mature protein has a theoretical molecular mass of 20,006 Da (13).
Figure 1.
Physicochemical characterization of recombinant rmIFNϵ. A, the supernatant from HEK293 cells transiently transfected with pCMV-Ifne-IRES-mCitrine showed the presence of a band detectable by an anti-mIFNϵ monoclonal antibody (clone H3) (left panel). Coomassie-stained SDS-PAGE (15%, v/v; right panel) revealed a band of ∼20 kDa immunoprecipitated from transiently transfected HEK293 cells using an anti-mIFNϵ monoclonal antibody (clone H3). The arrow indicates the presence of a band corresponding to the size of rmIFNϵ. B, identification of amino-terminal residues of purified rmIFNϵ showing homology with amino-terminal residues of mature rmIFNϵ protein (RefSeq accession number NP_796322). The upper line denotes the identified native rmIFNϵ signal peptide. C, Coomassie-stained reducing SDS-PAGE (15%, v/v) analysis of rmIFNϵ purified from insect cell culture supernatant (left panel) and Western blot analysis of the purified rmIFNϵ immunoblotted with an anti-IFNϵ mAb (clone H3; right panel). D, CD spectral analysis of purified rmIFNϵ compared with rmIFNβ and rmFNα1. A representative scan from three independent experiments is shown. ctrl, control; S/N, supernatant; MRE, mean residue ellipticity; deg, degrees; IP, immunoprecipitation.
Insect cell–expressed rmIFNϵ has an α-helical fold
For physicochemical and biological characterization, rmIFNϵ was produced in a baculovirus expression system and purified using an immunoaffinity chromatography column coupled with an anti-IFNϵ monoclonal antibody. Analysis of the purified protein by SDS-PAGE (Fig. 1C, left panel) and Western blotting (Fig. 1C, right panel) revealed the presence of a protein at the size expected for rmIFNϵ (∼20 kDa) that was detected with an anti-IFNϵ antibody (clone H3). The purified protein was subjected to circular dichroism (CD) spectral analysis to demonstrate the overall protein fold. As can be seen in Fig. 1D, the mean residue ellipticity showed minima at 208 and 222 nm, a profile characteristic of α-helical proteins, such as IFNα and IFNβ. These data suggest that the ∼20-kDa protein expressed and purified from insect cell culture had an α-helical fold typical of other type I IFNs.
rmIFNϵ has lower affinity for its cognate receptors
We used microscale thermophoresis (MST) to assess the binding affinities between rmIFNϵ and recombinant forms of the extracellular domains (ECDs) of mIFNAR1 and mIFNAR2 and compared these results with those obtained with other type I IFNs, rmIFNα1 and rmIFNβ (Table 1 and Fig. S1). Our results revealed that rmIFNϵ had a lower binding affinity for mIFNAR2-ECD than both rmIFNα1 and rmIFNβ. The affinity of the rmIFNα1-mIFNAR2-ECD interaction was 2.18 nm (mean of three independent experiments), similar to previously published studies (14), whereas the mIFNϵ-mIFNAR2-ECD interaction was measured to be 6.58 μm, showing ∼3000-fold lower affinity for mIFNAR2-ECD than rmIFNα1. For the interaction with mIFNAR1-ECD, we measured the affinity of rmIFNϵ to be 589 nm (mean of three independent experiments), which is around 46-fold lower compared with the rmIFNβ-mIFNAR1-ECD interaction at 12.7 nm (mean of three independent experiments) but ∼4-fold higher than the affinity of the rmIFNα1-mIFNAR1-ECD interaction. These results suggest that rmIFNϵ has different binding affinities for IFNAR1 and IFNAR2 compared with rmIFNα1 and rmIFNβ.
Table 1.
Specific activities and properties of rmIFNα1, rmIFNβ, and rmIFNϵ
The concentrations at which the IFNs exhibit 50% of the maximal response (EC50 or IC50 as indicated) for each of the antiviral, antiproliferative, and antibacterial responses on mouse cells are shown. The dissociation constant for binding to recombinant forms of mouse IFNAR1-ECD and IFNAR2-ECD are also given.
| Interferon | Specific activitya | Antiviral activityb (EC50) | Antiproliferative activityc (IC50) | Antibacterial activityc (IC50) | Affinity to IFNAR1-ECDd (KD) | Affinity to IFNAR2-ECDd (KD) |
|---|---|---|---|---|---|---|
| IU/mg | pmol/ml | pmol/ml | pmol/ml | nm | nm | |
| rmIFNα1 | 2.4 ± 0.2 × 107 | 3.17 ± 0.63 × 10−3 | 1.333 ± 0.243 | Not assessed | 2666.7 ± 665.8 | 2.18 ± 0.38 |
| rmIFNβ | 2.2 ± 0.6 × 108 | 0.39 ± 0.13 × 10−3 | 0.055 ± 0.076 | 3.16 ± 0.78 | 12.67 ± 5.03 | 1673.3 ± 424.4 |
| rmIFNϵ | 2.1 ± 0.3 × 105 | 214.7 ± 14.6 × 10−3 | 191.9 ± 93.51 | 382.1 ± 189.5 | 589.67 ± 125.9 | 6583.3 ± 1675.1 |
a Calculated by normalizing the amount of antiviral activity at the concentration of protein (mg/ml). Means ± S.D. are given.
b EC50 calculated by nonlinear regression (curve fit) using GraphPad Prism software (version 7.01). EC50 is shown as mean ± S.D. of duplicate independent experiments.
c IC50 calculated by nonlinear regression (curve fit) using GraphPad Prism software (version 7.01). IC50 is shown as mean ± S.D. of at least duplicate independent experiments.
d KD, the dissociation constant, was calculated using microscale thermophoresis by fitting the signal from thermophoresis + T-jump to the single binding model using the NT.Analysis software. Means ± S.D. of triplicate independent experiments are given for each IFN with each receptor component.
Signaling
Following receptor engagement, an early step in IFN signaling is activation of signal transducers and activators of transcription (STAT) proteins, which enter the nucleus to bind interferon-stimulated response elements (ISREs) in the promoters of interferon-regulated genes (IRGs). Therefore, we next investigated whether the rmIFNϵ would induce activation of STAT1 and whether STAT1 would bind ISRE and IRG promoter-driven signaling reporters.
rmIFNϵ induces STAT1 phosphorylation
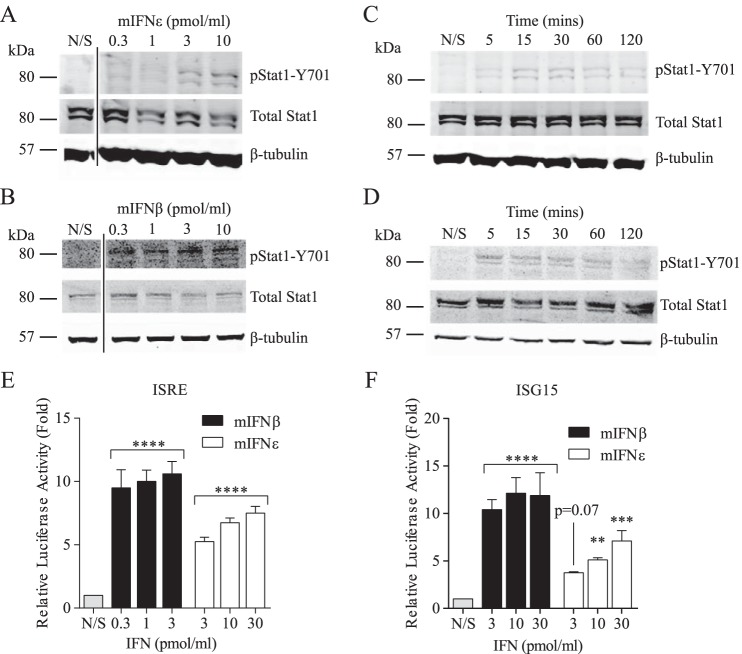
We sought to determine whether rmIFNϵ activated STAT1 like other type I IFNs. STAT1 phosphorylation on tyrosine 701 was apparent after stimulation of RAW264.7 cells with as little as 3 pmol/ml rmIFNϵ and was found to increase in a dose-dependent manner (Fig. 2A). That rmIFNβ induced phosphorylation of STAT1 at 0.3 pmol/ml, a lower dose than rmIFNϵ, suggested that rmIFNϵ is less active than rmIFNβ (Fig. 2B). To investigate whether or not the kinetics of STAT1 activation were different between rmIFNϵ and rmIFNβ, samples were taken 5, 15, 30, 60, and 120 min following stimulation with a 10 pmol/ml concentration of either IFN. STAT1 phosphorylation occurred as early as 5 min after rmIFNϵ stimulation, peaking 15–30 min after stimulation and decreasing after 60–120 min (Fig. 2C). Similarly, rmIFNβ stimulation resulted in peak STAT1 phosphorylation 5 min after treatment and was found to decrease by 120 min (Fig. 2D) as published previously (15). These results demonstrate that rmIFNϵ can induce the rapid activation of STAT1, although a higher dose is required to achieve a similar level of activation as seen following stimulation with rmIFNβ.
Figure 2.
rmIFNϵ induces STAT phosphorylation and activates canonical type I IFN signaling reporters. A–D, activation of STAT1 (Tyr701) phosphorylation by rmIFNϵ (A and C) and mIFNβ (B and D) in a dose- and time-dependent manner. RAW264.7 cells were treated for 60 min (A and B) or with a 10 pmol/ml concentration of either rmIFNϵ (C) or mIFNβ (D). STAT1 phosphorylation at Tyr701 (pStat1-Y701) total STAT1, and β-tubulin were detected in all whole-cell lysates. Data shown are representative of at least two independent experiments. E, measurement of luciferase activity in Ifnar1−/− MEFs transfected with an ISRE-luciferase reporter and stimulated with increasing doses of either rmIFNϵ or rmIFNβ. Data are representative of three independent experiments performed in technical triplicate. Means, with error bars representing S.D., are shown. Statistical analyses were performed using one-way ANOVA and represent significance of stimulated samples compared with unstimulated controls. F, measurement of luciferase activity in Ifnar1−/− MEFs transfected with a luciferase reporter under the control of 700 bp of ISG15 promoter and stimulated with increasing doses of either rmIFNϵ or rmIFNβ. Data are representative of three independent experiments performed in technical triplicate. Means, with error bars representing S.D., are shown. Statistical analyses were performed using one-way ANOVA and represent significance of stimulated samples compared with unstimulated controls (**, p < 0.01; ***, p < 0.001; ****, p < 0.0001). N/S, not stimulated.
rmIFNϵ can activate ISRE and IRG promoter-reporters
To determine the consequence of the aforementioned STAT phosphorylation, we used the STAT-dependent ISRE-luciferase reporter transfected into mouse embryonic fibroblasts (MEFs) to ascertain the relative ability of rmIFNϵ to drive conventional type I IFN–induced transcription. Our results show that rmIFNϵ induced an ISRE-luciferase response in a dose-dependent manner, consistent with its induction of STAT1 (Fig. 2E). To confirm this result, another luciferase reporter was utilized. This reporter consisted of a cloned promoter 700 bp upstream of the transcription start site of the IRG Isg15. rmIFNϵ was also able to achieve a similar dose-dependent luciferase induction with this construct, albeit more weakly than rmIFNβ (Fig. 2F). These data suggest that rmIFNϵ can induce STAT1 phosphorylation and signaling and the transcription of canonical IRGs via ISRE promoter elements.
Biological activities of rmIFNϵ
Type I IFNs are well-characterized in regard to their abilities to induce antiviral, antiproliferative, and immunoregulatory states in cells (16). We investigated the ability of rmIFNϵ to induce these responses in cells in comparison with rmIFNα1 or rmIFNβ in vitro.
rmIFNϵ demonstrates antiviral activity
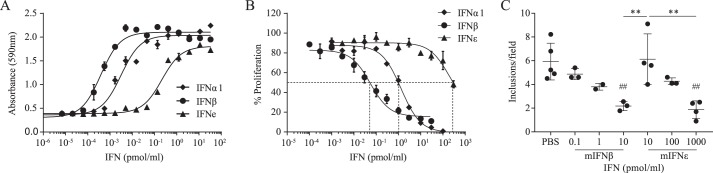
Because rmIFNϵ demonstrated characteristic type I IFN signaling, we sought to characterize whether it had the prototypic antiviral activity of the other type I IFNs using a cytopathic effect (CPE) inhibition assay (17). rmIFNϵ demonstrated robust antiviral activity with an EC50 of 214 × 10−3 pmol/ml (Table 1), albeit at ∼100- and 1000-fold less potency than rmIFNα1 (EC50 = 3.17 × 10−3 pmol/ml) and rmIFNβ (EC50 = 0.39 × 10−3 pmol/ml), respectively (Table 1 and Fig. 3A). The specific biological activity of type I IFNs is reported as international units (IU)/mg of protein. We determined the specific antiviral activity of rmIFNϵ to be 2.1 ± 0.3 × 105 IU/mg (Table 1). Again, this represents ∼100- and 1000-fold less potency than rmIFNα1 (2.4 × 107 IU/ml) and rmIFNβ (2.2 × 108 IU/ml), respectively (Table 1) (18).
Figure 3.
rmIFNϵ demonstrates antiviral, antiproliferative, and antibacterial activities in vitro. A, dose-response curves of the antiviral protection elicited by rmIFNϵ compared with mIFNα1 and mIFNβ on L929 cells following Semliki Forest virus infection. Data points represent means, with error bars representing S.D., from three independent experiments performed in technical duplicates. B, dose-response curves of the antiproliferative effect elicited by rmIFNϵ, mIFNα1, and mIFNβ on RAW264.7 cells. The dashed lines indicate the IC50, and data points represent means, with error bars representing S.D., from at least three independent experiments performed in technical duplicates. C, number of chlamydial inclusions observed in LA4 cells per field. Significance of responses was calculated using one-way ANOVA with Tukey's multiple comparison testing (**, p < 0.01 denotes significance from compared data sets; ##, p < 0.001 denotes significance from the PBS-treated control sample).
rmIFNϵ demonstrates antiproliferative effects on cells
We next determined the antiproliferative activity of rmIFNϵ on the mouse macrophage cell line RAW264.7. rmIFNϵ exhibited a dose-dependent antiproliferative effect with an IC50 of 191.9 pmol/ml (Table 1 and Fig. 3B). By contrast, rmIFNβ was about 200-fold (IC50 of 1.33 pmol/ml) and rmIFNα1 500-fold (IC50 of 0.055 pmol/ml) more potent at inhibiting cellular proliferation than rmIFNϵ.
rmIFNϵ demonstrates antibacterial activity against Chlamydia
Mice lacking IFNϵ have increased susceptibility to Chlamydia infection in the reproductive tract, and treatment with rmIFNϵ in vivo protects against this infection (10). To ascertain the ability of rmIFNϵ to directly (i.e. not via immune cell activation) exert antibacterial effects on epithelial cells, we treated a mouse epithelial cell line (LA4 cells) with rmIFNϵ or rmIFNβ before infecting the cells with Chlamydia muridarum in vitro. The proportion of cells with chlamydial inclusions was significantly reduced in a dose-dependent manner following pretreatment with rmIFNϵ and rmIFNβ (Fig. 3C). The IC50 for rmIFNϵ was 382 pmol/ml compared with the more potent rmIFNβ (IC50 = 3.156 pmol/ml). (Fig. 3C). Thus, rmIFNϵ has direct anti-Chlamydia activity, although it is ∼100-fold less potent than rmIFNβ as for its other properties examined above.
rmIFNϵ shows immunoregulatory activity on immune cells
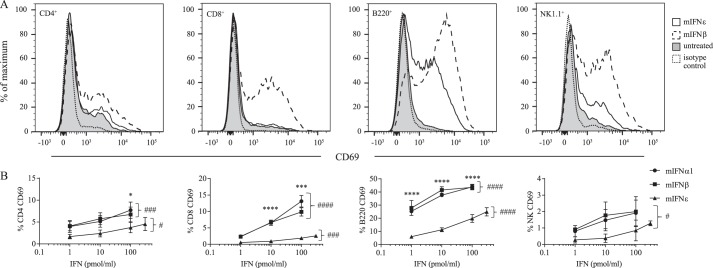
Type I IFNs have a well-documented ability to activate immune cells. They have been shown to induce the survival and proliferation of T cells, induce isotype switching of B cells, and potently activate NK cells (3–5). To investigate the immunoregulatory activities of rmIFNϵ, we stimulated spleen cells from mice ex vivo with increasing doses of rmIFNϵ, rmIFNα1, or rmIFNβ for 24 h and measured the cell-surface expression of the lymphocyte activation marker CD69 (19) on several immune cell types. All three IFNs showed a dose-dependent activation of CD4 and CD8 T cells, B cells, and NK cells (Fig. 4, A and B). The potency of rmIFNϵ to up-regulate the expression of CD69 on CD4+, CD8+, B220+, and NK1.1+ cells was 100–1000-fold less than rmIFNα1 and rmIFNβ, which were similar in potency to each other (Fig. 4B). Up-regulation of CD69 following stimulation by rmIFNϵ was greatest on B cells and NK cells (Fig. 4A).
Figure 4.
rmIFNϵ up-regulates the expression of CD69 on lymphocytes. A, histograms showing murine spleen cells either untreated or treated for 24 h with 100 pmol/ml mIFNβ or rmIFNϵ and surface-stained for CD4, CD8, B220, or NK1.1 and CD69 (or isotype control antibodies as indicated) as measured by flow cytometry. The histograms depict staining detected on untreated control cells (shaded histogram with gray outline), cells stained with an isotype control antibody (clear histogram with black dotted outline), rmIFNϵ-stimulated cells (clear histogram with solid black outline), and mIFNβ-stimulated cells (clear histogram with black dashed outline). Histograms are representative of two independent experiments performed in triplicate. B, murine spleen cells were either untreated or treated for 24 h with increasing doses (pmol/ml) of rmIFNα1, rmIFNβ, or rmIFNϵ. Live, single, CD45+ cells were further gated for CD4, CD8, B220, or NK1.1 expression and are double-positive for surface CD69 as measured by flow cytometry. Quadrant gates were set on isotype controls as indicated or using fluorescence-minus-one gating. Data are presented as means, with error bars representing S.D., from three independent experiments performed in technical triplicate. For each series of data, significance was calculated by an ordinary one-way ANOVA; for comparisons within each dose, significance was calculated using one-way ANOVA with Tukey's multiple comparison testing (*, p < 0.05; ***, p < 0.001; ****, p < 0.0001 denote significance between data points at equal concentrations of IFN. Significant differences in percentage of positive cells induced by the different doses of an IFN are indicated as follows: #, p < 0.05; ###, p < 0.001; ####, p < 0.0001.).
Murine IFNϵ demonstrates activity on human cells
One of the hallmarks of the conventional biological activities of type I IFNs is that they are highly species-specific (20) because the IFNs of one species do not bind to IFNAR receptors of other species. We therefore investigated whether rmIFNϵ was similarly restricted.
rmIFNϵ shows antiviral activity and induces an ISRE-luciferase reporter in human cells
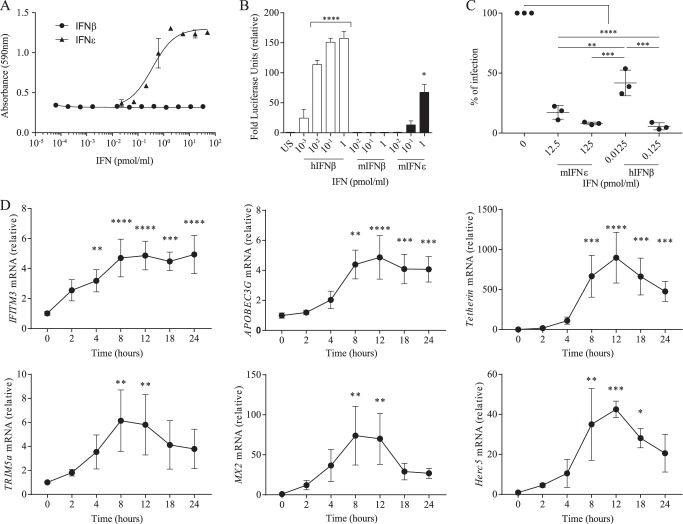
We analyzed the ability of rmIFNϵ and rmIFNβ to protect WISH (Wistar Institute, Susan Hayflick) cells from infection with encephalomyocarditis virus (EMCV) using the CPE inhibition assay. Surprisingly, rmIFNϵ demonstrated high antiviral activity in human cells, exhibiting an IC50 of 0.36 pmol/ml (Fig. 5A), equivalent to a specific antiviral activity of 2.4 ± 0.6 × 107 IU/mg on human cells and remarkably 100-fold higher than its activity on mouse cells. By contrast, rmIFNβ, as expected, had no detectable activity on human cells in the same assay (21). To further characterize the cross-species activity of rmIFNϵ, we next performed ISRE-luciferase assays in human HEK293 cells. rmIFNϵ stimulation was found to produce a strong luminescence signal that increased in a dose-dependent manner after 16 h of stimulation (Fig. 5B). As expected, rmIFNβ administration to the same cells did not result in a luminescence signal (Fig. 5B).
Figure 5.
rmIFNϵ demonstrates high biological activity on human cells. A, dose-response curves of the antiviral effect of rmIFNβ and rmIFNϵ in a WISH cell/EMCV antiviral assay. Absorbances were measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide staining of live cells as outlined under “Experimental procedures.” IC50 values were calculated to be: IFNϵ, 0.36 pmol/ml (95% confidence interval, 0.23–0.55). The IC50 of IFNβ could not be determined. Data are presented as means, with error bars representing S.D., of two independent experiments performed in duplicate. B, histograms demonstrating the ability of rmIFNϵ, hIFNβ, or rmIFNβ to induce the production of luciferase under the control of ISRE promoter elements in stably transfected HEK293T cells after 16-h stimulation. Data are presented as means, with error bars representing S.D., of three independent experiments performed in triplicate. Significance of responses was calculated using one-way ANOVA with Tukey's multiple comparison testing (****, p < 0.0001). C, rmIFNϵ anti-HIV activity in Sup-T1 cells using a luciferase reporter HIV-1. Cells were treated 24 h preinfection and at the time of infection with 12.5 or 125 nmol of rmIFNϵ or with 0.0125 or 0.125 nmol of hIFNβ. Infectivity levels were measured 48 h postinfection as luciferase readouts. Data shown are means of three independent experiments for each performed in duplicate (error bars represent S.D.). Significance of responses was calculated using one-way ANOVA with Tukey's multiple comparison testing (**, p < 0.01; ***, p < 0.001; ****, p < 0.0001). D, Sup-T1 cells were treated with 100 nmol of rmIFNϵ, collected, and lysed at 0, 2, 4, 8, 12, 18, and 24 h post-treatment. Quantitative real-time PCR was performed to determine gene expression of various ISGs (TRIM5α, MX2, HERC5, IFITM3, APOBEC3G, and BST2 (tetherin)) and normalized to 18S rRNA. Results were expressed as a relative change using the ΔΔCt method. The Sup-T1 cells data shown are a mean of three independent experiments for each performed in duplicate (error bars represent S.D.). No significant differences between cell lines were found at the gene expression baseline levels. Significance of responses was calculated using one-way ANOVA with Tukey's multiple comparison testing (*, p > 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). US, unstimulated.
Murine IFNϵ restricts HIV infection in human cells in vitro
Our in vivo mouse studies showed that IFNϵ protected the female reproductive tract from viral infection (10), and more recently, we have also shown that, in vitro, human IFNϵ restricts HIV infection at multiple stages of the viral cycle (22). Following from these results, in this study, we tested the ability of rmIFNϵ to protect human cells from HIV infection. First, we demonstrated the antiretroviral activity of this novel IFN using a reporter assay to demonstrate that rmIFNϵ inhibits HIV replication in a human Sup-T1 cell line (Fig. 5C) and in primary human peripheral blood lymphocytes (PBLs; Fig. S2A) in a dose-dependent manner. Next, we demonstrated the ability of rmIFNϵ to induce the expression of HIV restriction factors, namely TRIM5, IFITM3, MX2, APOBEC3G, HERC5, and BST2 (tetherin) mRNA, in a human Sup-T1 cell line (Fig. 5D) and primary human PBLs (Fig. S2B). Together, these data demonstrate that rmIFNϵ has high antiviral activity on human cells.
Discussion
The functions of the type I IFN family have been studied extensively over the last five decades to elucidate the pleiotropic biological roles of the prototypic α and β subtypes. However, the newest type I IFN to have been discovered, IFNϵ (9), is unique because it is constitutively expressed and hormonally regulated. These properties, together with our studies of IFNϵ-deficient mice, suggest a distinct function for IFNϵ. However, there were no data on the intrinsic properties of this protein. The data we present here represent critical specifications on the range and potency of the intrinsic properties of a novel cytokine, murine IFNϵ. These data will complement our interpretation of the functions of IFNϵ inferred from experiments in IFNϵ−/− mice. The availability of a recombinant murine form of this novel cytokine and knowledge of the nature and potency of its intrinsic actions will help elucidate its mechanism of action and potential therapeutic applications based on preclinical murine models of disease.
As murine IFNϵ had not been purified from biological tissues, it was important to determine the precise sequence of the mature protein. The only available information on the mature amino terminus was via prediction software (9, 23), which showed conflicting results in defining the IFNϵ signal peptide. We therefore expressed IFNϵ in mammalian cells and performed amino-terminal sequencing. Identification of leucine 22 as the first residue of the mature mIFNϵ is as predicted (23) and aligns with the amino-terminal residue identified for canine IFNϵ (24) and with the first residue of other mature IFNs, including IFNβ from numerous species. It is, however, different from IFNαs that tend to have a signal peptide of 23 residues in length and begin at residue 24 of the proprotein (Fig. S3). Having identified the amino-terminal sequence of mature mIFNϵ, we constructed a baculovirus expression construct for the production of a recombinant form of this protein. Antibody affinity chromatography yielded a preparation that was highly pure, endotoxin-free, and, according to CD spectral analysis, folded into an α-helical secondary structure in line with that of other type I IFN family members (25, 26).
The type I IFN system represents an interesting paradigm among cytokines whereby a myriad of biological activities are elicited by about 20 distinct but related proteins signaling via a common cell-surface receptor complex. The subtleties that govern these responses remain unclear (27–31). Sequence alignment of mIFNϵ with mIFNβ, mIFNα1, and hIFNα2 reveals the degree of homology of the IFNs within the known IFNAR1- and IFNAR2-binding interfaces, suggesting differences in the way these IFNs might interact with their receptors (Fig. S3). Analysis of this multiple alignment reveals that mIFNϵ is most similar at the amino acid level to mIFNβ and that overall, between the IFNs, residues are more conserved within the receptor interface regions than in other portions of the IFNs (Fig. S3). Although we have reported previously that mIFNϵ signaling required both IFNAR1 and IFNAR2, we did not determine direct interactions. In the present study, we assessed direct interactions and determined the binding affinity of IFNϵ for IFNAR components using MST. This analysis unexpectedly demonstrated that mIFNϵ bound mIFNAR1-ECD with higher affinity than mIFNAR2-ECD, different from data reported for the IFNα, which has a higher affinity for IFNAR2 (29). As expected, the affinity of the mIFNβ-IFNAR1 interaction measured by MST was consistent with our previous report using surface plasmon resonance (32). Notably, the affinity of the mIFNβ-IFNAR1 interaction was higher than and the mIFNβ-IFNAR2 interaction lower than these interactions reportedly measured using human proteins (29). In line with their secondary structure conservation, the relative affinities for IFNAR1 and IFNAR2 suggest that mIFNϵ is more similar to mIFNβ than to mIFNα. Nevertheless, we have reported a non-canonical IFN signaling pathway mediated by interactions between murine IFNβ and IFNAR1 in the absence of IFNAR2 (27), providing a precedent for selective interaction of IFNs with specific receptor chains.
These distinct properties of the IFNϵ-IFNAR interaction are also evidenced by the cross-species reactivity of rmIFNϵ we have demonstrated here on human cells. This feature contrasts with conventional type I IFNs whose interactions with their cognate receptors are such that murine IFNα and -β do not bind human IFNARs. This feature of IFNϵ is consistent with studies of canine IFNϵ, which was also shown to exhibit cross-species activities (24). This cross-species activity of murine IFNϵ might also have a practical advantage. For example, it may be useful in “humanized” mouse models established to study HIV, which have not been utilized to examine IFNϵ previously (33). To this end, we demonstrated that rmIFNϵ protects human PBLs and T cell lines from HIV infection. Consistent with this finding, this new IFN induced the expression of several HIV restriction factors that are active at different stages of the HIV replication cycle. Therefore, humanized mouse models could be used in the future to dissect the role of endogenous IFNϵ in the early stages of infection by HIV.
In addition to the unique differential interactions of rmIFNϵ with IFNAR1 and IFNAR2 components, these interactions exhibited 100–1000-fold lower affinity than interactions with rmIFNβ and rmIFNα, respectively. Although it may seem unusual that a biologically important protein had such a low affinity for receptors, there are parallels in IFNα subtypes that also vary in affinity and activity to a similar extent (29). The low affinity of IFNAR binding is consistent with the low biological activities of IFNϵ relative to conventional type I IFNα and/or -β (Table 1). All signaling and bioactivity measurements for rmIFNϵ, which we showed to activate classical JAK–STAT pathways, including STAT1 phosphorylation, IRG reporter transactivation, and antiviral, antibacterial, and immunoregulatory activities, were at least 100-fold lower than conventional IFNs (Table 1). This is consistent with the activities of human (11) and canine (29) IFNϵ expressed in bacterial systems, which were characterized to have similarly low specific activity.
Although IFNϵ activities are relatively low for a type I IFN, they are obviously sufficient to have protective effects against viral and bacterial infections in vivo (10). As such, it is important to note that we have demonstrated for the first time that murine IFNϵ indeed does have significant intrinsic activities to protect cells from viral or bacterial infection and activates T, B, and NK cells. Presumably, it is the combination of these actions of IFNϵ and/or its constitutive, compartmentalized expression that provides a unique, tissue-specific type I IFN profile sufficient for critical in vivo efficacy.
Indeed, the low affinity of receptor interaction for IFNϵ may be an advantage by limiting the potential toxicity associated with conventional IFNs. Furthermore, this low affinity may enable IFNe to be constitutively expressed without causing internalization of the IFNAR receptor that would render these cells refractory to conventional type I IFNs, a critical component of host defense. Such a distinct mechanism of action may be of particular biological importance in a site, such as the female reproductive tract, that must be protected from infection yet remain tolerant to implantation of a semiautologous embryo during reproduction. Indeed, IFNϵ expression is tightly regulated during the estrous cycle and pregnancy in mice and is lowest at the time of embryo implantation (10). It is therefore likely that the unique nature of IFNϵ engagement with the type I IFN receptor that we demonstrate here results in tailored biological activities that are appropriate in nature, strength, and duration for its location and functions.
Experimental procedures
Cell lines and recombinant IFNs
L929, RAW264.7, and Sup-T1 cells were purchased from the ATCC and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Pen/Strep; Life Technologies). HEK293 stably expressing ISRE-luciferase, HEK293, HeLa (ATCC), Ifnar1−/− MEF (34), and WISH (ATCC) cells were maintained in DMEM (Life Technologies) supplemented with 10% FBS and 1% Pen/Strep. Human PBLs from healthy blood donors (Red Cross Blood Bank Service, Melbourne, Australia) were isolated, phytohemagglutinin (PHA)-activated as described previously (22), and maintained in RPMI 1640 medium supplemented with 10% FBS and 1% Pen/Strep. All mammalian cells were maintained in a humidified incubator at 37 °C, 5% CO2. Spodoptera frugiperda (Sf9) and High FiveTM (BTI-TN-5B1-4 from Trichoplusia ni) cells were purchased from Life Technologies and maintained as described previously (35). Spleen cell homogenates were prepared from C57BL/6 mice by passage through a 70-μm sieve. Red blood cells were lysed in ammonium-chloride-potassium lysis buffer (Life Technologies), and cell concentration was adjusted to 1 × 107 cells/ml in RPMI 1640 medium supplemented with 10% FBS. pNL4-3 Luc Rev(−) construct was obtained from the National Institutes of Health AIDS Reagent Program. Recombinant rmIFNα1 and rmIFNβ were produced in-house as reported previously (35, 36). Human IFNβ (Rebif) was obtained from Merck Serono. The anti-mIFNϵ monoclonal antibody used throughout is as described previously (10).
Identification of amino-terminal cleavage site
The mouse Ifne1 gene was amplified from C57Bl/6 genomic cDNA and cloned downstream of a human CMV promoter to generate pCMV-Ifne-IRES-mCitrine. 2 μg of pCMV-Ifne1-IRES-mCitrine was transfected into 1 × 106 HEK293 cells using FuGENE 6 according to the manufacturer's instructions (Promega, Sydney) and incubated for 72 h. The culture supernatant was harvested, and endogenous immunoglobulins were cleared from the medium with Protein G beads (GE Healthcare). mIFNϵ was immunoprecipitated with 60 μg of anti-IFNϵ monoclonal antibody (clone H3; generated in-house; see below) and Protein G beads. PBS-washed Protein G beads were boiled in the presence of 5× Laemmli buffer, and proteins were separated by 15% (v/v) SDS-PAGE. Proteins were transferred to PVDF membrane (Merck Millipore) and stained with Coomassie Blue R-250 (Sigma-Aldrich). The 20-kDa protein band of interest was excised, and the amino-terminal sequence was determined by amino-terminal sequencing at the Monash University Proteomics Facility.
Plasmid construction for insect cell expression
The Ifne1 nucleotide sequence was codon-optimized for expression in insect cells and corresponds to amino acid residues 22–192 of murine IFNϵ (RefSeq accession number NP_796322). The 522-bp codon-optimized Ifne1 sequence was cloned into a modified pFastBacTM vector (Life Technologies) containing the honeybee melittin signal peptide (MSP), hereafter referred to as pFB-MSP. pFB-MSP was a kind gift from Kathryn Hjerrild (Hudson Institute of Medical Research). pFB-MSP-Ifne1 was transformed into JM109 cells, and colonies were screened for inserts using M13 forward (5′-GTACAATTGGAACCAAAGCGCA) and reverse (5′-GCAAGCTTTCATGGGTCAGGGTCT) primers. Positive clones were sequence-verified using the polyhedrin sequencing primer (5′-AAATGATAACCATCTCGC).
Generation of recombinant bacmid and baculovirus and expression of recombinant IFNϵ
The generation and PCR screening of recombinant bacmid and baculovirus were carried out as described previously (35). Briefly, PCR-positive colonies were expanded, and recombinant bacmid was isolated using an EndoFree Maxi-Prep kit according to the manufacturer's instructions (Qiagen). Recombinant baculovirus was generated by transfection of the purified bacmid into Sf9 insect cells, and high-titer baculovirus was generated as described previously (35). All recombinant protein expressions were carried out as described previously (35). The construct was designed so that rmIFNϵ would be expressed as a soluble protein and secreted into the culture medium.
Preparation of antibody affinity column
Standard laboratory protocols were used to scale up the monoclonal antibody production. The hybridoma clone, designated H3, was tested for potential Mycoplasma contamination, and upon confirming it was negative, the clones were adapted to low (<2%) FBS and Gibco Hybridoma serum-free medium (Thermo, catalog number 12045076). After sufficient cell density was achieved, a 10-liter working volume Wave bioreactor (GE Healthcare) was inoculated. Culture was scaled up to 5.5 liter, and feeding with glucose, GlutaMAX-I (Thermo, catalog number 35050061), and Phytone (BD Biosciences, catalog number 292450) was initiated. Culture was harvested when cell viability dropped below 50%. Cells and cell debris were removed by centrifugation followed by a 0.2-μm filtration. Monoclonal antibodies were captured on HiTrap MabSelect Xtra columns (GE Healthcare; 4 × 5-ml columns) with the bound protein eluting at low pH (0.1 m citrate, 0.1 m NaCl, pH 3) with immediate neutralization with 3 m Tris, pH 8.1. Analytical size exclusion chromatography indicated that the antibody eluted at the expected volume with no aggregate detected. Ten milligrams of the purified monoclonal antibody was then coupled to AminoLink Plus resin according to the manufacturer's instructions (Thermo Scientific). The resin was poured into the supplied column for use as a monoclonal antibody affinity chromatography column. Each column was used five times before being discarded and replaced with freshly coupled resin.
Purification of recombinant IFNϵ
Insect cell culture supernatants were clarified of cells by centrifugation as described (35) and supplemented with phenylmethanesulfonyl fluoride (PMSF) at a final concentration of 1 mm before dialysis against Tris-buffered saline (TBS; 10 mm Tris-HCl, 150 mm NaCl, pH 8.0) overnight at 4 °C using 12.5-kDa–cutoff dialysis tubing (Sigma-Aldrich). Particulates were removed by filtration of the dialysate through a 0.8-μm syringe-driven filter (Sartorius). The filtrate was applied to the anti-IFNϵ monoclonal antibody affinity chromatography column prepared above, and the column was washed with five column volumes (CVs) of TBS to remove nonspecifically bound proteins. Bound rmIFNϵ was eluted with 0.1 m glycine, pH 3.0, in 0.5× CV fractions. Collected fractions were immediately neutralized with 0.1 CV of 1 m Tris-HCl, pH 8.0, and buffer-exchanged by addition of 10× TBS (100 mm Tris-HCl, 1.5 m NaCl, pH 8.0). Protein-containing fractions, as determined by absorbance at 280 nm, were further supplemented with 10% (v/v) glycerol. Purified fractions were filter-sterilized and stored at 4 °C or snap frozen in liquid nitrogen for long-term storage at −80 °C.
Determination of protein concentration and endotoxin levels
Protein concentrations were determined by a standard Bradford colorimetric assay. Endotoxin levels were determined as described previously (35) using the ToxinSensor Endotoxin Assay kit according to the manufacturer's instructions (Genscript). Endotoxin concentrations were calculated as endotoxin units/μg of protein.
CD spectral analysis
The secondary structure of IFNs was determined on a Jasco J815 CD spectrophotometer at room temperature. Proteins were scanned at a concentration of 130 μg/ml in TBS, and triplicate scans between 190 and 260 nm were recorded. Data were converted to mean residue ellipticity by the equation of Correa and Ramos (37).
Microscale thermophoresis
For MST, mIFNAR1-ECD and rmIFNβ were expressed and purified from mammalian cell and insect cell culture, respectively, as described previously (35), and mIFNAR2-ECDC94S and rmIFNα1 were expressed and purified from mammalian cell culture also as described previously (27). Affinity measurements using MST were carried out with a Monolith NT.115 instrument (NanoTemper Technologies) as described previously (38, 39). mIFNAR1-ECD and mIFNAR2-ECDC94S were labeled using the NHS RED NanoTemper labeling kit according to the manufacturer's instructions. For the assay, 5 μl of labeled protein was mixed with 10 μl of the unlabeled IFNs (rmIFNα1, rmIFNβ, and rmIFNϵ) at various concentrations and 5 μl of 0.05% (w/v) Tween 20. All experiments were incubated for 30 min before applying samples to Monolith NT standard treated capillaries (NanoTemper Technologies). Thermophoresis was measured at 25 °C with laser off/on/off times of 5/30/5 s. Experiments were conducted at 20% light-emitting diode power and 40% MST IR laser power. Data from three independently performed experiments were fitted to the single binding model (NT.Analysis software version 1.5.41, NanoTemper Technologies) using the signal from thermophoresis + T-jump.
Flow cytometry
Flow cytometry was used to determine the cell-surface expression of CD69 on splenocytes. 2 × 106 spleen cells were stimulated for 24 h with the indicated doses of rmIFNα1, rmIFNβ, or rmIFNϵ; washed; and resuspended in PBS. Live-dead cell exclusion was determined using fixable viability dye (efluor506, eBioscience). Nonspecific antibody interactions were blocked with anti-CD16/CD32 antibody (clone p3; eBioscience; 1 μg/106 cells) in PBS containing 2% FCS. Antibodies were purchased from BD Biosciences: CD69-PE (clone H1.2F3), NK1.1-APC (clone PK136), B220-FITC (clone RA3-6B2), CD4-FITC (clone GK1.5), CD8-APC (clone 53-6.7), and IgG2a isotype control. Cells were stained for 30 min in the dark on ice. Data were acquired on a BD FACSCanto II (BD Biosciences) and analyzed using FlowJo software (TreeStar). Data are presented as percentage of double-positive cells (CD69+ and either CD4, CD8, B220, or NK) and are reported as mean ± S.D. of at least three independent biological replicates. Significance of responses was calculated using a one-way ANOVA with Dunnett's multiple comparison testing.
Luciferase assays
Luciferase assays were performed in immortalized Ifnar1−/− MEFs or in HEK293 cells that were either stably transfected with ISRE-luciferase reporter as described previously (34) or transiently transfected with an Isg15-luciferase reporter containing the upstream 700 bp of the Isg15 transcriptional start site. The Isg15-luciferase construct was kindly provided by Prof. Paula Pitha. Briefly, 2 × 104 cells/well in a 24-well plate were transfected with 30 ng of ISRE-luciferase, 100 ng of thymidine kinase-Renilla reporter, 1 ng of mIFNAR1, 1 ng of mIFNAR2 (for HEK293 cells only), and up to 0.5 μg of pEF-BOS with FuGENE 6 according to the manufacturer's instructions (Roche Applied Science). The cells were incubated for 24 h before being stimulated with IFN for 16 (HEK293 cells) or 8 h (Ifnar1−/− MEFs). Cells were lysed in reporter lysis buffer (Promega), and luminescence was measured with luciferase assay substrate (Promega) in a FLUOstar OPTIMA plate reader (BMG Labtech).
Antiviral assays
Antiviral assays were performed using the CPE inhibition assay as described previously on mouse L929 cells challenged with Semliki Forest virus (17) or WISH cells challenged with EMCV. Activities were normalized against National Institutes of Health reference standards (mouse, GU-02-901-511; or human, GA23-901-532) where 1 international unit (IU) is the amount of IFN required to provide protection to 50% of virus-exposed cells (IC50).
Antiproliferative assays
Antiproliferative assays were performed in either RAW264.7 mouse macrophage or HeLa cell lines using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich) as described previously (35). Absorbance was measured at 590 nm using a FLUOstar OPTIMA plate reader, and the percentage of proliferation was measured using the following formula: Percent proliferation = (Stimulated cells A590 − 2 × 103 cells A590)/(Unstimulated cells A590 − 2 × 103 cells A590) × 100. IC50 was calculated using GraphPad Prism software and is reported as pmol/ml of IFN required to inhibit cellular proliferation at 50% of the maximal response.
C. muridarum infection
LA4 (mouse lung epithelial) cells were infected as described previously (40). In brief, cells were plated at 3 × 104 on 10-mm glass coverslips in a 48-well culture plate in antibiotic-free Dulbecco's modified Eagle's medium/F-12 supplemented with 10% heat-inactivated FBS, 25 mm HEPES, and l-glutamine until >80% confluent (∼48 h). Cells were incubated for 24 h in the presence of various concentrations of rmIFNϵ, rmIFNβ, or PBS (untreated control). When monolayers were >80% confluent, cells were washed and then infected with Chlamydia (multiplicity of infection, 5/20) for 3 h. Cells were washed, and one of rmIFNϵ, rmIFNβ, or PBS was added again for a further 16 h. Chlamydial inclusions were stained using a Chlamydia Cel LPS kit according to the manufacturer's instructions (CelLabs). Intracellular chlamydial inclusions were labeled with fluorescein isothiocyanate, and cell nuclei were counterstained with rhodamine. The numbers of cell-associated inclusions per field were determined from each treatment with an average of 10 fields determined per coverslip and three to six coverslips per group at 40× magnification using a fluorescence microscope (Zeiss Axio Imager M2). Each condition was run in at least triplicate. IC50 was calculated using the nonlinear regression function in GraphPad Prism software and reported as pmol/ml of IFN required to inhibit formation.
HIV luciferase reporter infection of Sup-T1 cells and PHA-activated PBL cells
HIV luciferase reporter virus was produced by cotransfection of HIV envelope construct (pNLA1) and HIV pNL4-3 Luc Rev(−) construct at a ratio of 1:4 into 293T cells using polyethylenimine (PEI; Sigma). At 48 h post-transfection, viruses were harvested, cleared, and concentrated as described previously (22). Virus concentration was estimated with the HIV p24CA antigen capture assay according to the manufacturer's instructions (Xpress Bio). 24 h before infection, Sup-T1 cells or PHA-activated PBL cells were treated with different concentrations of rmIFNϵ or hIFNβ. 500 or 48 ng of HIV p24 capsid protein–equivalent virus particles were used to infect PBL cells (100,000/well) and Sup-T1 cells (100,000/well), respectively. 72 h postinfection, luciferase activity was measured using a Fluoroskan Ascent FL luminometer (Bright-Glo Luciferase Assay System, Promega). The amount of detectable luciferase activity reflected the relative levels of viral infectivity.
HIV infection of HeLa-based TZM-bl cells
TZM-bl cells were infected with NL4-3 WT virus produced via transfection of the pNL4-3 WT plasmid into 293T cells with PEI. Virus harvest, purification, concentration, and quantification were as described above. TZM-bl (10,000/well) cells were treated with 12.5 or 125 nmol of rmIFNϵ or 0.0125 or 0.125 nmol of hIFNβ, and luciferase activity was measured in a Fluoroskan Ascent FL luminometer (Bright-Glo Luciferase Assay System) 48 h postinfection (41).
Extraction of RNA and cDNA synthesis for quantitative real-time PCR
To evaluate gene expression by quantitative real-time PCR, following treatment with 100 nmol of rmIFNϵ, RNA was extracted using the RNeasy kit (Qiagen) from Sup-T1 or PBL cells at different time points (0, 2, 4, 8, 12, 18, and 24 h post-treatment). RNA was treated with DNase (Promega), and cDNA was synthesized using Moloney murine leukemia virus and random hexamers (Promega). Reverse transcription products of TRIM5α, MX2, HERC5, IFITM3, APOBEC3G, BST2 (tetherin), and 18S were quantified using previously published primers (22, 42). RT-quantitative PCR was performed using SYBR reagents (ABI). Results were normalized to 18S rRNA and expressed as relative change using the ΔΔCt method.
Author contributions
S. A. S. conceived the idea for the project, conducted most of the experiments, analyzed the data, and contributed to preparation of the manuscript. A. Y. M. prepared recombinant proteins and the antibody affinity columns and conducted experiments in antiviral assays. N. E. M. carried out flow cytometry experiments and contributed to preparation of the manuscript. K. Y. F. and A. D. generated and tested the pCMV-Ifne-IRES-mCitrine clone. M. D. T. carried out flow cytometry experiments and contributed to preparation of the manuscript. T. P. S. d. C. obtained and analyzed the MST experimental data and contributed to the preparation of the manuscript. D. H. and J. M. carried out and analyzed antibacterial assays. P. M. H. carried out antibacterial experiments and contributed to preparation of the manuscript. A. G. M. carried out the real-time PCR assays on human cells lines. S. G. E. carried out the anti-HIV assays on human cells lines. J. M. helped analyze the anti-HIV assays and real-time PCR assays on human cells lines. J. S. and G. L. conducted the large-scale purification of the monoclonal antibody and helped write the manuscript. N. A. d. and P. J. H. contributed to experimental planning, data analysis, and preparation of the manuscript.
Supplementary Material
Acknowledgments
We acknowledge the La Trobe University-Comprehensive Proteomics Platform for providing infrastructure. We thank Rebecca Smith for critical evaluation of the manuscript and Gideon Schreiber for helpful technical discussions.
This work was supported by National Health and Medical Research Council (NHMRC) New Investigator Grant APP1070732 (2014–2016) and NHMRC Project Grant APP1126524 (to N. D. W.), NHMRC Senior Principal Research Fellowship APP1117527 and NHMRC Project Grant APP1126524 (to P. J. H.), Australian Research Council Fellowship DP110103616 (to N. E. M.), NHMRC Fellowships 035733 and 1123319 (to M. D. T.), NHMRC Grants APP1003591 and APP1059242 (to J. C. H. and P. M. H.), a Deakin University scholarship (to A. G. M.), funding from the National Collaborative Research Infrastructure Strategy (to G. L.), Australian Centre for HIV and Hepatitis Virology Research Grants 2013-34 and 2015-12 (to P. J. H., N. E. M., and J. M.), and NHMRC Fellowship APP1091976 (to T. P. S. d. C.). Research at the Hudson Institute was supported by the Victorian Government's Operational Infrastructure Support Program. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- NK
- natural killer
- mIFN
- murine IFN
- rmIFN
- recombinant mIFN
- hIFN
- human IFN
- IFNAR
- IFNα/β receptor
- ECD
- extracellular domain
- ISRE
- interferon-stimulated response element
- IRG
- interferon-regulated gene
- CPE
- cytopathic effect
- EMCV
- encephalomyocarditis virus
- MST
- microscale thermophoresis
- PBL
- peripheral blood lymphocyte
- PHA
- phytohemagglutinin
- MSP
- honeybee melittin signal peptide
- TBS
- Tris-buffered saline
- CV
- column volume
- ANOVA
- analysis of variance
- MEF
- mouse embryonic fibroblast
- Pen/Strep
- penicillin/streptomycin
- APC
- allophycocyanin.
References
- 1. Pestka S., Krause C. D., and Walter M. R. (2004) Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32 10.1111/j.0105-2896.2004.00204.x [DOI] [PubMed] [Google Scholar]
- 2. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 3. Tough D. F. (2012) Modulation of T-cell function by type I interferon. Immunol. Cell Biol. 90, 492–497 10.1038/icb.2012.7 [DOI] [PubMed] [Google Scholar]
- 4. Le Bon A., Schiavoni G., D'Agostino G., Gresser I., Belardelli F., and Tough D. F. (2001) Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 14, 461–470 10.1016/S1074-7613(01)00126-1 [DOI] [PubMed] [Google Scholar]
- 5. Biron C. A., Nguyen K. B., Pien G. C., Cousens L. P., and Salazar-Mather T. P. (1999) Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17, 189–220 10.1146/annurev.immunol.17.1.189 [DOI] [PubMed] [Google Scholar]
- 6. Halloran P. F., Urmson J., Van der Meide P. H., and Autenried P. (1989) Regulation of MHC expression in vivo. II. IFN-α/βa inducers and recombinant IFN-α modulate MHC antigen expression in mouse tissues. J. Immunol. 142, 4241–4247 [PubMed] [Google Scholar]
- 7. Honda K., Sakaguchi S., Nakajima C., Watanabe A., Yanai H., Matsumoto M., Ohteki T., Kaisho T., Takaoka A., Akira S., Seya T., and Taniguchi T. (2003) Selective contribution of IFN-α/β signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. U.S.A. 100, 10872–10877 10.1073/pnas.1934678100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nguyen-Pham T. N., Lim M. S., Nguyen T. A., Lee Y. K., Jin C. J., Lee H. J., Hong C. Y., Ahn J. S., Yang D. H., Kim Y. K., Chung I. J., Park B. C., Kim H. J., and Lee J. J. (2011) Type I and II interferons enhance dendritic cell maturation and migration capacity by regulating CD38 and CD74 that have synergistic effects with TLR agonists. Cell. Mol. Immunol. 8, 341–347 10.1038/cmi.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardy M. P., Owczarek C. M., Jermiin L. S., Ejdebäck M., and Hertzog P. J. (2004) Characterization of the type I interferon locus and identification of novel genes. Genomics 84, 331–345 10.1016/j.ygeno.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 10. Fung K. Y., Mangan N. E., Cumming H., Horvat J. C., Mayall J. R., Stifter S. A., De Weerd N., Roisman L. C., Rossjohn J., Robertson S. A., Schjenken J. E., Parker B., Gargett C. E., Nguyen H. P., Carr D. J., et al. (2013) Interferon-ϵ protects the female reproductive tract from viral and bacterial infection. Science 339, 1088–1092 10.1126/science.1233321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tasker C., Subbian S., Gao P., Couret J., Levine C., Ghanny S., Soteropoulos P., Zhao X., Landau N., Lu W., and Chang T. L. (2016) IFN-ϵ protects primary macrophages against HIV infection. JCI Insight 1, e88255 10.1172/jci.insight.88255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng F. W., Duan Z. J., Zheng L. S., Xie Z. P., Gao H. C., Zhang H., Li W. P., and Hou Y. D. (2007) Purification of recombinant human interferon-ϵ and oligonucleotide microarray analysis of interferon-ϵ-regulated genes. Protein Expr. Purif. 53, 356–362 10.1016/j.pep.2006.12.013 [DOI] [PubMed] [Google Scholar]
- 13. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A (2005) Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press, New York [Google Scholar]
- 14. Jaitin D. A., Roisman L. C., Jaks E., Gavutis M., Piehler J., Van der Heyden J., Uze G., and Schreiber G. (2006) Inquiring into the differential action of interferons (IFNs): an IFN-α2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 26, 1888–1897 10.1128/MCB.26.5.1888-1897.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darnell J. E., Jr. (1997) STATs and gene regulation. Science 277, 1630–1635 10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- 16. Pestka S. (2007) The interferons: 50 years after their discovery, there is much more to learn. J. Biol. Chem. 282, 20047–20051 10.1074/jbc.R700004200 [DOI] [PubMed] [Google Scholar]
- 17. Hwang S. Y., Hertzog P. J., Holland K. A., Sumarsono S. H., Tymms M. J., Hamilton J. A., Whitty G., Bertoncello I., and Kola I. (1995) A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons α and β and alters macrophage responses. Proc. Natl. Acad. Sci. U.S.A. 92, 11284–11288 10.1073/pnas.92.24.11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knight E., Jr. (1975) Heterogeneity of purified mouse interferons. J. Biol. Chem. 250, 4139–4144 [PubMed] [Google Scholar]
- 19. Atzeni F., Schena M., Ongari A. M., Carrabba M., Bonara P., Minonzio F., and Capsoni F. (2002) Induction of CD69 activation molecule on human neutrophils by GM-CSF, IFN-γ, and IFN-α. Cell. Immunol. 220, 20–29 10.1016/S0008-8749(03)00002-9 [DOI] [PubMed] [Google Scholar]
- 20. Merigan T. C. (1964) Purified interferons: physical properties and species specificity. Science 145, 811–813 [DOI] [PubMed] [Google Scholar]
- 21. Samuel C. E., and Farris D. A. (1977) Mechanism of interferon action. Species specificity of interferon and of the interferon-mediated inhibitor of translation from mouse, monkey, and human cells. Virology 77, 556–565 10.1016/0042-6822(77)90481-0 [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Minambres A., Eid S. G., Mangan N. E., Pade C., Lim S. S., Matthews A. Y., de Weerd N. A., Hertzog P. J., and Mak J. (2017) Interferon ϵ promotes HIV restriction at multiple steps of viral replication. Immunol. Cell Biol. 95, 478–483 10.1038/icb.2016.123 [DOI] [PubMed] [Google Scholar]
- 23. Hermant P., Francius C., Clotman F., and Michiels T. (2013) IFN-ϵ is constitutively expressed by cells of the reproductive tract and is inefficiently secreted by fibroblasts and cell lines. PLoS One 8, e71320 10.1371/journal.pone.0071320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L., Xu L., Li Y., Li J., Bi Y., and Liu W. (2013) Molecular and functional characterization of canine interferon-ϵ. J. Interferon Cytokine Res. 33, 760–768 10.1089/jir.2013.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karpusas M., Nolte M., Benton C. B., Meier W., Lipscomb W. N., and Goelz S. (1997) The crystal structure of human interferon β at 2.2-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 94, 11813–11818 10.1073/pnas.94.22.11813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Senda T., Saitoh S., and Mitsui Y. (1995) Refined crystal structure of recombinant murine interferon-β at 2.15 Å resolution. J. Mol. Biol. 253, 187–207 10.1006/jmbi.1995.0544 [DOI] [PubMed] [Google Scholar]
- 27. de Weerd N. A., Vivian J. P., Nguyen T. K., Mangan N. E., Gould J. A., Braniff S. J., Zaker-Tabrizi L., Fung K. Y., Forster S. C., Beddoe T., Reid H. H., Rossjohn J., and Hertzog P. J. (2013) Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat. Immunol. 14, 901–907 10.1038/ni.2667 [DOI] [PubMed] [Google Scholar]
- 28. Lavoie T. B., Kalie E., Crisafulli-Cabatu S., Abramovich R., DiGioia G., Moolchan K., Pestka S., and Schreiber G. (2011) Binding and activity of all human α interferon subtypes. Cytokine 56, 282–289 10.1016/j.cyto.2011.07.019 [DOI] [PubMed] [Google Scholar]
- 29. Jaks E., Gavutis M., Uzé G., Martal J., and Piehler J. (2007) Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 366, 525–539 10.1016/j.jmb.2006.11.053 [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto K., Taniai M., Torigoe K., Yamamoto S., Arai N., Suemoto Y., Yoshida K., Okura T., Mori T., Fujioka N., Tanimoto T., Miyata M., Ariyasu H., Ushio C., Fujii M., et al. (2009) Creation of interferon-α8 (IFN-α8) mutants with amino acid substitutions against IFN-α receptor-2 (IFNAR-2)-binding sites using phage display system and evaluation of their biologic properties. J. Interferon Cytokine Res. 29, 161–170 10.1089/jir.2008.0038 [DOI] [PubMed] [Google Scholar]
- 31. Thomas C., Moraga I., Levin D., Krutzik P. O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S. E., Glenn J. S., Nolan G. P., Piehler J., Schreiber G., and Garcia K. C. (2011) Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632 10.1016/j.cell.2011.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Weerd N. A., Matthews A. Y., Pattie P. R., Bourke N. M., Lim S. S., Vivian J. P., Rossjohn J., and Hertzog P. J. (2017) A hot spot on interferon α/β receptor subunit 1 (IFNAR1) underpins its interaction with interferon-β and dictates signaling. J. Biol. Chem. 292, 7554–7565 10.1074/jbc.M116.773788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denton P. W., and García J. V. (2011) Humanized mouse models of HIV infection. AIDS Rev. 13, 135–148 [PMC free article] [PubMed] [Google Scholar]
- 34. Piganis R. A., De Weerd N. A., Gould J. A., Schindler C. W., Mansell A., Nicholson S. E., and Hertzog P. J. (2011) Suppressor of cytokine signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon α receptor (IFNAR1)-associated tyrosine kinase Tyk2. J. Biol. Chem. 286, 33811–33818 10.1074/jbc.M111.270207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stifter S. A., Gould J. A., Mangan N. E., Reid H. H., Rossjohn J., Hertzog P. J., and de Weerd N. A. (2014) Purification and biological characterization of soluble, recombinant mouse IFNβ expressed in insect cells. Protein Expr. Purif. 94, 7–14 10.1016/j.pep.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 36. Trajanovska S., Owczarek C. M., Stanton P. G., and Hertzog P. J. (2003) Generation and characterization of recombinant unmodified and phosphorylatable murine IFN-α1 in the methylotropic yeast Pichia pastoris. J. Interferon Cytokine Res. 23, 351–358 10.1089/107999003322226005 [DOI] [PubMed] [Google Scholar]
- 37. Correa D. H. A., and Ramos C. H. I. (2009) The use of circular dichroism spectroscopy to study protein folding, form and function. African J. Biochem. Res. 3, 164–173 [Google Scholar]
- 38. Soares da Costa T. P., Desbois S., Dogovski C., Gorman M. A., Ketaren N. E., Paxman J. J., Siddiqui T., Zammit L. M., Abbott B. M., Robins-Browne R. M., Parker M. W., Jameson G. B., Hall N. E., Panjikar S., and Perugini M. A. (2016) Structural determinants defining the allosteric inhibition of an essential antibiotic target. Structure 24, 1282–1291 10.1016/j.str.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 39. Christensen J. B., Soares da Costa T. P., Faou P., Pearce F. G., Panjikar S., and Perugini M. A. (2016) Structure and function of cyanobacterial DHDPS and DHDPR. Sci. Rep. 6, 37111 10.1038/srep37111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Asquith K. L., Horvat J. C., Kaiko G. E., Carey A. J., Beagley K. W., Hansbro P. M., and Foster P. S. (2011) Interleukin-13 promotes susceptibility to chlamydial infection of the respiratory and genital tracts. PLoS Pathog. 7, e1001339 10.1371/journal.ppat.1001339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montefiori D. C. (2005) Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12, Unit 12.11 10.1002/0471142735.im1211s64 [DOI] [PubMed] [Google Scholar]
- 42. Mous K., Jennes W., Camara M., Seydi M., Daneau G., Mboup S., Kestens L., and Van Ostade X. (2012) Expression analysis of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin in a Senegalese cohort of HIV-1-exposed seronegative individuals. PLoS One 7, e33934 10.1371/journal.pone.0033934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.