Abstract
The HLA-DRB1 locus is strongly associated with rheumatoid arthritis (RA) susceptibility, whereupon citrullinated self-peptides bind to HLA-DR molecules bearing the shared epitope (SE) amino acid motif. However, the differing propensity for citrullinated/non-citrullinated self-peptides to bind given HLA-DR allomorphs remains unclear. Here, we used a fluorescence polarization assay to determine a hierarchy of binding affinities of 34 self-peptides implicated in RA against three HLA-DRB1 allomorphs (HLA-DRB1*04:01/*04:04/*04:05) each possessing the SE motif. For all three HLA-DRB1 allomorphs, we observed a strong correlation between binding affinity and citrullination at P4 of the bound peptide ligand. A differing hierarchy of peptide-binding affinities across the three HLA-DRB1 allomorphs was attributable to the β-chain polymorphisms that resided outside the SE motif and were consistent with sequences of naturally presented peptide ligands. Structural determination of eight HLA–DR4–self-epitope complexes revealed strict conformational convergence of the P4-Cit and surrounding HLA β-chain residues. Polymorphic residues that form part of the P1 and P9 pockets of the HLA-DR molecules provided a structural basis for the preferential binding of the citrullinated self-peptides to the HLA-DR4 allomorphs. Collectively, we provide a molecular basis for the interplay between citrullination of self-antigens and HLA polymorphisms that shape peptide–HLA-DR4 binding affinities in RA.
Keywords: major histocompatibility complex (MHC), structural biology, arthritis, mass spectrometry (MS), X-ray crystallography
Introduction
Rheumatoid arthritis (RA)5 is an autoimmune disease of the synovial joints. A characteristic of RA is the presence of anti-citrullinated protein antibodies (ACPA) in sera, for which ∼70% of all patients are seropositive (1). ACPA target proteins that have undergone citrullination, a post-translational modification (PTM) process driven by a family of enzymes known as peptidyl-arginine deiminases (PAD), convert arginine to citrulline (2–4). PAD type 2 (PAD-2) and PAD-4 expression is closely associated with synovial joint inflammation in RA patients (5). Citrullination creates neo-self-antigens, and given the high rate of ACPA in RA patients, these antigens are considered the prime generators of the autoimmune CD4+ T cell response in ACPA+ RA. Citrulline-specific, antigen-experienced CD4+ T cells are found in HLA-DR4 (DRA1*01:01/HLA-DRB1*04:01) RA patients as well as in HLA-DR4 (DRA1*01:01/HLA-DRB1*04:01) transgenic mice primed with citrullinated self-antigens (6–11). Furthermore, citrulline-specific Th1 and Th17 cells are increased in the HLA-DRB1*04:01+ RA patients, and pro-inflammatory cytokines are produced by CD4+ T cells in response to citrullinated self-antigens (8–11). Sources for the primary RA-associated autoantigens may be from the site of disease, including articular cartilage and synovial fluids (12–14), but others may be derived from blood plasma or surrounding mucosal tissues that are susceptible to inflammation (2). These proteins could undergo PTMs during numerous physiologic processes, including infection, apoptosis, and cellular stress. Some of the best-characterized autoantigens that bind ACPAs are citrullinated vimentin, fibrinogen, α-enolase, and type II collagen, which are present at high levels in the joint synovium (3, 15).
One of the key inherited risk factors that contribute to ACPA-positive RA is the human leukocyte antigen (HLA) class II loci, namely HLA-DRB1, which encodes the HLA class II antigen-presenting molecules (16–21). The antigen-binding groove of the HLA class II molecule can accommodate peptide ligands that vary in length, but the main pockets that interact most strongly with the bound peptide are P1, P4, P6, P7, and P9, which can accommodate the side chains of the peptide residues 1, 4, 6, 7, and 9 (22, 23). A conserved amino acid sequence QKRAA, QRRAA, or RRRAA in position 70–74 of the HLA-DRB1 chain, known as the shared epitope (SE) motif, is highly prevalent (∼90%) among ACPA seropositive patients (11, 16). This SE motif defines the P4 pocket of the high-risk HLA-DRB1 RA-associated allomorphs. Subsequent genome-wide association studies have shown that two polymorphisms encoding β-chain residues at positions 11 and 13 at the base of the P4 pocket are also strongly associated with RA susceptibility (16). We have previously reported the structural basis for the association of the SE, citrullination, and RA by showing the size and charge of the P4 pocket accommodate antigens with a citrulline residue in the P4 position but prevents the binding of peptide ligands with the natively-encoded positively-charged P4-Arg (24).
Although citrullinated peptides have generally shown enhanced affinity to several SE HLA-DRB1 allomorphs relative to native peptides (25), the affinity of individual peptides has not been compared, with respect to polymorphisms outside the SE motif. Here, we show the affinity of citrullinated peptides across three different HLA-DRB1 allomorphs and reveal the key interactions between RA-associated antigens with RA-susceptible HLA-SE allomorphs. The crystals structures of eight citrullinated peptides bound to HLA-DRB1*04:01/*04:04/*04:05 demonstrate the close convergence of the binding modes of the P4-Cit, with polymorphisms outside this pocket accounting for the hierarchy of binding of citrullinated self-peptides to the three HLA-DR4 allomorphs. These specificities were also revealed in the natural repertoire of bound peptides, suggesting similar selective pressure operates on the self-peptide repertoire (immunopeptidome). Accordingly, we provide a molecular understanding of the interplay between the citrullination of self-antigens and HLA polymorphism that shape peptide-HLA binding affinity in RA.
Results
P1 and P9 specificities distinguish the shared epitope-positive HLA-DR4 peptidomes
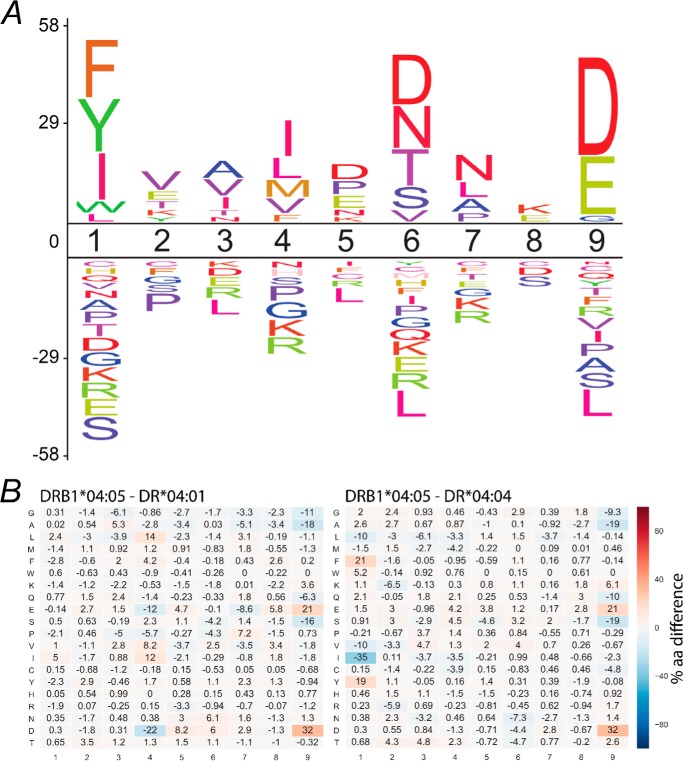
The extent to which polymorphisms outside the P4 pocket contribute to the natural selection of peptides was established using monoallelic HLA-DR4+ antigen-presenting cells. For this purpose, we generated transfectants of the T2 cell line (class II-deficient) that expressed HLA-DM and either HLA DRB1*04:01, 04:04, or 04:05. We have previously reported the immunopeptidomes of the HLA-DRB1*04:01+ and 04:04+ cells (24). Each of these datasets contained over 1000–3000 unique and high confidence peptides, and although earlier reports have identified some HLA-DRB1*04:05 peptide ligands (26), here we report a large dataset of peptide ligands (n = 2935) from the same parental T2 cell line (Table S1). These endogenous peptide sequences determined from multiple peptide elution experiments were identified with high confidence using strict bioinformatic criteria that includes the removal of common contaminants (27). The motif for HLA-DRB1*04:05 generated using this approach (Fig. 1A) was in general agreement with previously determined motifs (28, 29) and, like the other SE+ DR4 allomorphs, disfavored Arg at P4 and showed strong preferences at P1 (Phe, Tyr, and Ile), P6 (Asp, Asn, and Thr), and P9 (Asp and Glu). To compare these preferences to the other SE+ HLA-DR4 allomorphs examined in this study, we generated a heat map of difference matrices to highlight differences in their naturally selected ligands (Fig. 1B). These heat maps show the difference in amino acid usage at each position of the core 9-mer of peptides bound to HLA-DRB1*04:05 compared with HLA-DRB1*04:01 or HLA-DRB1*04:04. Of note, HLA-DRB1*04:05 selected peptides with similar P1 usage to HLA-DR*04:01 but a quite different selection of peptides with acidic P9 residues. In contrast, P1 and P9 differences were observed when HLA-DR*04:05-bound peptides were compared with HLA-DR*04:04-bound peptides, with an increase in aromatic amino acid selection at P1 and increased acidic residue selection at P9.
Figure 1.
Motif analysis of naturally presented peptides. DRB1*04:05 peptide repertoire analysis reveals amino acids that are more (positive values) or less (negative values) represented in the DRB1*04:05 9-mer cores (n = 659) compared with frequencies found in human proteome (A). Heat map depicting position-specific (columns 1–9) amino acid (rows) differences in 9-mer cores of the DRB1*04:05 compared with *04:01 and *04:04 on a scale of −100% (decreased in DRB1*04:05) to 100% (increased in DRB1*04:05) (B). A marked increase in Asp (21%), and Glu (32%) at P9 was observed in DRB1*04:05 (n = 659) compared with either *04:01 (n = 241) or *04:04 (n = 265).
RA autoantigen peptide affinity for HLA-DRB1 molecules
Given the peptide preferences for the given HLA-DR4 allomorphs identified by mass spectrometry analyses, we next aimed to determine the propensity of self-antigens implicated in RA to bind to HLA-DRB1 molecules possessing the SE motif. To establish this, we modified and undertook a fluorescence polarization assay across 34 peptides (Table 1 and Fig. 2) to measure the IC50 of each peptide bound to HLA-SE+ allomorphs, namely HLA-DRB1*04:01, HLA-DRB1*04:04, and HLA-DRB1*04:05. The HLA-DRB1*04:04 molecule varies from HLA-DRB1*04:01 at two polymorphic positions, K71R and G86V, whereas HLA-DRB1*04:05 also differs from HLA-DRB1*04:01 at two positions, namely K71R and D57S within the antigen-binding cleft (Fig. 3A). The predicted core (P1–P9) registers for the HLA–peptide interaction are underlined in Table 1, generally with a hydrophobic P1-anchor residue and a P4-Arg or P4-Cit. Arginine residues located outside the P4 pocket were also modified to citrulline and assayed for binding to the HLA-DRB1 allomorphs. The 34 peptides are predicted to place the citrulline in the P4 pocket, with the exception of the following: aggrecan-93Cit(89–103) and collagen type II-1240Cit(1237–1249) with a P2-Cit; histone-2B-73,80Cit(68–82) with a P3-Cit; and CILP-305Cit(297–309) with a P7-Cit. Four peptides possessed more than one citrullinated residue, namely aggrecan-93,95Cit(89–103), CILP-988,991,994Cit(983–995), vimentin-64,69,71Cit(59–71), and fibrinogen β-72,74Cit(69–81).
Table 1.
IC50 values in (μm) candidate RA associated autoantigens bound to HLA-SE alleles: DRB1*04:01, DRB1*04:04 and DRB1*04:05
Underlined are the predicted core binding region from P1 to P9 pocket. Citrulline is labelled as X. Mean values represent IC50 obtained from three independent experiments. Binding affinity of >250 μm is considered no binding (NB).
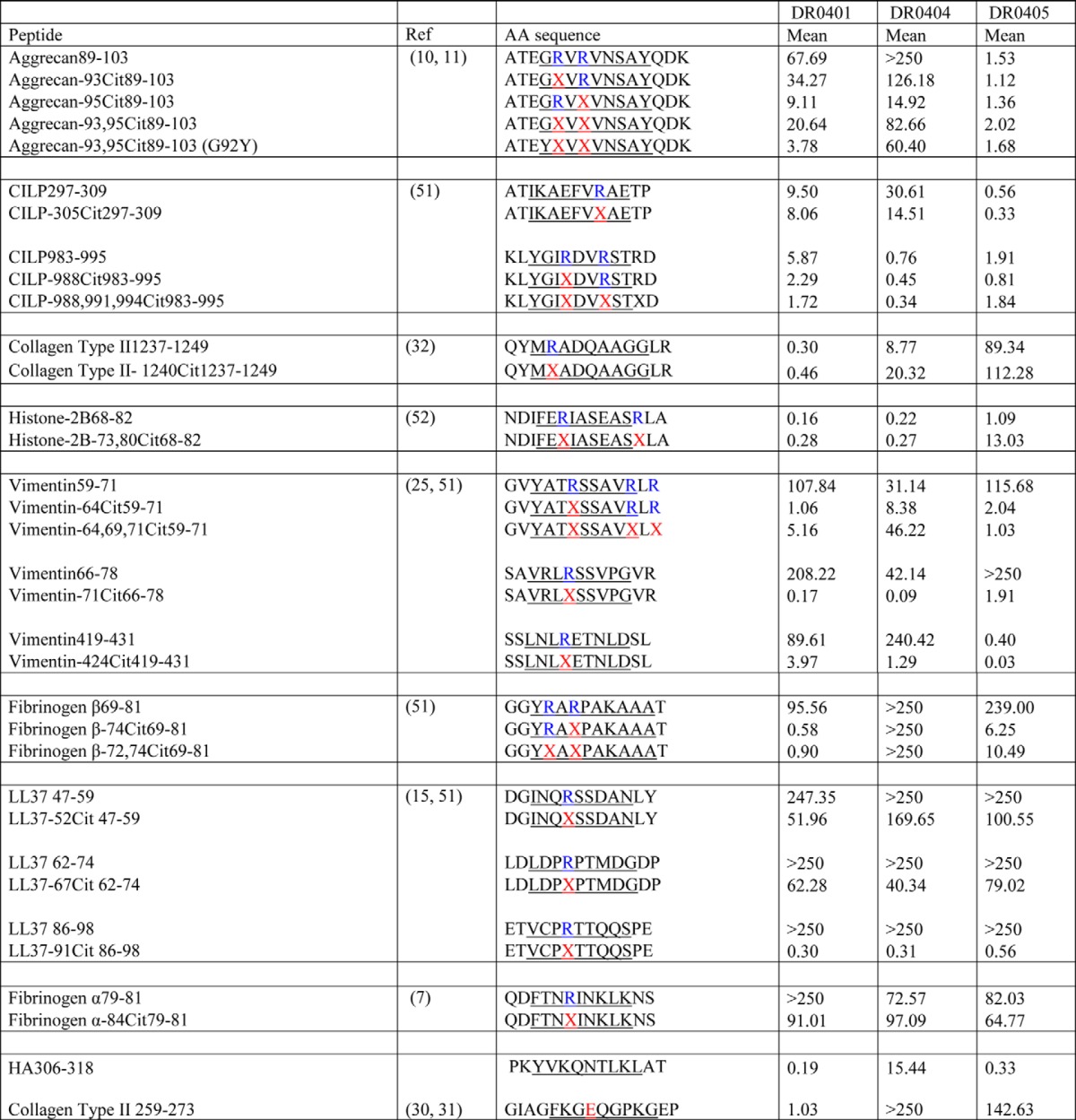
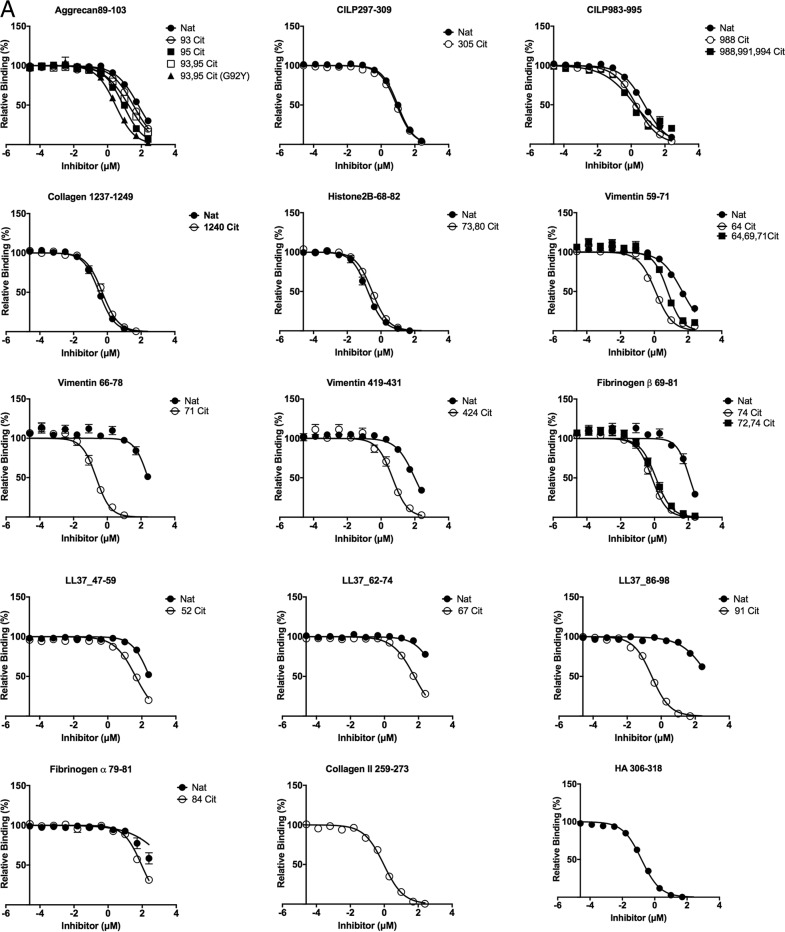
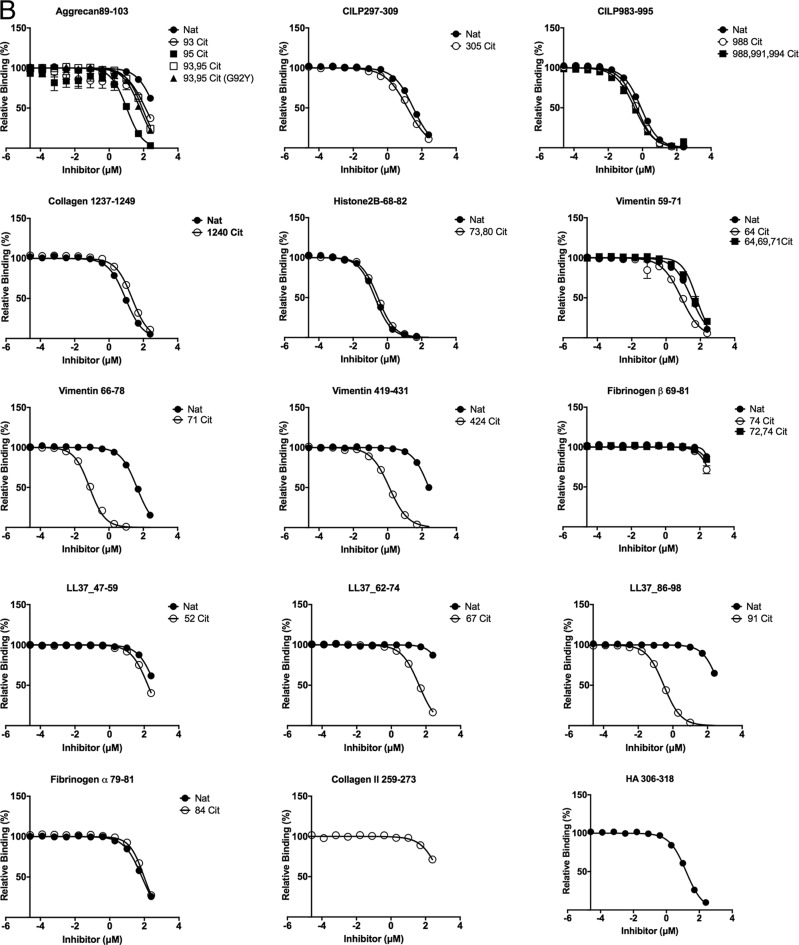
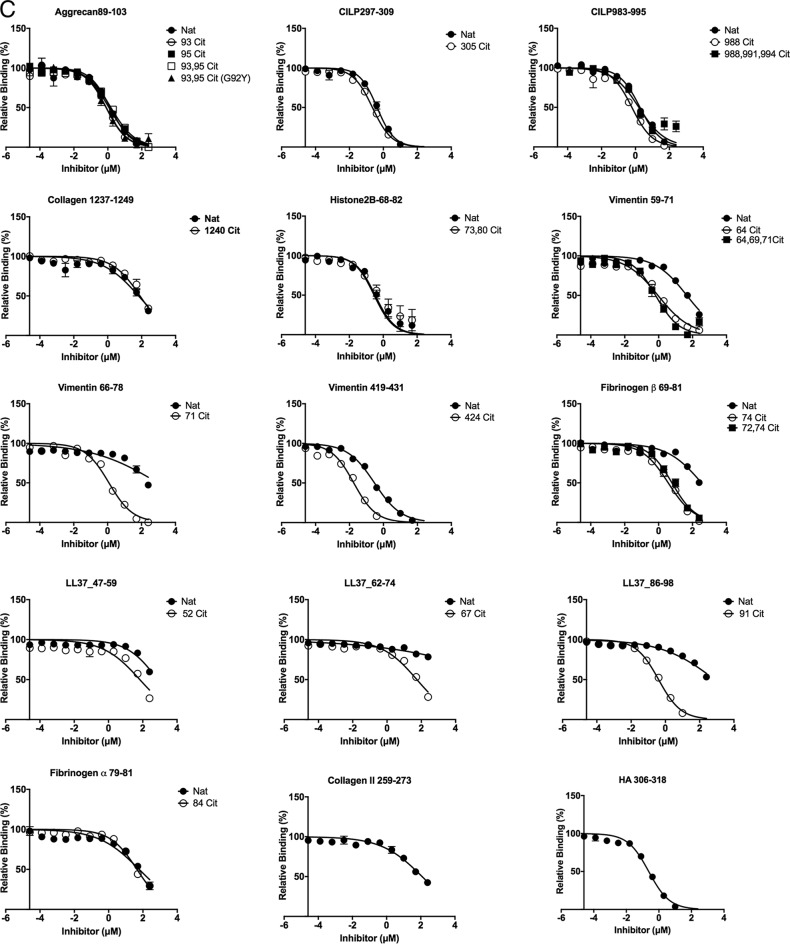
Figure 2.
Titration curve of self-peptide binding reflected by displacement of reporter fluorescent peptide in HLA-DRB1*04:01 (A), HLA-DRB1*04:04 (B), and HLA-DRB1:04:05 (C). Each data point represents normalized relative binding (in percentage) from three independent experiments. Mean values are plotted, and error bar showed SE Nat, native peptide without citrulline; Cit, citrullinated peptide. Curve fit for each set of peptides for all three allomorphs HLA-DRB1*04:01, HLA-DRB1*04:04, and HLA-DRB1:04:05 are displayed in Table S2.
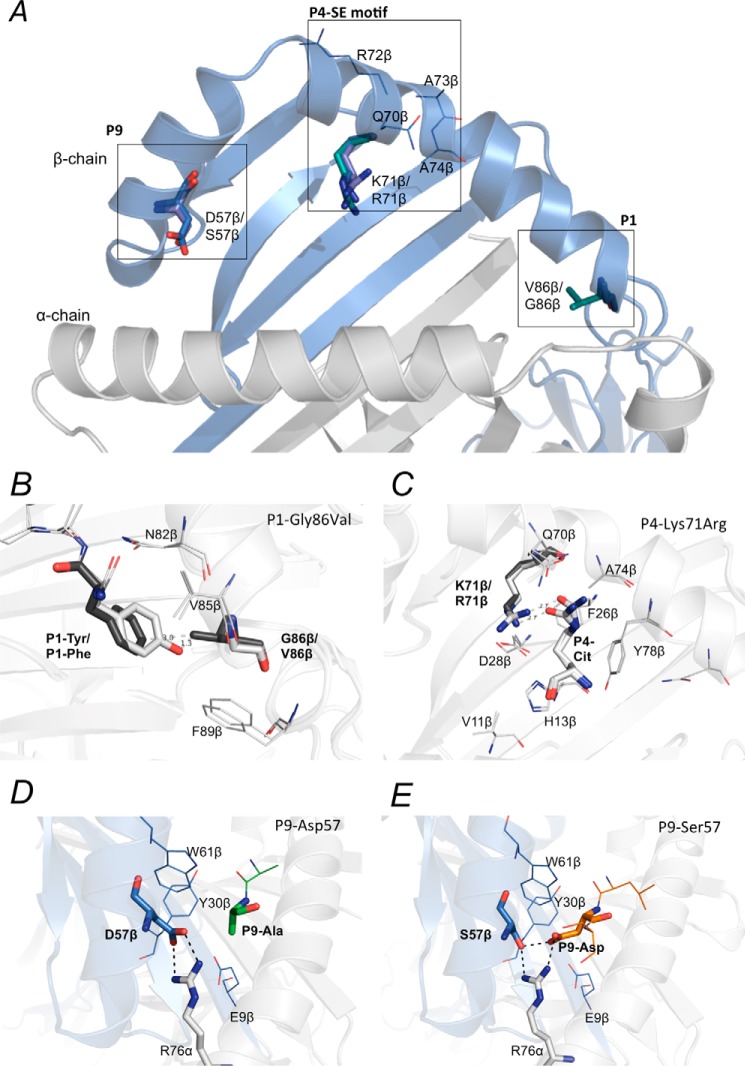
Figure 3.
Polymorphic residues in the peptide-binding cleft of HLA-SE molecules. Polymorphic residues in the P1, P4, and the P9 pocket of HLA-DRB1*04:01, DRB1*04:04, DRB1*04:05, and the shared-epitope motif are shown (A). The G86V variation on the DRβ chain of HLA-DRB1*04:04 may obstruct binding of hydrophobic residues such as Tyr in the P1 pocket (B). The P4-Cit forms a conserved hydrogen bond with the Lys-71β/Arg-71β, a polymorphic residue in the P4 pocket between HLA-DRB1*04:01 and DRB1*04:04/*04:05 (C). The P9 pocket of HLA-DRB1*04:01 and DRB1*04:04 consists of a Asp-57β that shapes the P9 pocket of these two alleles by forming a salt bridge with the conserved Arg-76α (D). The D57S variation on the DRβ chain of HLA-DRB1*04:05 forms extensive hydrogen bonds with P9-Asp in the peptide (E). The P9 pocket of HLA-DRB1*04:05 with a Ser-57β that has a shorter side chain has been shown to have a preference for negatively charged residue in the P9 position of peptides.
Peptides with relative IC50 >250 μm and <5 μm were considered non-binding and moderate binders to the HLA allomorphs, respectively. Peptides with relative IC50 <1 μm (Table 1) for the HLA allomorphs were considered to have high affinity. The fluorescence polarization values were converted to percentage of binding using the values for fully bound (∼150–250 mP) and free fluorescence (∼50 mP). Our aim was to determine the following: (a) whether the enhanced capacity of HLA-SE allomorphs to present self-antigens is restricted to P4-Cit or whether additional positions influence HLA binding; (b) whether a citrullinated epitope could bind to all HLA-SE with high affinity despite the polymorphisms in the peptide-binding cleft; and (c) whether there is a conserved structural basis for binding of citrullinated peptides to all three HLA allomorphs.
P4-citrulline-enhanced peptide affinity for HLA-DRB1*SE allomorphs
The binding affinity of aggrecan(89–103), vimentin(59–71), vimentin(66–78), vimentin(419–431), fibrinogen β(69–81), LL37(47–59), LL37(62–74), and LL37(86–98) possessing P4-Cit was increased by at least 4-fold in comparison with P4-Arg in the native sequence when bound to HLA-DRB1*04:01 (Table 1 and Fig. 2A). The P4-Cit peptide-mediated affinity enhancement for HLA-DRB1*04:01 was significant (p value < 0.05; one-way analysis of variance) in comparison with the P4-Arg counterpart or peptides with a single citrulline residue at the P2, P3, or P7 positions. The peptide affinity for native and citrullinated collagen type II(1237–1249) (Cit1240 in position P2) in HLA-DRB1*04:01 did not differ significantly, with relative IC50 values of 0.30 and 0.46 μm, respectively. The DRB1*04:01 peptide affinity for both native and citrullinated histone 2B(68–82) (Cit73 in position P3) were similar with relative IC50 values of 0.16 and 0.28 μm, and CILP(297–309) (Cit305 in position P7) with relative IC50 values of 9.50 and 8.06 μm. These data indicate that citrullination outside the P4 pocket does not impact the peptide affinity for the given HLA allomorphs.
The overall affinity of the peptide for DRB1*04:01/*04:04/*04:05 was influenced by the P1-anchor residue in the peptide. For example, peptide antigens with a P1 residue such as Gly, Ile, and Leu in aggrecan(89–103) (ATEGRVRVNSAYQDK), LL37(47–59) (DGINQRSSDANLY), and LL37(62–74) (LDLDPRPTMDGDP) showed lower affinity for HLA-DRB1*04:01 despite the presence of P4-Cit (Table 1 and Fig. 2A). In contrast, HLA-DRB1*04:01 exhibited a higher affinity for aggrecan-93,95Cit(89–103) (G92Y) that possessed a Tyr instead of Gly in the P1 position. However, a P1-Tyr is disfavored in HLA-DRB1*04:04 due to the presence of the G86V polymorphism near the P1 pocket (Fig. 3A). For example, HLA-DRB1*04:04 binds poorly to aggrecan-93,95Cit(89–103) (G92Y) (ATEYXVXVNSAYQDK), vimentin-64Cit(59–71) (GVYATXSSAVRLR), fibrinogen β-74Cit(69-81) (GGYRAXPAKAAAT), and the reference peptide HA(306–318) (PKYVKQNTLKLAT) with a relative IC50 >8 μm. This observation is consistent with the previous finding that HLA-DRB1*04:04 did not bind to the immunodominant CII(259–273) peptide, which is immunodominant in DRB1*0101 and *0401 mice with collagen-induced arthritis (30, 31). In the absence of a large P1-anchor, HLA-DRB1*04:05 binds with higher affinity (relative IC50 <1 μm) to peptides that are predicted to possess a negatively-charged Glu or Asp in the P9 pocket, including CILP-305Cit(297–309) (ATIKAEFVRAETP) and vimentin-424Cit(419–431) (SSLNLXETNLDSL). HLA-DRB1*04:05 also showed a similar affinity for native and citrullinated aggrecan(89–103), which suggests the peptide-binding register may have shifted to accommodate an aspartate residue within the P9 pocket. Like HLA-DRB1*04:04, HLA-DRB1*04:05 bound poorly to the CII(259–273) peptide. This is presumably due to the suboptimal nature of the P9-Gly residue, which is much less represented in the peptidome of HLA-DRB1*04:05 than the other two allomorphs (Fig. 1B) and is likely attributable to the D57S polymorphism (Fig. 3, D and E). Accordingly, although P4-Cit enhances binding to the HLA-DRB1 allomorphs, peptide residues outside the P4 pocket impact on the peptide-binding affinity in a manner that is shaped by HLA polymorphism.
Overlapping sequences of vimentin bind to HLA-SE allomorphs
Citrullinated vimentin peptides stimulate a dose-dependent response in PBMCs from RA patients (6) as well as CD4+ T cells isolated from immunized HLA-DR4 transgenic mice (25). Here, we tested three regions of vimentin: positions 59–71 (GVYATRSSAVRLR), 66–78 (SAVRLRSSVPGVR), and 419–431 (SSLNLXETNLDSL). HLA-DRB1*04:01 binds to vimentin-71Cit(66–78) with relatively high affinity (IC50 0.17 μm), compared with vimentin-64Cit(59–71) at 1.06 μm and vimentin-64,69,71Cit(59–71) at 5.16 μm (Table 1, Fig. 2A). The HLA-DRB1*04:04 allomorph exhibited a preference for vimentin-71Cit(66–78) (IC50 = 0.09 μm) in comparison with vimentin-64Cit(59–71) (IC50 = 8.38 μm) and vimentin-64,69,71Cit(59–71) (IC50 = 46.22 μm) (Table 1 and Fig. 2B). In contrast, HLA-DRB1*04:05 had a strong preference for vimentin-424Cit(419–431) with a relative IC50 of 0.03 μm compared with vimentin-64Cit(59–71) and vimentin-71Cit(66–78), with IC50 values of 2.04 and 1.91 μm, respectively (Table 1 and Fig. 2C). These differing binding preferences suggest that the HLA-DR β-chain polymorphism can impact antigen hierarchies in disease development.
Increased citrullination of self-antigens do not improve binding affinity for HLA-SE
Fibrinogen β-72,74Cit(69–81) showed lower binding affinity for HLA-DRB1*04:01 and HLA-DRB1*04:05 in comparison with fibrinogen β-74Cit(69–81) (Table 1, Fig. 2, A and C). Similarly, CILP-988,991,994Cit(983–995) binds with lower affinity compared with a single P4-Cit residue when bound to HLA-DRB1*04:05. Furthermore, HLA-DRB1*04:05 binds with slightly weaker affinity (IC50 of 1.03 μm) to vimentin-64,69,71Cit(66–78) when compared with a single citrullinated residue at the P4 position. Although the aggrecan(89–103) peptide was a poor binder to the HLA-SE allomorphs, two citrulline residues at positions P2 and P4 (i.e. aggrecan-93Cit(89–103) or aggrecan-95Cit(89–103) versus aggrecan-93,95Cit(89–103)) did not enhance peptide affinity. Accordingly, increased citrullination of self-antigens does not increase affinity for the HLA-DRB1 allomorphs.
Structural comparison of HLA-DR4 with RA-associated autoantigens
Next, we ascertained whether the presence of multiple citrullinated residues in a given epitope caused a register shift when bound to a given HLA-DRB1 allomorph and how the polymorphisms of the HLA-SE allomorphs impact peptide binding. To address this, we solved the crystal structures of HLA-DRB1*04:01 with a collagen type II-1240Cit(1237–1249) (P2-Cit), fibrinogen β-74Cit(69–81), and fibrinogen β-72,74Cit(69–81) (P2, P4-Cit); HLA-DRB1*04:01 and HLA-DRB1*04:04 with the same peptide LL37–91Cit(86–98); HLA-DRB1*04:04 with both native and citrullinated histone2B(68–82); and HLA-DRB1*04:05 with vimentin424Cit(419–431) (Tables 2 and 3).
Table 2.
Data collection and refinement statistics for DRB1*04:01 in complex with self-antigens
Statistics for the highest-resolution shell are shown in parentheses. r.m.s.d. means root mean square deviation.
| DR401_Fib74Cit, PDB code 6BIL | DR401_Fib72,74Cit Fib72, 74Cit, PDB code 6BIJ | DR401_CII1240Cit, PDB code 6BIN | DR0401_LL37_Cit91, PDB code 6BIV | |
|---|---|---|---|---|
| Data collection | ||||
| Resolution range | 32.32–2.40 (2.49–2.40) | 31.4–2.10 (2.18–2.10) | 39.07–2.50 (2.59–2.50) | 45.08–2.90 (3.00–2.90) |
| Space group | C 2 2 21 | C 2 2 21 | C 2 2 21 | C 2 2 21 |
| Unit cell a, b, c (Å) | 67.41, 184.12, 77.30 | 67.01, 177.71, 77.03 | 67.35, 183.71, 77.19 | 66.86, 180.30, 73.67 |
| Unit cell α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Total reflections | 38,304 (3691) | 54,582 (5354) | 33,116 (3281) | 20,482 (2016) |
| Unique reflections | 19,170 (1851) | 27,291 (2677) | 16,901 (1667) | 10,241 (1008) |
| Multiplicity | 7.1 (6.9) | 7.2 (7.3) | 5.7 (5.7) | 6.9 (7.0) |
| Completeness (%) | 99.58 (98.09) | 99.94 (100.00) | 99.88 (100.00) | 100.00 (100.00) |
| Mean I/σ(I) | 11.46 (3.28) | 13.21 (2.93) | 6.15 (2.13) | 7.87 (1.71) |
| R-merge | 0.0495 (0.2418) | 0.04824 (0.2562) | 0.1082 (0.3974) | 0.06883 (0.4452) |
| CC1/2 | 0.996 (0.808) | 0.997 (0.784) | 0.983 (0.49) | 0.992 (0.664) |
| Wilson B-factor | 25.46 | 21.84 | 27.37 | 47.37 |
| Refinement | ||||
| Reflections used in refinement | 19,169 (1851) | 27,287 (2677) | 16,975 (1667) | 10,241 (1008) |
| Reflections used for R-free | 984 (80) | 1368 (148) | 890 (82) | 494 (51) |
| R-work | 0.1890 (0.2324) | 0.1807 (0.2082) | 0.2006 (0.2476) | 0.2010 (0.2911) |
| R-free | 0.2431 (0.3120) | 0.2386 (0.2646) | 0.2380 (0.2868) | 0.2484 (0.3151) |
| r.m.s.d. (bonds) | 0.002 | 0.009 | 0.004 | 0.003 |
| r.m.s.d. (angles) | 0.66 | 0.92 | 0.78 | 0.93 |
| Ramachandran favored (%) | 97.3 | 97.55 | 97.58 | 97.3 |
| Ramachandran allowed (%) | 2.7 | 2.45 | 2.42 | 2.7 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
| Average B-factor | 26.23 | 26.75 | 26.73 | 44.01 |
| Macromolecules | 25.95 | 26.15 | 26.19 | 43.64 |
| Ligands | 38.54 | 37.28 | 54.28 | 66.62 |
| Solvent | 28.99 | 31.63 | 25.19 | 33.13 |
Table 3.
Data collection and refinement statistics for DRB1*04:04/*04:05 in complex with self-antigens
Statistics for the highest-resolution shell are shown in parentheses. r.m.s.d. means root mean square deviation.
| DR0404_LL37_Cit91, PDB code 6BIX | DR0404_His2B, PDB code 6BIY | DR0404_His2Bcit, PDB code 6BIZ | DR0405_Vim424Cit, PDB code 6BIR | |
|---|---|---|---|---|
| Data collection | ||||
| Resolution range | 39.22–2.20 (2.279–2.20) | 39.22–2.05 (2.12–2.05) | 39.09–2.10 (2.18–2.10) | 39.34–2.3 (2.38–2.30) |
| Space group | C 2 2 21 | C 2 2 21 | C 2 2 21 | C 2 2 21 |
| Unit cell a, b, c (Å) | 67.34, 183.06, 78.43 | 67.83, 182.48, 76.81 | 68.19, 182.60, 76.84 | 67.46, 183.01, 77.08 |
| Unit cell α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Total reflections | 50,100 (4930) | 59,256 (5892) | 56,781 (5590) | 42,840 (4064) |
| Unique reflections | 25,066 (2466) | 30,214 (2983) | 28,423 (2799) | 21,517 (2081) |
| Multiplicity | 5.5 (5.6) | 4.4 (4.5) | 7.2 (6.8) | 6.8 (6.3) |
| Completeness (%) | 100.00 (100.00) | 99.51 (99.93) | 100.00 (100.00) | 99.31 (97.61) |
| Mean I/σ(I) | 8.58 (4.27) | 10.20 (2.73) | 18.75 (5.06) | 9.45 (2.49) |
| R-merge | 0.03499 (0.1193) | 0.05276 (0.2877) | 0.03453 (0.191) | 0.05655 (0.2767) |
| CC1/2 | 0.997 (0.954) | 0.989 (0.739) | 0.998 (0.88) | 0.995 (0.817) |
| Wilson B-factor | 19.6 | 24.92 | 24.07 | 25.56 |
| Refinement | ||||
| Reflections used in refinement | 25,064 (2466) | 30,210 (2983) | 28,422 (2799) | 21,511 (2081) |
| Reflections used for R-free | 1275 (112) | 1536 (131) | 1416 (155) | 1102 (100) |
| R-work | 0.1692 (0.1749) | 0.1825 (0.2410) | 0.1856 (0.2404) | 0.1858 (0.2347) |
| R-free | 0.2130 (0.2634) | 0.2319 (0.2786) | 0.2284 (0.3165) | 0.2380 (0.2502) |
| r.m.s.d. (bonds) | 0.007 | 0.007 | 0.008 | 0.007 |
| r.m.s.d (angles) | 0.98 | 0.86 | 0.92 | 0.96 |
| Ramachandran favored (%) | 98 | 97.59 | 98 | 96.71 |
| Ramachandran allowed (%) | 1.9 | 2.41 | 1.9 | 3.29 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
| Average B-factor | 22.41 | 28.64 | 26.16 | 25.6 |
| Macromolecules | 21.81 | 27.95 | 25.58 | 25.3 |
| Ligands | 36.68 | 44.58 | 40.05 | 43.44 |
| Solvent | 27.41 | 33.98 | 29.92 | 27.03 |
Conserved orientation of citrulline in the P4 pocket of HLA-SE
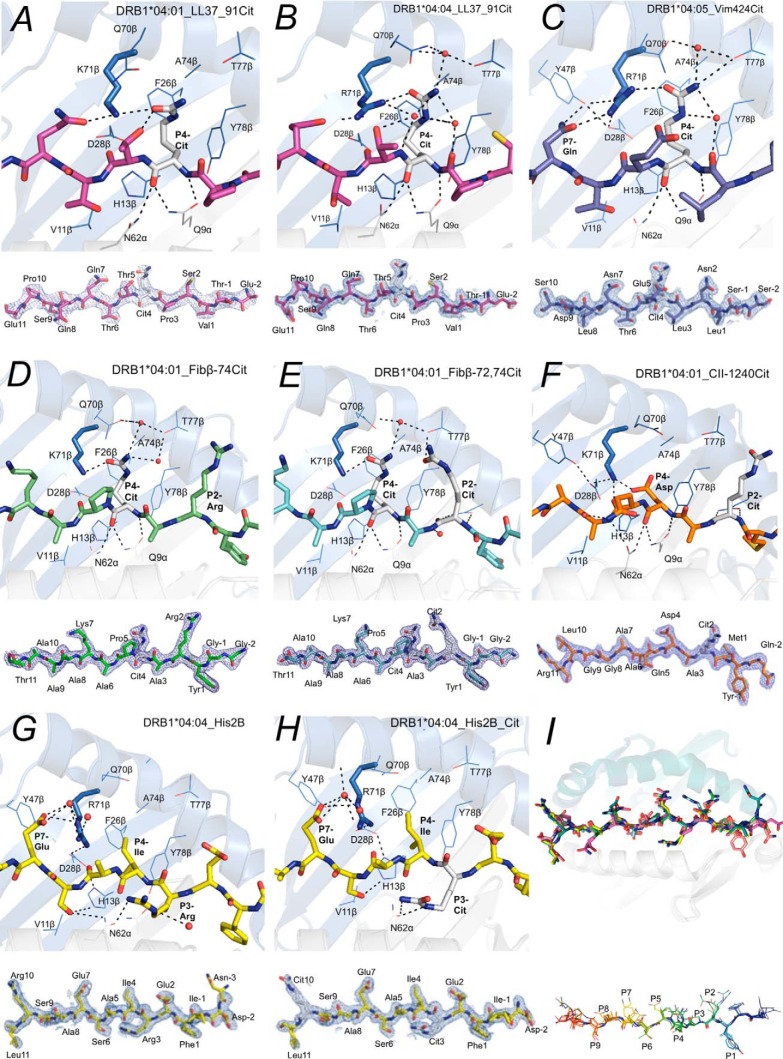
The HLA-DRB1*04:01/*04:04/*04:05 allomorphs bind partially overlapping, but not identical, sets of peptides; thus the polymorphic residues must be involved in determining the varying binding specificities (Table 1 and Fig. 2). To explore the role of the polymorphic site, Lys-71β/Arg-71β, in the SE motif, we solved crystal structures of HLA-DRB1*04:01 and HLADRB1*04:04 in complex with the same peptide, LL37–91Cit(86–98) (Fig. 4, A and B). Superposition of these two structures revealed that they bind in a similar location and adopt a closely related conformation. The P4-Cit side chain of LL37–91Cit(86–98) bent upward and leaned against Phe-26β and Tyr-78β in the P4 pocket of HLA-DRB1*04:01 and hydrogen bonded to Lys-71β and Gln-70β. The P4-Cit of LL37–91Cit(86–98) in HLA-DRB1*04:04 also packed against Phe-26β and Tyr-78β and formed a hydrogen bond with Arg-71β, the latter of which was stabilized by a hydrogen bond with Asp-28β and Tyr-47β and P7-Gln in the peptide. The crystal structure of HLA-DRB1*04:05 with vimentin-424Cit(419–431) also showed the P4-Cit adopted a conserved U-shaped upward conformation as seen in DRB1*04:01 and DRB1*0404 (Fig. 3C). Here, the P4-Cit of vimentin-424Cit(419–431) hydrogen bonds with Arg-71β, which was further stabilized by forming contacts with Asp-28β, Tyr-47β, and the P7-Gln. Overall, the P4-Cit is presented in a highly conserved manner in the three HLA-DRB1*04:01/*04:04/*04:05 allomorphs (Fig. 4I).
Figure 4.
Crystal structures of HLA-SE in complex with RA-associated antigens. The HLA-DRB1* molecule is represented as a schematic, whereas the RA-associated peptides are shown as sticks, and the electron density maps of these peptides are shown as 2Fo − Fc maps contoured at 1σ. Both HLA-DRB1*04:01 (A) and DRB1*04:04 (B) bound to the LL37–91Cit86–98 and interact with the peptide P4-Cit in a highly conserved manner despite Lys-71β/Arg-71β polymorphism in the P4 pocket. C, Arg-71β of DRB1*04:05, like the HLA-DRB1*04:01/*04:04, forms a conserved hydrogen bond with the P4-Cit in peptide vimentin-424Cit419–431. Crystal structures of HLA-DRB1*04:01 in complex with either fibrinogen-β-74Cit69–81 (D) or fibrinogen-β-72,74Cit69–81 (E), the same epitope with additional citrulline in the P2 pocket, showed no register shift upon binding to the DR molecule. The P2-Cit of both fibrinogen-β-72,74Cit69–81 and F, collagen type II-1240Cit1237–1249 bound to HLA-DRB1*04:01 are projected outward from the peptide-binding groove and are accessible by TCRs. The P4-Asp, in collagen type II-1240Cit1237–1249, like the P4-Cit, forms a hydrogen bond with the Lys-71β in HLA-DRB1*04:01. The presence of P3-Arg (G) or P3-Cit (H) in the histone2B68–82 peptide does not change the affinity of the peptide for the HLA-DRB1*04:04 and interacts with Asn-62α. The P4-Ile in the histone2B68–82 peptide liberates the Arg-71β in DRB1*04:04 to form a hydrogen bond with P7-Glu and Asp-28β. I, overlay of the eight structures showed no significant conformational changes in the HLA molecules, and the peptide alignment is highly conserved. Citrulline in the P4 pocket is presented upward in a highly conserved manner.
Next, we asked whether having multiple citrullinated residues in a single epitope could cause a register shift when bound to a given HLA-DRB1 allomorph. To address this, we determined the structure of HLA-DRB1*04:01 in complex with fibrinogen β-74Cit(69–81) (IC50 = 0.58 μm), as well as fibrinogen β-72,74Cit(69–81) (IC50 = 0.90 μm) (Fig. 4, D and E, and Tables 2 and 3). The P4-Cit of fibrinogen β-74Cit(69–81) and fibrinogen β-72,74Cit(69–81) forms a direct hydrogen bond with Lys-71β and a water-mediated hydrogen bond with Thr-77β in HLA-DRB1*04:01. The P2-Arg of fibrinogen β-74Cit(69–81) pointed away from the peptide-binding groove and was freely accessible for TCR engagement. The P2-Cit of fibrinogen β-72,74Cit(69–81) is also oriented away from the peptide-binding groove and hydrogen bonds to the carbonyl oxygen of Thr-77β. Accordingly, no register shift in either citrullinated peptide was observed upon binding to HLA-DRB1*04:01.
Citrulline in position P2 and P3 has minimal impact on peptide binding
The presence of a citrulline in the P2 position does not affect peptide affinity for the HLA-SE tested (Table 1 and Fig. 2). Indeed, the P2 pocket of the HLA-DRB1*04:01 could accommodate either arginine or citrulline as reflected in the fluorescence polarization data (Table 1 and Fig. 2). To address how an epitope with a P2-Cit could be accommodated, we determined the crystal structure of the HLA-DRB1*04:01 in complex with CII-1240Cit(1237–1249) that has a P2-Cit (Fig. 4F). The refined structure was very similar to that of HLA-DRB1*04:01 in complex with native CII(1237–1249) peptide (32). The bound CII peptide is extended across the peptide-binding site in a linear manner with Met-1239 occupying the P1 non-polar pocket, Asp-1242 in the P4 pocket, and Ala-1244 and Gly-1247 in the P6 and P9 pockets, respectively. The side chain of the P2-Cit in peptide CII-1240Cit(1237–1249), like the P2-Arg, projects away from the peptide-binding site of HLA-DRB1*04:01 and is freely accessible for TCR recognition.
Next, we determined the structures of both native and citrullinated peptide histone2B(68–82) (P3-Cit) in complex with HLA-DRB1*04:04 (Fig. 4, G and H). These histone2B peptides bound to the HLA-DRB1*04:04 with similar IC50 of 0.22 and 0.27 μm. The P3-Cit of the peptide histone2B-73,80Cit (68–82) hydrogen bonds with Asn-62α in the HLA-DRB1*04:04, which is further stabilized by P6-SerO in the peptide that forms hydrogen bonds with Asn-62α and His-13β. The side chain of the P3-Arg is also solvent-exposed in the HLA-DRB1*04:04-histone2B(68–82) complex, in which the guanidinium group hydrogen bonds to the carbonyl oxygen of the same Asn-62α.
Collectively, citrulline in position P2 or P3 in an epitope has minimal influence on peptide affinity for HLA-SE as the side chains of these residues project outward from the peptide-binding groove, and thus both arginine and citrulline can be accommodated without changing the overall peptide affinity.
Presentation of peptides with amino acid residues other than citrulline in the P4 pocket of HLA-SE
The SE motif creates an electropositive P4 pocket encompassing a positively charged residue at position 71β. Consistent with our previous findings, these SE allomorphs allowed binding of autoantigen-derived peptides with polar or acidic residues at P4 but disallowed peptides with a P4 arginine due to electrostatic repulsion (24). In the HLA-DRB1*04:01–CII-1240Cit(1237–1249) complex, the side chain of the negatively charged P4-Asp, a preferred P4-anchor residue in the natural repertoire, is oriented upward similar to P4-Cit and forms a salt bridge with Lys-71β in HLA-DRB1*04:01 (Fig. 4F). In the HLA-DRB1*04:04 crystal structures in complex with either the native or citrullinated histone2B(68–82), Ile was placed in the P4 pocket of the HLA-DRB1*04:04 molecule. Subsequently, the Arg-71β in the HLA-DRB1*04:04 becomes available to form a salt bridge with Asp-28β and water-mediated hydrogen bond with the P7-Glu of the peptide (Fig. 4, G and H). Alignment of Cα backbone of the peptides bound to HLA-SE allomorphs highlights the conserved feature of citrulline and other non-positively charged residues. Thus, there is sufficient plasticity within the P4 pocket to accommodate differing amino acids.
HLA polymorphism dictates peptide-binding affinity
The crystal structures of the HLA-DR4–peptide complexes show that the peptides bind as a straight extended chain within the binding groove with no extensive conformational changes in the HLA-SE molecule itself. Three side chains of the self-peptides are accommodated by polymorphic P1, P4, and P9 pockets in the HLA-DRB1*04:01/*04:04/*04:05-binding site. These pockets determine the peptide specificity of the HLA-DRB1*04:01/*04:04/*04:05.
The fibrinogen β-74Cit(69–81) (GGYRAXPAKAAAT) peptide provides an opportunity to examine the effect of polymorphism in the P1, P4, and P9 pockets on peptide binding affinity for each HLA-DR4 allomorph. Fibrinogen β(69–81) only binds to HLA-DRB1*04:01 and HLA-DRB1*04:05 when there is a P4-Cit (Table 1) suggesting that the Lys-71β/Arg-71β in the SE motif is the key discriminating factor preventing binding of the native peptide (Fig. 3C). However, the fibrinogen β-74Cit(69–81) peptide did not bind to HLA-DRB1*04:04. HLA-DRB1*04:04 differs from 04:01 at two amino acids on the β-chain: G86V and K71R that are located near the P1 and P4 pockets, respectively (Fig. 3A). Gly-86β as visualized in the DRB1*04:01–fibrinogen β-74Cit(69–81) complex (Figs. 3D and 4D) allows the binding of large aromatic and non-polar side chains such as Tyr. The substitution of Gly-86β for Val-86β places the Val side chain such that it would sterically hinder the binding of the P1-Tyr of this peptide to HLA-DRB1*04:04 (Fig. 3B).
In contrast, HLA-DRB1*04:05 bound to the fibrinogen β-74Cit(69–81) peptide but with lower affinity compared with HLA-DRB1*04:01. HLA-DRB1*04:05 differs from HLA-DRB1*04:01 and HLA-DRB1*04:04 at two positions: D57S and G86V on the β-chain, located near the P1 and P9 pockets, respectively. Arising from our immunopeptidome studies (Fig. 1), HLA-DRB1*04:05 has a very strong preference for peptides with Asp or Glu in the P9 position. Indeed, soluble recombinant HLA-DRB1*04:05 was only expressed with a modified CLIP (as described under “Experimental procedures”) but not with the invariant CLIP in HEK293S cells. The Ser-57β in the P9 pocket of HLA-DRB1*04:05, as observed in the structure of HLA-DRB1*04:05–vimentin-424Cit(419–431) complex, forms extensive contacts with the Asp in the P9 position of the peptide (Fig. 3, D and E). Therefore, the lower binding affinity of fibrinogen β-74Cit(69–81) to HLA-DRB1*04:05 compared with HLA-DRB1*04:01 is likely due to the unfavorable Ala at P9. This D57S variant in HLA-DRB1*04:05 may form a less stable complex with class II-associated invariant chain peptide compared with HLA-DRB1*04:01 and HLA-DRB1*04:04, which have Asp-57β that normally forms a salt bridge with Arg-86α (33). Although there is a strong correlation between the ACPA titer in RA patient sera and the SE motif, the binding of citrullinated peptides is fine-tuned by distinct P1 and P9 specificities of the different SE + HLA-DR4 allomorphs.
Discussion
Although HLA-DR4 molecules can present a range of citrullinated self-peptides that are associated with RA, the binding hierarchy of such peptides against a given HLA-SE molecule was unclear. A PTM peptide may generate a novel T cell epitope in two ways: (i) increased MHC class II binding affinity by modifying a key anchor residue, or (ii) modification of a non-anchor but solvent-exposed residue for TCR contact (34). A recent study on RA-associated HLA-DRB1*14:02 that encodes two polymorphic residues at the base of the P4 pocket (V11S and H13S) showed that, in contrast to DRB*04:01, both citrullinated and non-citrullinated peptides can be accommodated within the HLA-DRB1*14:02 molecule and stimulate a T cell response among Indigenous Native American RA patients (21). Here, we addressed the impact of HLA polymorphism and peptide citrullination on the HLA-SE binding hierarchy of self-peptides implicated in RA.
The shared epitope residues are critical in selecting specific amino acids at position P4 of peptide that will bind to DR4 as reflected in the fluorescent polarization assay carried out across 34 RA-related self-peptides. In general, P4-Cit increased peptide affinity for the majority of the citrullinated peptides for HLA-DRB1*04:01/*04:04/*04:05 and is accommodated in a highly conserved manner in the P4 pocket of the HLA-SE as shown in the series of crystal structures solved here and in earlier studies (11, 24). Our data also showed that not all HLA allomorphs bind to citrullinated antigens with the same affinity and that citrulline outside of the P4 pocket has minimal impact on the peptide-HLA affinity compared with their native arginine counterpart.
The HLA-DRB1*04:01/*04:04/*04:05 bound to an overlapping but not an identical set of citrullinated peptides. This is most likely attributable to polymorphic residues within the peptide-binding cleft of these HLA-SE allomorphs and is also reflected in the selection of naturally presented peptides by these different HLA-DR4 allomorphs. Moreover, the majority of these self-antigens were identified in studies utilizing either HLA-DRB1*04:01 RA patients PBMC or DR4/IE transgenic mice (Table 1), which may explain why certain citrullinated self-peptides bound better to HLA-DRB1*04:01 but not HLA-DRB1*04:04/*04:05 (35). Thus, there is a need for further epitope discovery in the context of these other HLA-DR4-SE allomorphs. It also appears that each HLA allomorph has its own binding hierarchy for these RA-associated T cell epitopes. Whether the hierarchy of peptides reflects the immunodominant epitopes responsible for RA disease development in patients carrying specific RA-susceptible alleles will depend on T cell studies in vivo and structural analysis with the TCR. There are some parallels with other autoimmune-like diseases, such as celiac disease, where PTM peptides could bind to HLA allomorphs with higher affinity and subsequently form more stable peptide–MHC complexes and prolong antigen presentation to the autoreactive T cells at the site of inflammation (36–39). Further studies aimed at determining whether a correlation between the rank order of RA-associated epitopes and disease pathogenesis exists will help to identify key autoantigens involved in RA development.
Experimental procedures
Peptides
Panels of 13–15-mer peptides with overlapping sequences spanning sections of the vimentin, aggrecan, fibrinogen, cartilage-intermediate proteins (CILP), collagen type II, and LL37 were synthesized by GL Biochem (Shanghai, China). Arginine in the peptides was systematically replaced with citrulline where it was appropriate (Table 1). For competition binding assay, the N-terminally acetylated HA(306–318) analog Ac-PRFVK(Tamra)QNTLRLAT was labeled with TAMRA through the primary amine Lys-310 (GL Biochem, Shanghai, China). This peptide was shown to bind to HLA-DRB1* alleles in a fluorescent polarization assay (40). Lysine to arginine modification at positions 307 and 315 was made to the peptide to limit fluorescent labeling occurring at Lys-310 only. Each peptide was dissolved in water, TBS (10 mm Tris-Cl, pH 8, 150 mm NaCl), or DMSO at 1 mm and subsequently diluted as needed. Peptide concentration was determined by 215 nm peptide carbon backbone measurements.
Fluorescence polarization assay
Various concentrations of each test peptide (range from 500 to 0 μm) were incubated in competition with 20 nm fluorescent reference peptides to bind with 100 nm recombinant HLA-DR protein in the presence of 20 nm HLA-DM in Assay Buffer (100 mm trisodium citrate, pH 5.4, 50 mm NaCl, 5 mm EDTA), as described previously (40). Fluorescent polarization was measured after 24 or 72 h of incubation at 37 °C using PHERAstar microplate reader (BMG LABTECH). Peptide-binding curves were simulated by non-linear regression with Prism software (Version 7.01, GraphPad Software Inc.) using a sigmoidal dose-response curve. IC50 binding values were calculated as the peptide concentration needed for 50% inhibition of reference peptide binding. Relative binding values (%) were calculated as the IC50 value of the substituted peptide divided by the IC50 value of the non-substituted peptide at a given concentration (×100).
Expression and purification of HLA-DR4
The cDNA encoding extracellular domains of HLA-DRB1*04:01/-HLA-DRB1*04:02/-HLA-DRB1*04:05 were separately cloned into the pHLsec expression vector with a sequence encoding an N-terminal covalently linked factor Xa-cleavable, Strep-tag MHC class II-associated invariant chain peptide (Strep-CLIP; WSHPQFEKGAPVSKMRMATPLLMQA) (24). To express HLA-DRB1*04:05 recombinant protein, a methionine to aspartic acid (underlined; WSHPQFEKGAPVSKMRMATPLLDQA) was introduced to enhance CLIP affinity for HLA-DRB1*04:05 and subsequently heterodimer stability for expression and downstream purification. Soluble recombinant HLA-DRB1* proteins were expressed in HEK293S (GnTI−/−) cells (41) and purified with a three-step strategy using immobilized metal affinity (nickel-Sepharose 6 Fast Flow; GE Healthcare), size-exclusion (Superdex 200; GE Healthcare) and anion-exchange (HitrapQ; GE Healthcare) chromatography as described previously (24).
Peptide loading of HLA-DR4
HLA-DRB1*-Strep-CLIP was incubated with factor Xa (New England Biolabs) in the presence of 2 mm CaCl2 for 6 h at room temperature to remove the covalent linked Strep-CLIP peptide. This invariant peptide was exchanged with test peptides in solution at 20-fold molar excess, catalyzed by HLA-DM at 1:5 DM/HLA-DRB1* ratio. The exchange was done between 24 and 72 h at 37 °C in 50 mm tri-sodium citrate, pH 5.4, and 5 mm EDTA. The peptide-loaded HLA-DRB1* was separated from unloaded HLA-DRB1*-CLIP and HLA-DM using StrepTactin-Sepharose (IBA, Göttingen, Germany). The unbound fraction, containing HLA-DRB1* loaded with test peptides, was concentrated for structural studies.
Protein crystallization and structural determination
For crystallization, HLA-DRB1* was buffer-exchanged to 25 mm Tris, pH 7.6, 50 mm NaCl, and 2 mm CaCl2. The Fos/Jun zippers were removed by an overnight incubation with enterokinase (New England Biolabs) at room temperature. Subsequently, the extracellular domain of the heterodimer HLA-DRB1* loaded with the peptide of interest was purified using anion-exchange chromatography (HitrapQ, GE Healthcare). HLA-DRB1* in complex with peptides was crystallized in the presence of 16–30% PEG3350, 200 mm potassium nitrate, 100 mm Bistris propane, pH 7.6. Plate-like crystals were exposed to cryoprotectant, 16% ethylene glycol for 30 s before being flash-cooled in liquid nitrogen. A Morpheus crystallization condition (42) containing 100 mm imidazole/MES, pH 6.5, 20 mm NPS mix, 12.5% MPD, 8–12% PEG3350, 12.5% PEG1000 were used for the HLA-DRB1*–peptide complex that did not crystallize in the previous condition described. Data sets were collected on the MX1 or MX2 beamline of the Australian Synchrotron.
Data were integrated with XDS or iMOSFLM and scaled and merged in Aimless. Phases were obtained using molecular replacement in PhaserMR, CCP4 suite (43). The PDB entry 4MDI (chain A and B, without glycans or peptides) was used as search model for HLA-DRB1* for molecular replacement. The structures were built and refined in Coot and REFMAC/PHENIX (44). Final model was validated in Molprobity (45).
Repertoire analysis of HLA-DR4 allomorphs
T2-HLA-DRB1*04:05 cells expressing HLA-DM were generated via retroviral transduction of the parental T2 line as described previously (46). Cells were expanded in RPMI 1640 medium, 10% FCS and pellets of 109 cells snap-frozen in liquid nitrogen. Cells were ground under cryogenic conditions and resuspended in lysis buffer (0.5% IGEPAL, 50 mm Tris, pH 8, 150 mm NaCl, and protease inhibitors) as described previously (47, 48). Cleared lysates were passed over a protein A pre-column followed by an affinity column cross-linked with a monoclonal antibody specific for HLA-DR (LB3.1). Peptide–MHC complexes were eluted from the column by acidification with 10% acetic acid. Peptides were isolated using reversed-phase HPLC (Chromolith C18 Speed Rod, Merck) on an Akta Ettan HPLC system (GE Healthcare). Fractions were concentrated and run on an AB SCIEX 5600+ triple TOF high-resolution mass spectrometer as described (27). Acquired data were searched against the human proteome (Uniprot/SwissProt version 2016_12) using ProteinPilotTM software version 5 (AB SCIEX). The resulting peptide identities were subject to strict bioinformatic criteria, including the use of a decoy database to calculate the false discovery rate (FDR). A 5% FDR cutoff was applied, and the filtered dataset was further analyzed manually to exclude redundant peptides and known contaminants. To generate motifs, the minimal core sequences found within nested sets were extracted, and the resulting list of peptides were aligned using MEME, where motif width was set to 9–15 and motif distribution set to “one per sequence” (49). Peptides derived from HLA or immunoglobulin molecules were not included in the final motif analysis. Motifs were submitted to iceLogo for visualization using the frequencies of amino acids in the human proteome as a reference set (50). Comparison was made to previously published HLA-DR*04:01 and 04:04 datasets using standard statistical tools (24).
Author contributions
Y. T. T., S. H. R., and S. W. S. formal analysis; Y. T. T., S. H. R., S. W. S., K. L. L., A. W. P., H. H. R., and J. R. investigation; Y. T. T. and J. R. writing-original draft; Y. T. T., S. H. R., S. W. S., K. L. L., R. T., A. S., D. G. B., A. W. P., H. H. R., and J.R. writing-review and editing; J. P., A. W. P., H. H. R., and J. R. supervision; J. P. and K. L. L. methodology; R. T., A. S., D. G. B., H. H. R., and J. R. conceptualization; A. S. and J. R. resources; A. S., D. G. B., and J. R. funding acquisition.
Supplementary Material
Acknowledgments
We thank Nadine Dudek for contributing to the peptide elution study, Keshia Kim and Mai Tran for technical assistance, the Monash Macromolecular crystallization facility staff for assistance with crystallization, and the staff at the Australian Synchrotron for assistance with data collection.
The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Tables S1 and S2.
- RA
- rheumatoid arthritis
- SE
- shared epitope
- ACPA
- anti-citrullinated protein antibody
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- PTM
- post-translational modification
- PAD
- peptidyl-arginine deiminase
- Cit
- citrulline
- TCR
- T cell response
- PBMC
- peripheral blood mononuclear cell
- FDR
- false discovery rate
- TAMRA
- 6-carboxy-N,N,N′,N′-tetramethylrhodamine.
References
- 1. El-Gabalawy H. (2009) The preclinical stages of RA: lessons from human studies and animal models. Best Pract. Res. Clin. Rheumatol. 23, 49–58 10.1016/j.berh.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 2. Malmström V., Catrina A. I., and Klareskog L. (2017) The immunopathogenesis of seropositive rheumatoid arthritis: from triggering to targeting. Nat. Rev. Immunol. 17, 60–75 10.1038/nri.2016.124 [DOI] [PubMed] [Google Scholar]
- 3. Trouw L. A., Rispens T., and Toes R. E. (2017) Beyond citrullination: other post-translational protein modifications in rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 331–339 10.1038/nrrheum.2017.15 [DOI] [PubMed] [Google Scholar]
- 4. Van Steendam K., Tilleman K., and Deforce D. (2011) The relevance of citrullinated vimentin in the production of antibodies against citrullinated proteins and the pathogenesis of rheumatoid arthritis. Rheumatology 50, 830–837 10.1093/rheumatology/keq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foulquier C., Sebbag M., Clavel C., Chapuy-Regaud S., Al Badine R., Méchin M. C., Vincent C., Nachat R., Yamada M., Takahara H., Simon M., Guerrin M., and Serre G. (2007) Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 56, 3541–3553 10.1002/art.22983 [DOI] [PubMed] [Google Scholar]
- 6. Feitsma A. L., van der Voort E. I., Franken K. L., el Bannoudi H., Elferink B. G., Drijfhout J. W., Huizinga T. W., de Vries R. R., Toes R. E., and Ioan-Facsinay A. (2010) Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum. 62, 117–125 10.1002/art.25059 [DOI] [PubMed] [Google Scholar]
- 7. Hill J. A., Bell D. A., Brintnell W., Yue D., Wehrli B., Jevnikar A. M., Lee D. M., Hueber W., Robinson W. H., and Cairns E. (2008) Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J. Exp. Med. 205, 967–979 10.1084/jem.20072051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerstner C., Dubnovitsky A., Sandin C., Kozhukh G., Uchtenhagen H., James E. A., Ronnelid J., Ytterberg A. J., Pieper J., Reed E., Tandre C., Rieck M., Zubarev R. A., Ronnblom L., Sandalova T., et al. (2016) Functional and structural characterization of a novel HLA-DRB1*04:01-restricted α-enolase T cell epitope in rheumatoid arthritis. Front. Immunol. 7, e494 10.3389/fimmu.2016.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snir O., Rieck M., Gebe J. A., Yue B. B., Rawlings C. A., Nepom G., Malmström V., and Buckner J. H. (2011) Identification and functional characterization of T cells reactive to citrullinated vimentin in HLA-DRB1*0401-positive humanized mice and rheumatoid arthritis patients. Arthritis Rheum. 63, 2873–2883 10.1002/art.30445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. von Delwig A., Locke J., Robinson J. H., and Ng W. F. (2010) Response of Th17 cells to a citrullinated arthritogenic aggrecan peptide in patients with rheumatoid arthritis. Arthritis Rheum. 62, 143–149 10.1002/art.25064 [DOI] [PubMed] [Google Scholar]
- 11. Law S. C., Street S., Yu C. H., Capini C., Ramnoruth S., Nel H. J., van Gorp E., Hyde C., Lau K., Pahau H., Purcell A. W., and Thomas R. (2012) T-cell autoreactivity to citrullinated autoantigenic peptides in rheumatoid arthritis patients carrying HLA-DRB1 shared epitope alleles. Arthritis Res. Ther. 14, R118 10.1186/ar3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Beers J. J., Schwarte C. M., Stammen-Vogelzangs J., Oosterink E., Božič B., and Pruijn G. J. (2013) The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and β-actin. Arthritis Rheum. 65, 69–80 10.1002/art.37720 [DOI] [PubMed] [Google Scholar]
- 13. Snir O., Widhe M., Hermansson M., von Spee C., Lindberg J., Hensen S., Lundberg K., Engström A., Venables P. J., Toes R. E., Holmdahl R., Klareskog L., and Malmström V. (2010) Antibodies to several citrullinated antigens are enriched in the joints of rheumatoid arthritis patients. Arthritis Rheum. 62, 44–52 10.1002/art.25036 [DOI] [PubMed] [Google Scholar]
- 14. Biswas S., Sharma S., Saroha A., Bhakuni D. S., Malhotra R., Zahur M., Oellerich M., Das H. R., and Asif A. R. (2013) Identification of novel autoantigen in the synovial fluid of rheumatoid arthritis patients using an immunoproteomics approach. PLoS One 8, e56246 10.1371/journal.pone.0056246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kinloch A., Tatzer V., Wait R., Peston D., Lundberg K., Donatien P., Moyes D., Taylor P. C., and Venables P. J. (2005) Identification of citrullinated α-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res. Ther. 7, R1421–R1429 10.1186/ar1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raychaudhuri S., Sandor C., Stahl E. A., Freudenberg J., Lee H. S., Jia X., Alfredsson L., Padyukov L., Klareskog L., Worthington J., Siminovitch K. A., Bae S. C., Plenge R. M., Gregersen P. K., and de Bakker P. I. (2012) Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 10.1038/ng.1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Padyukov L., Seielstad M., Ong R. T., Ding B., Rönnelid J., Seddighzadeh M., Alfredsson L., Klareskog L., and Epidemiological Investigation of Rheumatoid Arthritis (EIRA) study group. (2011) A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis. 70, 259–265 10.1136/ard.2009.126821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R. R., Manoharan A., Ortmann W., Bhangale T., Denny J. C., et al. (2014) Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Beers J. J., Willemze A., Stammen-Vogelzangs J., Drijfhout J. W., Toes R. E., and Pruijn G. J. (2012) Anti-citrullinated fibronectin antibodies in rheumatoid arthritis are associated with human leukocyte antigen-DRB1 shared epitope alleles. Arthritis Res. Ther. 14, R35 10.1186/ar3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koning F., Thomas R., Rossjohn J., and Toes R. E. (2015) Coeliac disease and rheumatoid arthritis: similar mechanisms, different antigens. Nat. Rev. Rheumatol. 11, 450–461 10.1038/nrrheum.2015.59 [DOI] [PubMed] [Google Scholar]
- 21. Scally S. W., Law S. C., Ting Y. T., Heemst J. V., Sokolove J., Deutsch A. J., Bridie Clemens E., Moustakas A. K., Papadopoulos G. K., van der Woude D. V., Smolik I., Hitchon C. A., Robinson D. B., Ferucci E. D., Bernstein C. N., et al. (2017) Molecular basis for increased susceptibility of indigenous North Americans to seropositive rheumatoid arthritis. Ann. Rheum. Dis. 76, 1915–1923 10.1136/annrheumdis-2017-211300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- 23. Adams E. J., and Luoma A. M. (2013) The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31, 529–561 10.1146/annurev-immunol-032712-095912 [DOI] [PubMed] [Google Scholar]
- 24. Scally S. W., Petersen J., Law S. C., Dudek N. L., Nel H. J., Loh K. L., Wijeyewickrema L. C., Eckle S. B., van Heemst J., Pike R. N., McCluskey J., Toes R. E., La Gruta N. L., Purcell A. W., Reid H. H., et al. (2013) A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 210, 2569–2582 10.1084/jem.20131241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill J. A., Southwood S., Sette A., Jevnikar A. M., Bell D. A., and Cairns E. (2003) Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J. Immunol. 171, 538–541 10.4049/jimmunol.171.2.538 [DOI] [PubMed] [Google Scholar]
- 26. Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., and Stevanovič S. (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 10.1007/s002510050595 [DOI] [PubMed] [Google Scholar]
- 27. Dudek N. L., Tan C. T., Gorasia D. G., Croft N. P., Illing P. T., and Purcell A. W. (2012) Constitutive and inflammatory immunopeptidome of pancreatic beta-cells. Diabetes 61, 3018–3025 10.2337/db11-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Friede T., Gnau V., Jung G., Keilholz W., Stevanovič S., and Rammensee H. G. (1996) Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. Biochim. Biophys. Acta 1316, 85–101 10.1016/0925-4439(96)00010-5 [DOI] [PubMed] [Google Scholar]
- 29. Kinouchi R., Kobayasi H., Sato K., Kimura S., and Katagiri M. (1994) Peptide motifs of HLA-DR4/DR53 (DRB1*0405/DRB4*0101) molecules. Immunogenetics 40, 376–378 10.1007/BF01246679 [DOI] [PubMed] [Google Scholar]
- 30. Fugger L., and Svejgaard A. (2000) Association of MHC and rheumatoid arthritis: HLA-DR4 and rheumatoid arthritis–studies in mice and men. Arthritis Res. 2, 208–211 10.1186/ar89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anderson K. M., Roark C. L., Portas M., Aubrey M. T., Rosloniec E. F., and Freed B. M. (2016) A molecular analysis of the shared epitope hypothesis: binding of arthritogenic peptides to DRB1*04 alleles. Arthritis Rheumatol. 68, 1627–1636 10.1002/art.39636 [DOI] [PubMed] [Google Scholar]
- 32. Dessen A., Lawrence C. M., Cupo S., Zaller D. M., and Wiley D. C. (1997) X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity 7, 473–481 10.1016/S1074-7613(00)80369-6 [DOI] [PubMed] [Google Scholar]
- 33. Patil N. S., Pashine A., Belmares M. P., Liu W., Kaneshiro B., Rabinowitz J., McConnell H., and Mellins E. D. (2001) Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J. Immunol. 167, 7157–7168 10.4049/jimmunol.167.12.7157 [DOI] [PubMed] [Google Scholar]
- 34. Petersen J., Purcell A. W., and Rossjohn J. (2009) Post-translationally modified T cell epitopes: immune recognition and immunotherapy. J. Mol. Med. 87, 1045–1051 10.1007/s00109-009-0526-4 [DOI] [PubMed] [Google Scholar]
- 35. Sidney J., Becart S., Zhou M., Duffy K., Lindvall M., Moore E. C., Moore E. L., Rao T., Rao N., Nielsen M., Peters B., and Sette A. (2017) Citrullination only infrequently impacts peptide binding to HLA class II MHC. PLoS One 12, e0177140 10.1371/journal.pone.0177140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Henderson K. N., Tye-Din J. A., Reid H. H., Chen Z., Borg N. A., Beissbarth T., Tatham A., Mannering S. I., Purcell A. W., Dudek N. L., van Heel D. A., McCluskey J., Rossjohn J., and Anderson R. P. (2007) A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 27, 23–34 10.1016/j.immuni.2007.05.015 [DOI] [PubMed] [Google Scholar]
- 37. Kim C. Y., Quarsten H., Bergseng E., Khosla C., and Sollid L. M. (2004) Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 101, 4175–4179 10.1073/pnas.0306885101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tye-Din J. A., Stewart J. A., Dromey J. A., Beissbarth T., van Heel D. A., Tatham A., Henderson K., Mannering S. I., Gianfrani C., Jewell D. P., Hill A. V., McCluskey J., Rossjohn J., and Anderson R. P. (2010) Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci. Transl. Med. 2, 41ra51 [DOI] [PubMed] [Google Scholar]
- 39. Hardy M. Y., and Tye-Din J. A. (2016) Coeliac disease: a unique model for investigating broken tolerance in autoimmunity. Clin. Transl. Immunology 5, e112 10.1038/cti.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yin L., and Stern L. J. (2014) Measurement of peptide binding to MHC class II molecules by fluorescence polarization. Curr. Protoc. Immunol. 106, 5.10.11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reeves P. J., Callewaert N., Contreras R., and Khorana H. G. (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 99, 13419–13424 10.1073/pnas.212519299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gorrec F. (2009) The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 42, 1035–1042 10.1107/S0021889809042022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen V. B., Arendall W. B. 3rd., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for ma cromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pang S. S., Berry R., Chen Z., Kjer-Nielsen L., Perugini M. A., King G. F., Wang C., Chew S. H., La Gruta N. L., Williams N. K., Beddoe T., Tiganis T., Cowieson N. P., Godfrey D. I., Purcell A. W., et al. (2010) The structural basis for autonomous dimerization of the pre-T-cell antigen receptor. Nature 467, 844–848 10.1038/nature09448 [DOI] [PubMed] [Google Scholar]
- 47. Illing P. T., Vivian J. P., Dudek N. L., Kostenko L., Chen Z., Bharadwaj M., Miles J. J., Kjer-Nielsen L., Gras S., Williamson N. A., Burrows S. R., Purcell A. W., Rossjohn J., and McCluskey J. (2012) Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 10.1038/nature11147 [DOI] [PubMed] [Google Scholar]
- 48. Dudek N. L., Croft N. P., Schittenhelm R. B., Ramarathinam S. H., and Purcell A. W. (2016) A systems approach to understand antigen presentation and the immune response. Methods Mol. Biol. 1394, 189–209 10.1007/978-1-4939-3341-9_14 [DOI] [PubMed] [Google Scholar]
- 49. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., and Noble W. S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 10.1038/nmeth1109-786 [DOI] [PubMed] [Google Scholar]
- 51. James E. A., Rieck M., Pieper J., Gebe J. A., Yue B. B., Tatum M., Peda M., Sandin C., Klareskog L., Malmström V., and Buckner J. H. (2014) Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol. 66, 1712–1722 10.1002/art.38637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sohn D. H., Rhodes C., Onuma K., Zhao X., Sharpe O., Gazitt T., Shiao R., Fert-Bober J., Cheng D., Lahey L. J., Wong H. H., Van Eyk J., Robinson W. H., and Sokolove J. (2015) Local joint inflammation and histone citrullination provides a murine model for the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis Rheumatol. 67, 2877–2887 10.1002/art.39283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.