Abstract
The [URE3] yeast prion is the self-propagating amyloid form of the Ure2 protein. [URE3] is cured by overexpression of several yeast proteins, including Ydj1, Btn2, Cur1, Hsp42, and human DnaJB6. To better understand [URE3] curing, we used real-time imaging with a yeast strain expressing a GFP-labeled full-length Ure2 construct to monitor the curing of [URE3] over time. [URE3] yeast cells exhibited numerous fluorescent foci, and expression of the GFP-labeled Ure2 affected neither mitotic stability of [URE3] nor the rate of [URE3] curing by the curing proteins. Using guanidine to cure [URE3] via Hsp104 inactivation, we found that the fluorescent foci are progressively lost as the cells divide until they are cured; the fraction of cells that retained the foci was equivalent to the [URE3] cell fraction measured by a plating assay, indicating that the foci were the prion seeds. During the curing of [URE3] by Btn2, Cur1, Hsp42, or Ydj1 overexpression, the foci formed aggregates, many of which were 0.5 μm or greater in size, and [URE3] was cured by asymmetric segregation of the aggregated seeds. In contrast, DnaJB6 overexpression first caused a loss of detectable foci in cells that were still [URE3] before there was complete dissolution of the seeds, and the cells were cured. We conclude that GFP labeling of full-length Ure2 enables differentiation among the different [URE3]-curing mechanisms, including inhibition of severing followed by seed dilution, seed clumping followed by asymmetric segregation between mother and daughter cells, and seed dissolution.
Keywords: amyloid, molecular chaperone, prion, protein folding, yeast
Introduction
Prions are infectious proteins that have two stable conformations: a properly folded conformation and a misfolded infectious amyloid conformation. Thus far, approximately a dozen prions have been identified in yeast; the best studied of these are [PSI+], [URE3], and [RNQ+] (1). To maintain prion propagation from generation to generation, there must be severing of the prion seeds by Hsp104 and transmission of the seeds from mother to daughter cells during cell division. To cure yeast of the misfolded amyloid form of the prion, the prion seeds must be eliminated, which can occur by several different mechanisms. One mechanism of curing is inactivation of Hsp104, thereby inhibiting severing of the prion seeds (2–4). Another mechanism of curing yeast prions is aggregation of the prion seeds followed by asymmetric segregation of the seeds between mother and daughter cells, thus reducing the number of seeds transmitted to the daughter cells (5–7). Finally, yeast prions can be cured by dissolution of the seeds, which can occur either by dissociating monomers from the ends of the seeds or by inhibiting the growth of the seeds (8, 9). When seed growth is blocked, the severing activity of Hsp104 reduces the size of the seeds until the yeast are cured.
To understand the mechanism of yeast prion curing under different conditions, laboratories have used biochemical, genetic, and cell biological approaches. These include measuring the kinetics of curing, determining the relative number of monomers in the aggregates and soluble pool, and live cell fluorescence imaging of fluorescent-labeled prions. In the earliest imaging studies, GFP was fused to the prion domain rather than to the full-length prion protein; this was done with the prion domain of Sup35 (10). The GFP-labeled prion domain forms aggregates in cells with the amyloid form of the prion, but not in cells when the prion protein is properly folded. However, when just the prion domain was expressed in cells with the amyloid prion form, the cells did not show the numerous foci that are thought to represent the prion seeds.
In later imaging studies, full-length prion proteins rather than just the prion domains were labeled. However, labeling of the full-length prion protein poses challenges; the labeled prion protein must retain its native activity when its properly folded, and at the same time, the labeling must not interfere with the conversion to and propagation of the amyloid conformation. This has been successfully achieved for Sup35, which in its amyloid form is the [PSI+] prion. Sup35 is a multidomain protein, consisting of an N-terminal domain, an M or middle domain and a C-terminal domain. When GFP was inserted between the N-terminal and M domains of Sup35, the GFP-labeled Sup35 (NGMC) protein became an excellent way to monitor the Sup35 conformation (11, 12). Importantly, NGMC shows diffuse fluorescence and retains the activity of the parent protein when properly folded, whereas it forms numerous foci representing the prion seeds when it is converted to the [PSI+] prion. Studies using this construct have been very useful in understanding how different [PSI+] variants, which differ in their relative number of prion seeds and amount of soluble Sup35, pass [PSI+] from mother to daughter cells and also in understanding the different mechanisms for curing [PSI+] (8, 13–15). With the use of this construct, we have determined that, in addition to the severing activity of Hsp104 that reduces the size and increases the number of seeds, Hsp104 has a trimming activity that reduces the size of the seeds but does not affect the number of seeds (15). The trimming activity of Hsp104 has been shown to be required for the curing of [PSI+] by Hsp104 overexpression (8).
In contrast to [PSI+], a full-length Ure2 labeled GFP has not yet been constructed that maintains [URE3] propagation. This was the case regardless of whether Ure2 was labeled at its C-terminal end or internally labeled after residue 65 (16). The Wickner laboratory found that, unfortunately, expression of these GFP-labeled Ure2 constructs from a URE2 promoter on a centromeric plasmid cured the [URE3] prion (16). Instead of a full-length labeled Ure2 protein, they used an N-terminal Ure2 prion domain labeled at the C terminus with GFP to monitor [URE3]. When the latter construct was expressed in [URE3] yeast, the cells had fluorescent clumps instead of foci (16). A functional full-length GFP-labeled Ure2 that stably propagated [URE3] would be very useful for understanding the mechanisms by which overexpression of different proteins cure [URE3], including Btn2, Cur1, and Hsp42 and two different J-domain proteins, Ydj1 and human DnaJB6 (6, 9, 17, 18). Imaging studies employing such a functional GFP-labeled Ure2 would complement genetic, biochemical, and cell biological approaches that have been used to understand how these various proteins cure [URE3].
Thus far, no mechanism has been proposed for how Hsp42 overexpression cures [URE3]. On the other hand, asymmetric segregation has been proposed as the mechanism by which Btn2 overexpression cures [URE3], based on the observation that clumps of GFP-labeled Ure2-prion domain partially colocalized with Btn2 (6). As for overexpression of Cur1 and Ydj1, curing has been attributed to inhibition of fragmentation because of insufficient Sis1, which is required for Hsp104 severing (19, 20). Cur1 sequesters Sis1 in the nucleus, whereas Ydj1 appears to compete with Sis1 for Ssa1, which reduces the concentration of the Sis1-Ssa1, a complex that binds and activates Hsp104. An alternative model for the curing of [URE3] by Ydj1 overexpression is that Ydj1 binds to soluble Ure2 and inhibits the addition of the Ure2 monomers to the end of the prion seeds that then continue to shorten as they are severed by Hsp104 (21). A similar mechanism has been proposed for the curing of [URE3] by human DnaJB6b, another member of the Hsp40 family. Like Ydj1, DnaJB6 inhibits nucleation of the Ure2 protein in biochemical assays using purified proteins (9) and also other amyloids such as amyloid β42 and polyglutamine peptides (22–24). On this basis, the Masison laboratory proposed that human DnaJB6 cures [URE3] by binding to the prion seeds and inhibiting their growth (9).
In this study, we engineered a full-length GFP-labeled Ure2, which, unlike other GFP-labeled reporters of full-length Ure2, stably maintained the [URE3] prion. This was done by expressing in [URE3] yeast relatively low levels of the labeled Ure2 protein, in which the GFP was inserted after residue 90, along with endogenous levels of unmodified Ure2. Curing of [URE3] by guanidine inactivation of Hsp104 showed that the percentage of cells with foci correlated with the percentage of [URE3] cells measured using a plating colony assay. When yeast overexpressed Btn2, Cur1, Hsp42, or Ydj1, the foci assembled into aggregates, which reduced the number of prion seeds, ultimately curing [URE3]. In contrast to Ydj1, the human DnaJB6 caused a loss of detectable foci prior to curing, which is consistent with DnaJB6 blocking the growth of the amyloid seeds, but not their severing.
Results
Establishment of a GFP-labeled Ure2 strain
Our aim was to make a GFP-labeled Ure2 protein to use in imaging the curing of [URE3] in real time. Expression in of a C-terminal GFP-labeled full-length Ure2 construct made by the Wickner laboratory in the 1076 yeast, a strain of yeast that is [URE3] and [rnq−], produced cells with aggregates as well as cells with diffuse fluorescence (16). Similar results were obtained when we expressed a Ure2 construct internally labeled with GFP at residue 65. Because expression of these constructs has been shown to cure [URE3], the cells with diffuse fluorescence presumably are cured ([ure-o]) yeast. As an alternative, we engineered a full-length GFP-labeled Ure2 by inserting GFP at residue 90, the terminal residue of the protease-resistant fibril core of the Ure2 amyloid (25, 26). When this GFP–Ure2 fusion protein was expressed from the URE2 promoter on a centromeric plasmid in 1076[URE3] yeast, confocal imaging showed numerous foci, as well as clumps and, importantly, no cells with diffuse fluorescence. Unfortunately, however, plating assays showed that this construct, like the constructs made by the Wickner laboratory (16), cured the [URE3] prion. Furthermore, this construct did not stably maintain [URE3] even when integrated into the URE2 chromosomal locus. To circumvent this problem, we expressed relatively low levels of this GFP–Ure2 fusion protein by using a plasmid with the RNQ1 promoter instead of the URE2 promoter. Based on the number of Ure2 and Rnq1 molecules produced per yeast cell as measured by the Weissman laboratory (27), only ∼10% of the Ure2 molecules per cell should then be labeled with GFP. After transforming the plasmid with the RNQ1 promoter into 1076[URE3] yeast, confocal imaging showed numerous foci in the yeast, and importantly, the red/white colony assay showed only white colonies, indicating that all of the yeast was still [URE3] (25).
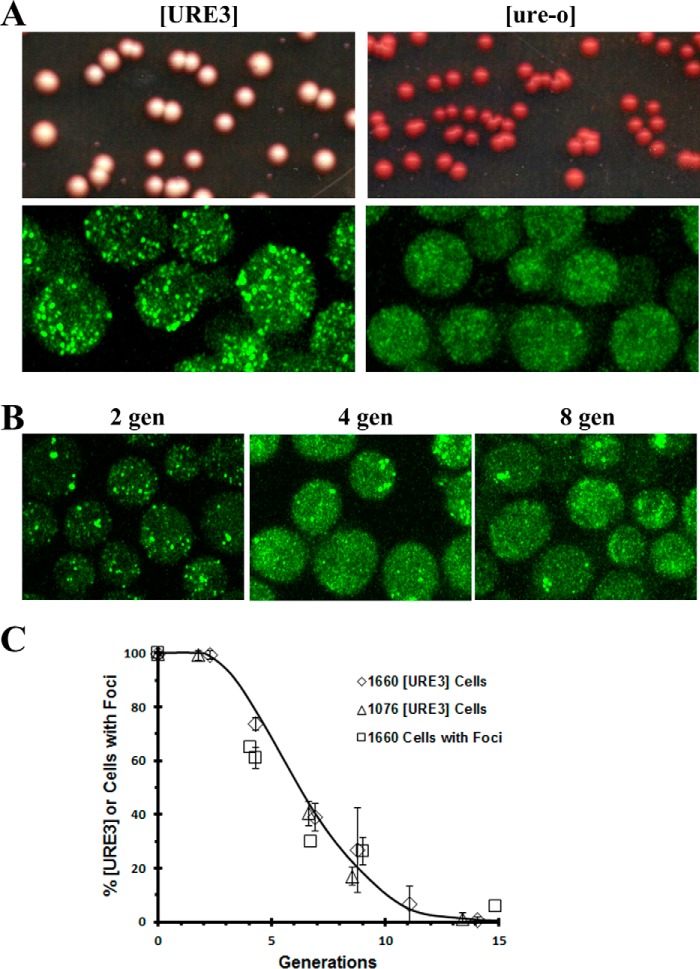
We therefore integrated the GFP–Ure2 fusion gene into the RNQ1 chromosomal locus of the 1075 yeast strain to make the 1660 yeast strain. Fluorescent imaging showed numerous foci in the 1660 [URE3] yeast cells, whereas no foci were present after this strain was cured in guanidine (Fig. 1A). When the 1660[URE3] strain was assayed for [URE3] on ½ YPD plates (28), all the colonies were white, indicating that [URE3] was stably maintained, whereas as expected, all the colonies were red after curing the 1660[URE3] strain in guanidine (Fig. 1A). As the 1660[URE3] yeast were cured in guanidine, the maximum Z-stack projections show a progressive decrease in foci number with increasing incubation time in guanidine (Fig. 1B), consistent with guanidine inhibiting the severing of the prion seeds so that the seeds are diluted out by cell division. These results are very different from the results that were observed using GFP-labeled Sup35 (NGMC) where we observed that during the curing of [PSI+] in guanidine, there was a loss of detectable foci prior to curing (15). These latter results revealed that, in addition to the well-characterized severing activity of Hsp104, Hsp104 has another activity, which we called trimming, that reduces the size of the seeds but does not affect the number of seeds.
Figure 1.
Characterization of the 1660 [URE3] strain. A, the 1660[URE3] and the cured 1660[ure-o] yeast strains were imaged and plated. The 1660[URE3] strain was cured in 5 mm guanidine to make the 1660 [ure-o]. These strains were imaged on the Zeiss 880 microscope and plated on ½ YPD plates. Images are maximized projections of Z-stack confocal images. B, images of 1660 [URE3] strain during curing in guanidine. Confocal images were obtained using the Zeiss 880 microscope after incubation in guanidine for two, four, or eight generations (gen), respectively. The images are the maximum Z-stack projections. The same settings were used in all images. C, the kinetics of curing the 1660[URE3] and 1076[URE3] yeast strains in 5 mm guanidine was measured using the red/white colony assay. The cells at the indicated time were plated on ½ YPD plates, and only completely red colonies were counted as cured. The data represent the averages and standard deviation obtained from three independent experiments. The cells with foci were measured in 1660 [URE3] yeast following the addition of 5 mm guanidine (>300 cells were counted/time point). The percentage of cells with foci as a function of generation time are plotted on the same graph as the curing curves.
Next, we determined whether the expression of the GFP–Ure2 fusion protein affected the rate of [URE3] curing by guanidine. This was done by measuring the rate of [URE3] curing in both the 1660 strain and 1076 strains. Fig. 1C shows that the rate of [URE3] curing as determined by the red/white colony assay is the same in these two strains, which establishes that expression of the GFP–Ure2 fusion protein does not significantly affect [URE3] curing by guanidine. To validate the use of the 1660[URE3] strain as a real-time reporter of [URE3] curing, we next examined whether the GFP-labeled foci are representative of the entire population of prion seeds. If so, then during the curing of [URE3] by guanidine, the percentage of cells with GFP-labeled foci should be equivalent to the percentage of [URE3] cells. Therefore, we determined the percentage of cells with GFP-labeled foci (>300 cells/time point) at different times after addition of guanidine, and this percentage was plotted as a function of generation time. Fig. 1C shows that these data are nearly superimposable on the curing curve, indicating that the cells with GFP-labeled foci are equivalent to [URE3] cells. Therefore, the 1660[URE3] strain is a new tool for real-time imaging of the curing of [URE3] under different conditions.
Curing of [URE3] by overexpression of Btn2, Cur1, or Hsp42
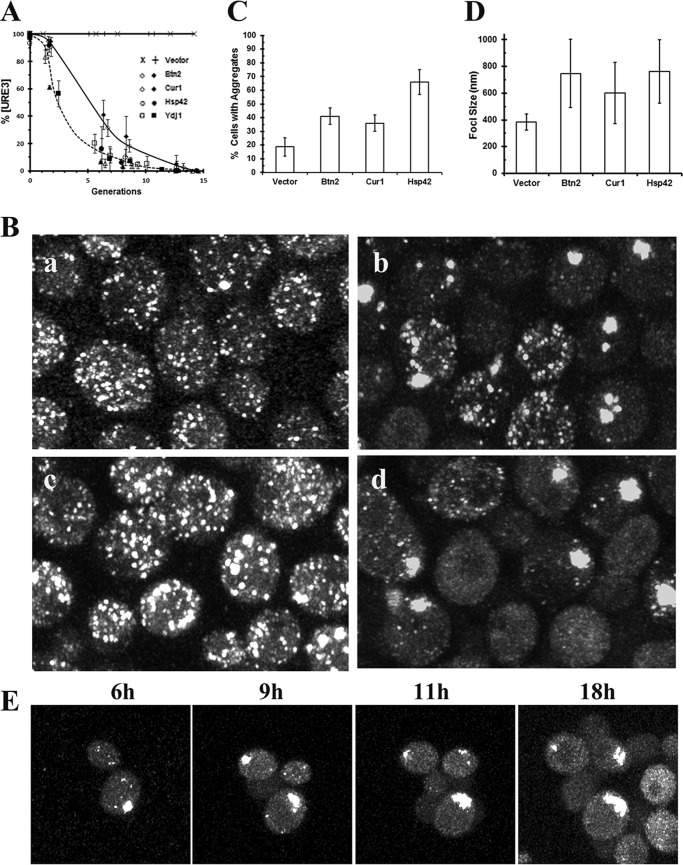
Because [URE3] is cured by overexpression of Btn2, Cur1, or Hsp42 (6, 17, 18), we wanted to determine the basic mechanisms of curing by these three proteins by visualizing the curing as it occurs in the 1660[URE3] strain. First, we measured the rate of [URE3] curing using the red/white colony assay when these proteins were overexpressed from the GAL1 promoter in the 1660 and 1076 [URE3] strains. As shown in Fig. 2A, overexpression of Btn2 and Hsp42 cured [URE3] faster than overexpression of Cur1 in both the 1660 and 1076 yeast strains; as expected, the vector control, which was treated the same as the other samples, showed no curing. To elucidate the basic mechanisms of curing by these three different proteins, concomitantly with plating the yeast, we followed the curing by real-time Z-stack confocal imaging. As we observed for the curing of [URE3] by guanidine, the imaging data correlated with the curing curves. All cells started with numerous foci, and as the prion was cured, there was an increase in the fraction of cells without foci until all the cells were devoid of foci. However, unlike the curing in guanidine, imaging showed that cells formed large aggregates prior to curing.
Figure 2.
The curing of the 1660[URE3] by overexpression of Btn2, Cur1, or Hsp42. A, the curing [URE3] as a function of time was measured in the 1660 (open symbols) and 1076 (closed symbols) yeast strains overexpressing Btn2, Cur1, or Hsp42. The vector control is also shown for the 1660 (×) and 1076 (+) [URE3] strains. Yeast was grown overnight in raffinose medium prior to addition of galactose to induce expression from the GAL1 promoter. The extent of curing was measured on ½ YPD plates. The vector control was treated the same as the other samples. The data represent the averages and standard deviation obtained from three independent experiments. B, confocal images of 1660 [URE3] strain in the vector control (panel a) or in yeast overexpressing Btn2 (panel b), Cur1 (panel c), or Hsp42 (panel d). The images were taken two generations after induction of overexpression by addition of galactose on the Zeiss 880 microscope. The images are maximized projections of Z-stack confocal images. C, the percentage of cells with aggregates was measured in the vector control and in cells overexpressing the indicated proteins for two generations. For each sample, >200 cells were analyzed to determine whether they contained aggregates >0.3 μm. The values represent the averages and standard deviation. D, the average size of the aggregates was measured in the vector control and in cells overexpressing the indicated proteins for two generations. Only aggregates of >0.3 μm were measured. The values represent the averages and standard deviation (n > 150 aggregates/sample). E, images from a time-lapse movie showing formation of aggregates in the 1660 [URE3] strain overexpressing Hsp42 for the indicated time in galactose medium. Imaging was performed on the Zeiss 880 confocal microscope. The images are maximized projections of Z-stack confocal images.
Fig. 2B shows the aggregation of the foci in yeast grown two generations in galactose medium. These maximum Z-stack confocal images were obtained using the Zeiss Airyscan detector, which has a lateral resolution of ∼140 nm in the lateral axis. We measured aggregates >300 in the vector control (Fig. 2B, panel a), and in yeast overexpressing Btn2, Cur1 or Hsp42 (Fig. 2B, panels b–d). In the vector control, ∼20% of the cells had an aggregate with an average size of 380 nm, whereas there were more cells with aggregates and the size of the aggregates were larger in yeast overexpressing Btn2, Cur1, or Hsp42. Specifically, more than one-third of the cells had an aggregate with an average size ranging from 0.5 to 0.8 μm in yeast overexpressing Btn2, Cur1, or Hsp42 (Fig. 2, C and D). Consistent with the slower curing of [URE3] by overexpression of Cur1, the percentage of cells with aggregates and the foci size were smaller than in cells overexpressing either Btn2 or Hsp42. The formation of these aggregates would cause a marked reduction in their mobility compared with the diffraction limited size of the seeds present in the 1660[URE3] control. In addition, some of the yeast had aggregates greater than 1 μm regardless of the protein being overexpressed. Such large aggregates would have trouble passing through the bud neck of dividing cells, and certainly the rapid aggregate formation is consistent with the kinetics of curing by overexpression of these three different proteins. By imaging the same field of 1660[URE3] cells induced in galactose to overexpress Hsp42, the GFP–Ure2 aggregates showed a progressive increase in size, and new aggregates continue to form in the daughter cells (Fig. 2E). Therefore, overexpression of Btn2, Cur1, or Hsp42 causes aggregation of the prion seeds, which in turn reduces the seed number and restricts their transmission to the daughter cells.
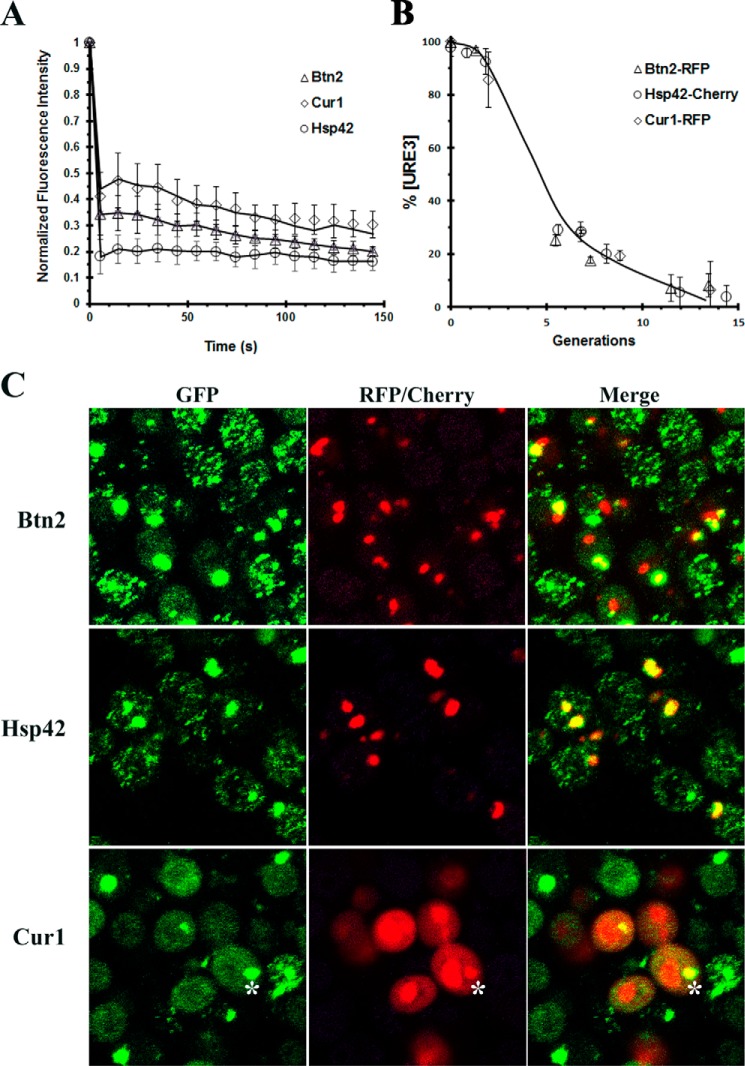
Next, we examined whether the GFP–Ure2 was immobilized in the large aggregates by measuring the fluorescence recovery after photobleaching in cells incubated for two generations in galactose medium. Unlike the Ure2–GFP foci, which moved rapidly throughout the cell, the large aggregates showed very limited movement. These large aggregates were photobleached, and the recovery of fluorescence was measured. As shown in Fig. 3A, there is no significant fluorescence recovery of the GFP–Ure2 in the aggregates, indicating that the GFP–Ure2 in the aggregates is not exchanging with the soluble pool of GFP–Ure2.
Figure 3.
Characterization of the aggregates formed by 1660[URE3] overexpressing Btn2, Hsp42, or Cur1. A, the fluorescence recovery after photobleaching of aggregates present in 1660[URE3] yeast overexpressing Btn2, Cur1, and Hsp42. A minimum of five aggregates were photobleached for each sample on the Nikon A1R confocal microscope. The data were normalized by setting the initial fluorescence value to 1.0. The average value and a standard deviation are plotted for each time point. Photobleaching experiments were performed on the Nikon A1R confocal microscope. B, the time course of curing of 1660[URE3] by overexpression of Btn2–RFP, Cur1–RFP, and Hsp42–Cherry. The data represent the averages and standard deviation obtained from three independent experiments. C, colocalization of GFP–Ure2 with Btn2, Hsp42, and Cur1. Btn2–RFP or Hsp42–Cherry was overexpressed for two generations. Cur1–RFP was overexpressed for five generations. The asterisk shows colocalization of Ure2–GFP aggregate with Cur1–RFP. The maximum Z-stack projections of confocal images obtained on the Nikon A1R confocal microscope.
Finally, we examined the colocalization of the GFP-labeled Ure2 aggregates with Btn2, Cur1, or Hsp42 with the use of dual color imaging. To do this, we used Btn2, Cur1, or Hsp42 that was labeled with a red fluorophore (either RFP or Cherry). First, we checked that the fluorescent labeling of these proteins did not significantly affect their ability to cure the 1660[URE3] yeast. Comparing Fig. 3B with Fig. 2A shows that fluorescent labeling of Btn2 and Hsp42 reduced the rate of [URE3] curing by two or three generations but did not significantly affect the rate of curing by Cur1. When we imaged the colocalization of GFP–Ure2 with Btn2, the cytosolic GFP–Ure2 aggregates colocalized with the Btn2 clumps (Fig. 3C). However, most cells had several Btn2 clumps, and only some of these clumps were associated with GFP–Ure2 aggregates. This is reflected in our analysis of colocalization of these proteins (in 200 cells); >90% of the GFP–Ure2 aggregates colocalized with the Btn2 clumps, whereas ∼50% of the Btn2 clumps colocalized with the GFP–Ure2 aggregate. Another protein that formed cytosolic clumps is Hsp42, but in general, there were fewer clumps of Hsp42 per cell than clumps of Btn2. The GFP–Ure2 aggregates showed extensive colocalization with the Hsp42 clumps; there was >90% colocalization of GFP–Ure2 aggregates with the Hsp42 clumps. Furthermore, in contrast to the Btn2 clumps, >90% of the Hsp42 clumps colocalized with the GFP–Ure2 aggregates. Cur1, a paralog of Btn2 that predominantly has a nuclear localization (17, 29), also produced cytosolic GFP–Ure2 aggregates. We did observe a low level of Cur1 in the cytoplasm, and occasionally, there was colocalization of GFP–Ure2 with the cytosolic Cur1 (see asterisk in Fig. 3C). Therefore, even though these proteins have very different cellular functions and localizations, they all caused aggregation of the GFP-labeled prion seeds, which would lead to curing of [URE3] by asymmetric segregation.
Mechanism of curing of [URE3] by Cur1
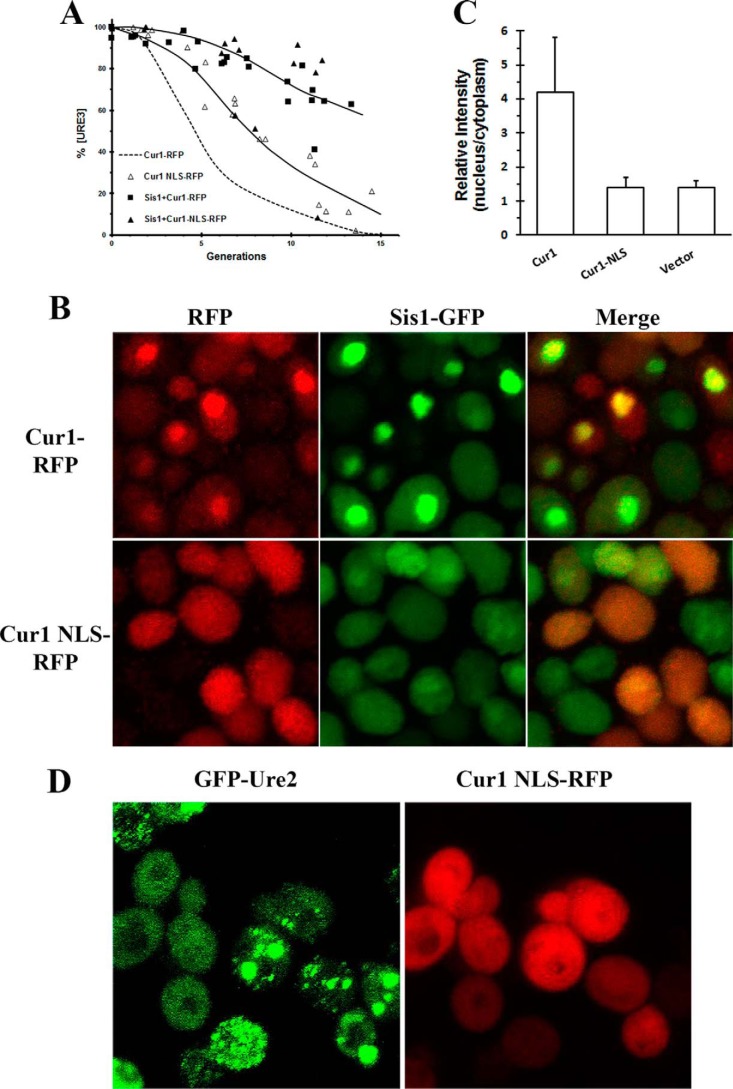
It has been proposed by the Zhouravleva laboratory (20) that overexpression of Cur1 cures [URE3] by recruiting Sis1 into the nucleus, thereby depleting it from the cytosol. This is based on their finding that when they overexpressed a Cur1 fragment (Cur1Δ3–30) that does not localize to the nucleus, there was neither recruitment of Sis1 to the nucleus nor curing of [URE3]. Unexpectedly, we observed curing of [URE3] when we overexpressed a mutant of Cur1 (Cur1–NLS), which contains two point mutations at its nuclear localization site, Cur1 (K26A,K27A), that presumably would prevent it from localizing to the nucleus. As shown in Fig. 4A, overexpression of Cur1–NLS cures [URE3], although at a slightly slower rate than wildtype Cur1. These results are consistent with the finding from the study by Alberti and co-workers (29) in which they examined curing of an artificial yeast prion by overexpression of a Cur1 ΔNLS mutant.
Figure 4.
Mechanism of curing [URE3] by Cur1 and Cur1–NLS. A, the curing of 1660[URE3] yeast was measured as a function of generation time in yeast overexpressing RFP-labeled Cur1–NLS or in yeast coexpressing Sis1 and RFP-labeled Cur1 or in yeast coexpressing Sis1 and RFP-labeled Cur1–NLS. The dashed line is the data shown in Fig. 3B for the curing of 1660[URE3] by overexpression of RFP-labeled Cur1. The Sis1 and the RFP-labeled Cur1 constructs were both overexpressed from the GAL1 promoter. B, colocalization of Sis-GFP with RFP-labeled Cur1 or Cur1–NLS. The cells were imaged after five generations in galactose medium to induce expression of the different Cur1 constructs. C, the intensity of GFP-labeled Sis1 in the nucleus compared with its intensity in the cytoplasm was measured in vector control cells or cells overexpressing either RFP-labeled Cur1 or Cur1 NLS. After fixing the cells with 4% paraformaldehyde, the nucleus was stained with 4′,6′-diamino-2-phenylindole to delineate the nucleus. The ImageJ program was used to measure the GFP fluorescence intensity in the nuclear and cytoplasmic compartments. A minimum of 30 cells were measured for each condition. The average values and standard deviation are plotted for each condition. D, images of 1660[URE3] yeast overexpressing RFP-labeled Cur1–NLS for five generations. The maximum Z-stack projections of confocal images were obtained on the Nikon A1R confocal microscope.
Previous studies have shown that wildtype Cur1 recruits Sis1 to the nucleus, but not when Cur1 is mutated to prevent its nuclear localization (20, 29). To verify these results, we coexpressed the different RFP-labeled Cur1 constructs with GFP-labeled Sis1 (Fig. 4B), and then cells were fixed, and the nucleus was stained with 4′,6′-diamino-2-phenylindole to quantify the localization of Cur1 and Sis1 in the nuclear and cytoplasmic compartments. There was approximately three times more wildtype Cur1 in the nucleus than in the cytoplasm, whereas there was no significant difference in the intensity of Cur1–NLS between the nucleus and the cytoplasm. Similarly, there was approximately four times more Sis1 in the nucleus than in the cytoplasm in yeast overexpressing Cur1, whereas there was no significant difference in relative Sis1 intensity between the nucleus and the cytoplasm in cells overexpressing mutant Cur1–NLS or in the vector control (Fig. 4C). Therefore, Sis1 has a very different localization depending on whether Cur1 or Cur1–NLS is overexpressed.
Interestingly, however, even though overexpression of Cur1–NLS does not sequester Sis1 into the nucleus, it still cures [URE3]; imaging of the 1660[URE3] yeast showed aggregation of the GFP-labeled Ure2 foci in cells overexpressing Cur1–NLS (Fig. 4D), just as we observed in yeast overexpressing wildtype Cur1. To explain this result, we made use of the observation that Cur1 binds directly to Sis1 (29). On this basis, we hypothesized that even though overexpression of Cur1–NLS does not recruit Sis1 to the nucleus, it reduces the cytoplasmic pool of Sis1 by forming a Cur1–NLS–Sis1 complex. This was tested by determining whether overexpression of Sis1 antagonizes the curing of [Ure3] by overexpression of Cur1–NLS, as well as by wildtype Cur1. As shown in Fig. 4A, overexpression of Sis1 markedly reduced the rate of curing of 1660[URE3] by the Cur1–NLS, as well as by the wildtype Cur1. Occasionally, we observed a colony in which overexpression of Sis1 did not antagonize the curing by either wildtype Cur1 or mutant Cur1, but we also occasionally observed curing in the vector control as well. Therefore, these data are consistent with overexpression of either Cur1 or Cur1–NLS curing [URE3] by depleting the free pool of cytoplasmic Sis1. Furthermore, these data suggest that a reduction in the free pool of Sis1 causes aggregation of the GFP-labeled Ure2 foci, thereby curing [URE3] by asymmetric segregation.
Curing of [URE3] by overexpression by other molecular chaperones
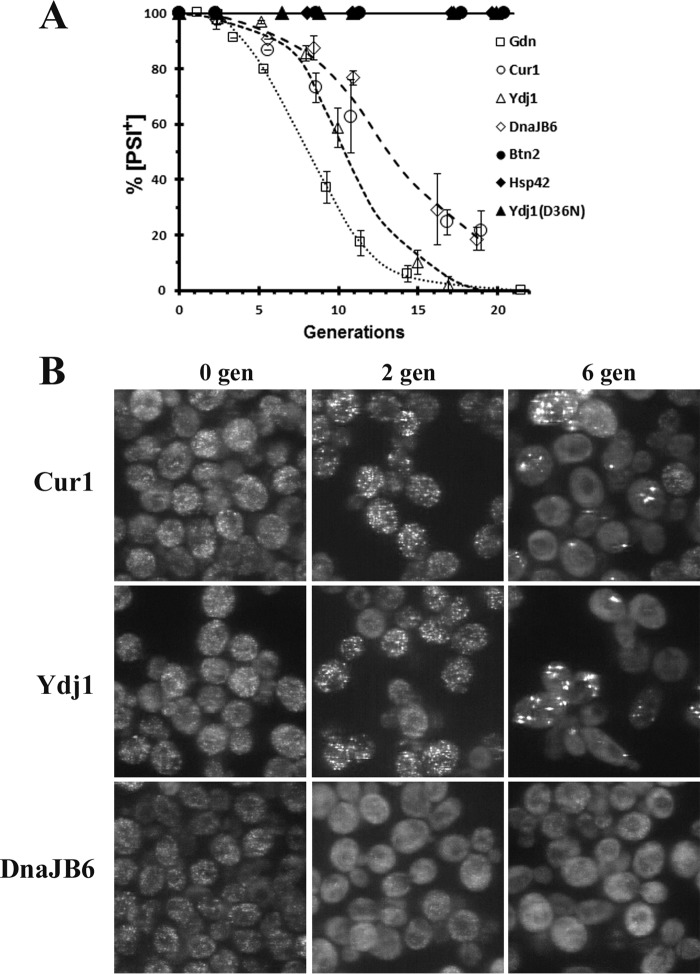
Overexpression of two different J-domain proteins, Ydj1 and human DnaJB6, has been shown to cure [URE3], but with very different kinetics (9). In agreement with this study, we also found that overexpression of Ydj1 cures 1660[URE3] much faster than overexpression of DnaJB6 when curing was measured using the red/white colony assay. Furthermore, the shapes of the curing curves are very different (Fig. 5A), which suggests that these J-domain proteins cure [URE3] by different mechanisms. As yeast were cured, we simultaneously imaged the GFP-labeled foci. Like the results obtained with overexpression of Btn2, Cur1, and Hsp42, overexpression of Ydj1 caused aggregation of the foci. As shown in Fig. 5B, after two generations in galactose medium, the Ure2–GFP foci formed aggregates, which persisted until the yeast were cured. Analysis of the images (200 cells) showed that ∼60% of the cells had multiple aggregates with an average size of ∼500 nm (470 ± 125 nm). After four generations in galactose medium, a time when the yeast were ∼50% cured, images showed there were cells with no foci and cells with multiple aggregates that were not significantly larger than those observed after two generations (average size 480 ± 100 nm). As observed previously (30), the curing of [URE3] by overexpression of Ydj1 requires a functional J-domain. Specifically, there was neither curing nor large aggregate formation of the 1660[URE3] strain overexpressing Ydj1 (D36N), which has a point mutation inhibiting its binding to members of the Hsp70 family. Therefore, overexpression of Ydj1 with a functional J-domain causes aggregation of the prion seeds, which cures [URE3] when the aggregates are diluted out by cell division.
Figure 5.
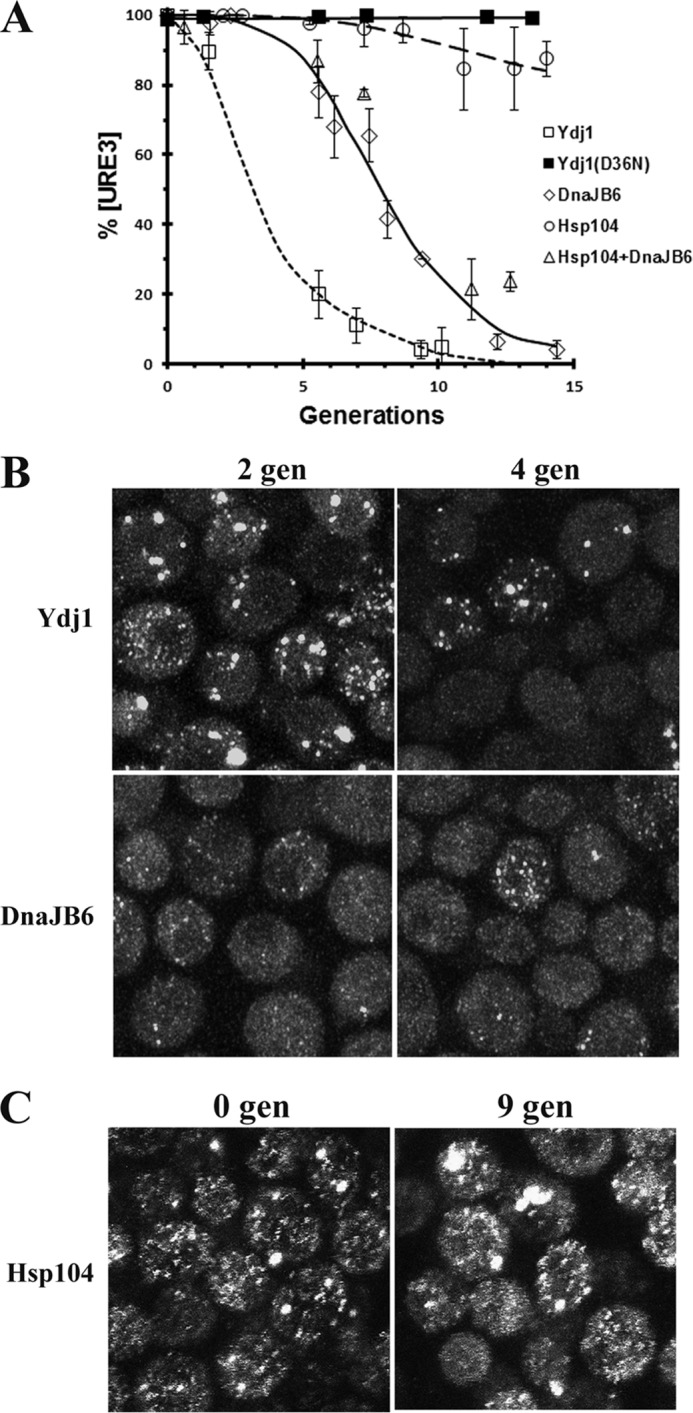
The curing of 1660[URE3] by other molecular chaperones. A, the rate of curing of 1660[URE3] was measured using the red/white colony assay for yeast overexpressing Ydj1, Yjd1(D36N), DnaJB6, Hsp104, or coexpressing Hsp104 and DnaJB6. All proteins were overexpressed from the GAL1 promoter. The data points represent the averages and standard deviation obtained from three independent experiments. B, images of 1660[URE3] overexpressing either Ydj1 or DnaJB6 for either two generations or four generations. The maximum Z-stack projections of confocal images were obtained on the Zeiss 880 confocal microscope. C, images of 1660[URE3] prior to overexpression and overexpressing Hsp104 for nine generations from the GAL1 promoter. The images are maximized projections of Z-stack confocal images obtained on the Nikon A1R confocal microscope.
In contrast to overexpression of Ydj1, overexpression of human DnaJB6 did not cause aggregation of the Ure2 foci. Instead, in some yeast overexpressing DnaJb6 for two generations, the foci were difficult to resolve above the background fluorescence even though plating assays showed that the yeast were still more than 95% [URE3] (Fig. 5B). After four generations in galactose, at which time the [URE3] yeast were ∼10% cured, the background fluorescence had increased along with a concomitant loss of detectable foci, but there were still cells with detectable foci. As expected, no foci were detected after all the yeast were cured. Taken together, these data suggest that Ydj1 cures [URE3] by asymmetric segregation of the seeds, whereas DnaJB6 cures [URE3] by dissolution of the seeds.
To help clarify whether overexpression of Hsp104 cures [URE3] (6, 18), we also examined the effect of overexpressing Hsp104 in the 1660[URE3] strain. Following Hsp104 overexpression, the rate of curing was measured by the red/white colony assay. In agreement with the results of Kryndushkin et al. (6), overexpression of Hsp104 does cure [URE3], but curing occurs at a very slow rate. As shown in Fig. 5A, following induction of Hsp104 overexpression from the GAL1 promoter, only ∼15% of the 1660[URE3] cells were cured in 15 generations. This slow rate of curing is very different from the relatively rapid rate of curing of [PSI+] by Hsp104 overexpression (31). Another difference from [PSI+] curing by Hsp104 overexpression is that prior to curing, the 1660[URE3] yeast showed no loss of detectable foci because of trimming of the prion seeds by overexpressed Hsp104. Instead, the cells had foci, and some also had aggregates after nine generation in galactose medium (Fig. 5C). Finally, we examined whether overexpression of Hsp104, which presumably would increase the rate of severing of the prion seeds, affected curing of 1660[URE3] by DnaJB6 overexpression. As shown in Fig. 5A, the rate of curing of [URE3] by DnaJB6 overexpression was not affected by overexpression of Hsp104. These results suggest that increasing the severing of the seeds does not increase the rate of curing by DnaJB6 overexpression assuming, of course, that severing of the prion seeds increases at higher expression levels of Hsp104.
Effect of overexpressing Btn2, Cur1, Hsp42, Ydj1, or DnaJB6 in [PSI+] yeast
Next, we examined how the curing of [PSI+] yeast compares with the curing of [URE3]. Specifically, we examined whether overexpression of these different curing agents cured the L2888 weak [PSI+] variant. In L2888 [PSI+] yeast, GFP-labeled Sup35 (NGMC) is expressed from the SUP35 chromosomal locus, enabling us to image the curing of [PSI+]. Among [PSI+] variants, L2888[PSI+] is a weak variant (32), and it cures much more readily than a strong [PSI+] variant (33). However, compared with 1660[URE3] prion, the L2888[PSI+] variant has many more seeds, as evident by comparing the rate of curing of these two prions in guanidine (compare Fig. 6A and Fig. 1C). Despite the difference in seed number, overexpression of Cur1, Ydj1, or DnaJB6 from the GAL1 promoter cured L2888[PSI+] (Fig. 6A), but at a slower rate than they cured 1660[URE3]. Overexpression of Btn2 and Hsp42 did not cure L2888[PSI+], nor did overexpression of Ydj1(D36N), which has a point mutation disrupting its interaction with members of the Hsp70 family.
Figure 6.
The effect of overexpression of Btn2, Cur1, Hsp42, Ydj1, or DnaJB6 in the L2888[PSI+] strain. A, the curing of [PSI+] as a function of generation time was measured in L2888[PSI+]. Yeast was either incubated in 5 mm guanidine or in grown in galactose medium to overexpress Btn2, Cur1, Hsp42, Ydj1, Ydj1(D36N), or DnaJB6. The cells were plated on ½ YPD plates, and curing was measured using the red/white colony assay. The data represent the averages and standard deviation obtained from three independent experiments. B, confocal images of L2888[PSI+] overexpressing Cur1 or Ydj1 or DnaJB6 prior to induction, two generations (gen) in galactose medium, and six generations in galactose medium. The images are maximized projections of Z-stack confocal images obtained on the Zeiss Live confocal microscope. The same settings were used for all images.
The L2888[PSI+] yeast were imaged to determine the changes in the GFP-Sup35 fluorescence during curing. As shown in Fig. 6B, the fluorescent foci became larger and then aggregated with time when either Cur1 or Ydj1 was overexpressed when imaged either at two or six generations in galactose. On the other hand, overexpression of DnaJB6 caused a loss of detectable foci in cells that still plated as [PSI+]. Therefore, it appears that these proteins produced a similar change in the fluorescent foci in both 1660[URE3] and L2888[PSI+] yeast, although the former were cured more readily than the latter. It should be noted that the weak 1758[PSI+] variant, which expresses the unmodified Sup35 protein, was only cured by overexpression of DnaJB6 and not by overexpression of either Cur1 or Ydj1 unlike L2888[PSI+]. This difference is probably due to expression of NGMC in [PSI+] yeast, which makes for a weaker [PSI+] variant, as was observed previously (32). None of these different proteins when overexpressed from the GAL1 promoter cured the strong SY80[PSI+] variant.
Discussion
This study examined the curing of [URE3] in real time by using the 1660[URE3] yeast strain that expresses GFP-labeled Ure2 from the RNQ1 promoter. To cure [URE3], the seeds must be eliminated, which was achieved in this study by three different mechanisms, each of which led to a very different appearance of the GFP-labeled prion protein. These mechanisms are 1) inhibition of severing followed by diluting out the seeds, 2) clumping of the seeds followed by asymmetric segregation between mother and daughter cells, and 3) dissolution of the seeds. Regardless of the method used to cure [URE3], when curing was complete, none of the cells had fluorescent foci. Furthermore, the curing of the 1660[URE3] strain by these different mechanisms was not significantly different from the 1076[URE3] strain, which does not express GFP–Ure2. These results contrast with studies from the Cullin laboratory (34) in which they found that the properties of the [URE3] prion propagated by a Ure2–GFP construct, which consisted of the full-length Ure2 protein labeled with GFP at its C-terminal, were very different from that of the endogenous Ure2 protein. For example, the [URE3] yeast expressing Ure2–GFP were not cured by guanidine. Surprisingly, there were no foci associated with the presence of the [URE3] prion, which lead them to propose that Ure2 exists in a soluble state rather than in an aggregated state (35). They did observe Ure2–GFP aggregates, but these were dead-end aggregates rather than prion seeds (34).
We first examined the curing of [URE3] by guanidine-induced inhibition of Hsp104 severing activity. As expected, inhibition of severing caused a progressive loss of seeds concomitant with an increase in properly folded soluble cytosolic Ure2. Importantly, in the 1660[URE3] strain, there was an excellent correlation between the percentage of cells that visually retained foci and the percentage of cells that were still [URE3] as determined by the plating assay, which strongly suggests that the foci are the prion seeds. The second mechanism of curing that we examined was asymmetric segregation of the seeds, which occurred when Btn2, Cur1, Hsp42, or Ydj1 was overexpressed. These proteins induced the formation of aggregates, which both reduced the number of seeds and impeded their transmission to the daughter cells. Finally, when DnaJB6 was overexpressed, we observed curing by dissolution of the prion seeds; in this case, the seeds appeared to decrease in size until they were all eliminated.
Does our imaging data agree with the mechanisms previously proposed for the curing of [URE3]? No mechanism was previously proposed for the curing of [URE3] by overexpression of Hsp42, whereas the Wickner laboratory (16) proposed that overexpression of Btn2 cures by asymmetric segregation. Our results agree with their proposed mechanism, which was based on their observation that aggregates of overexpressed Btn2 colocalized with the GFP-labeled prion domain of Ure2, which, by itself, tends to aggregate. In addition, our conclusion that the mechanism of curing by overexpression of human DnaJB6 in dissolution of the seeds is consistent with the proposal of the Masison laboratory that DnaJB6 blocks the growth of the prion seeds (9). This proposal was based on in vitro data showing that DnaJB6 arrested the growth of amyloid fibers composed of the prion-forming domains of either Ure2 or Sup35 and also the growth of amyloid fibers composed of either amyloid β fragments or polyglutamine fragments (9, 22–24). Blocking the growth of the prion seeds, but not the severing of the seeds by Hsp104, would cause a gradual reduction in the size of the seeds as they are repeatedly severed, followed by their ultimate dissolution. However, this proposed mechanism is not consistent with our observation that overexpression of Hsp104 did not affect the rate of curing of [URE3] by DnaJB6, assuming there is greater severing of the prion seeds at the higher levels of Hsp104 expression. Evidently, more work is needed to understand the curing of yeast prions by DnaJB6.
In contrast to our imaging data for Btn2 and DnaJB6, which agree with previously proposed mechanisms of curing by these agents (6, 9), our imaging data showing large GFP-labeled Ure2 aggregates in yeast overexpressing either Cur1 or Ydj1 do not fit with proposed models for the curing of [URE3] by these proteins. It has previously been suggested that overexpression of Cur1 and Ydj1 cures by inhibiting severing of the prion seeds because of a reduction in Sis1 levels. Instead, our imaging data of 1660[URE3] show that overexpression of these proteins causes aggregation of the Ure2 foci, which in turn would cause curing by asymmetric segregation. The data are consistent with overexpression of either wildtype Cur1 or the Cur1–NLS mutant reducing cytosolic Sis1 activity, but wildtype Cur1 achieves this by binding and sequestering Sis1 in the nucleus, whereas the mutant Cur1–NLS achieves this by binding and sequestering Sis1 in the cytosol. In contrast to our results, the Zhouravleva laboratory (20) found that [URE3] was not cured by overexpression of a Cur1 fragment (Cur1Δ3–30), which includes deletion of the nuclear localization sequence, residues 25–29 (29). One possibility for the difference in results is that the truncated fragment binds with lower affinity to Sis1 than the Cur1–NLS mutant, but our studies also differ in the [URE3] variant and the genetic background of the yeast strain.
Like Cur1, overexpression of Ydj1 was proposed to cure [URE3] by inhibiting the severing of the prion seeds (19), a proposal based on competition between Ydj1 and Sis1 for Ssa1. Consistent with this proposal, overexpression of Ydj1 does not cure [URE3] if mutated in its J-domain to disrupt its binding to members of the Hsp70 family (30). The competition for Ssa1 between these Hsp40 proteins reduces the amount of Sis1-Ssa1 complex bound to Hsp104, which would reduce the severing activity of Hsp104 (36, 37). However, our imaging data of 1660[URE3] overexpressing Ydj1 suggest that reducing Sis1 activity in the cytosol causes aggregate formation, although inhibition of severing may also be contributing to the curing of [URE3]. These results suggest that in addition to the role of Sis1 in propagating prions, Sis1 may function, perhaps as a cochaperone with Ssa1, to inhibit the aggregation of the prion seeds.
Although overexpression of Hsp42, Btn2, Cur1, and Ydj1 all cured [URE3] by a mechanism involving asymmetric segregation, we found that only the latter two proteins cure [PSI+]. This is the first time that overexpression of Cur1 has been shown to cure [PSI+] yeast, and in fact, a recent report using a different yeast strain found that overexpression of Cur1, rather than curing [PSI+], made it a stronger [PSI+] variant (20). Overexpression of Ydj1 has not previously been shown to cure [PSI+], but it has been shown to cure an artificial [PSI+] strain that expresses a chimeric Sup35 composed of the N-terminal domain from Pichia methanolica and the M and C-terminal domains from Saccharomyces cerevisiae (38). Like the images obtained with the 1660[URE3] yeast strain, imaging of the L2888[PSI+] strain showed fewer, brighter foci in yeast when either Cur1 or Ydj1 was overexpressed, indicative of aggregation of the prion seeds. These proteins cured the weak L2888[PSI+] variant, but not the strong SY80[PSI+] variant. However, apparently there is not a direct relationship between seed number and curing of yeast prions by overexpression of Ydj1 based on the observation that overexpression of Ydj1 cured only some variants of the yeast [RNQ+] prion (39), even though all of the [RNQ+] variants evidently have much fewer seeds than [PSI+] yeast based on the observation that the number of Rnq1 molecules per cell is one-hundredth the number of Sup35 molecules per cell (27). This suggests that the structure of the prion must also affect the curing of prions by overexpression of Ydj1. For example, although we observed no curing of [PSI+] by overexpression of Hsp42, the Shorter laboratory reported that overexpression of Hsp42 cured [PSI+] (40). This difference between our results may be due to the [PSI+] variant or the genetic background of the yeast.
We also observed that overexpression of DnaJB6 cured the L2888[PSI+] strain and caused a change in the appearance of the GFP-labeled Sup35 foci like that observed in the 1660[URE3] yeast. Specifically, there was a loss of detectable foci in the yeast even in cells that were still [PSI+]. In our previous studies, we observed dissolution of the prion seeds in [PSI+] overexpressing Hsp104 (8, 31). Although the GFP-labeled Sup35 seeds had a similar appearance to that caused by overexpression of DnaJB6, the mechanism by which Hsp104 overexpression causes dissolution is very different. Curing caused by Hsp104 overexpression is caused by dissociation of Sup35 monomers from the seeds in an Hsp104-dependent reaction that we termed trimming (15). The trimming that we observed by overexpression of Hsp104 has different properties than curing by overexpression of DnaJB6. Specifically, overexpression of Hsp104 cures both weak and strong [PSI+] variants (33) and is dependent on the cochaperone Sti1 (41, 42).
Even though overexpression of Hsp42, Btn2, Cur1, and Ydj1 all cure [URE3] by asymmetric segregation, studies using deletion mutants have revealed differences in the detailed mechanisms by which these proteins aggregate the prion seeds. For example, Hsp42 is required for [URE3] curing caused by overexpression of Btn2, but not for [URE3] curing caused by overexpression of Cur1 (17). On the other hand, Cur1, but not Btn2, is required for [URE3] curing caused by overexpression of Hsp42 (17), which illustrates that the curing of [URE3] involves complex interactions between these various proteins. Despite these interactions, these curing agents share the common feature that increasing Sis1 levels antagonize their ability to cure [URE3] (17, 19), possibly either by Sis1 binding directly to the seeds or by acting as a cochaperone with Ssa1 to inhibit their aggregation (17).
In conclusion, our results show that overexpression of Hsp42, Btn2, Ydj1, and Cur1 cure [URE3] by asymmetric segregation of the prion seeds. Interestingly, Hsp42, Btn2, Ydj1, and Cur1, expressed at the levels induced by stress conditions, all function in yeast protein quality control; i.e. they mediate the sequestration of misfolded non-amyloid proteins to different cellular deposition sites, thus reducing their toxicity (29, 43–45). Future work using the 1660[URE3] strain should help elucidate the detailed mechanisms by which these four agents aggregate the prion seeds and how these mechanisms relate to their ability to sequester misfolded non-amyloidogenic proteins.
Experimental procedures
Yeast strains
Yeast strain 1075[URE3][RNQ+] (MATα, kar1-1, PDAL5::ADE2, his3Δ202, leu2Δ1, trp1Δ63, ura3-52) (46) was cured of the [RNQ+] prion to make yeast strain 1076[URE3] strain. Strain 1660 has URE2–GFP regulated by the RNQ1 promoter integrated at the RNQ1 genomic locus in place of the RNQ1 coding region. It was made by transforming strain 1075 with a 3.1-kilobase pair KpnI–SalI fragment from plasmid pDCM144 (see below) and selecting HIS+ transformants.
pDCM144 carries a URE2–GFP allele with GFP between URE2 codons 90 and 91 (see below for description of insertion of the GFP). There are an additional 500 bp downstream of the URE2 stop codon followed by Schizosaccharomyces pombe HIS5, which complements S. cerevisiae his3. These sequences are flanked by the RNQ1 promoter (500 base pairs upstream of the initiator codon) and terminator (500 base pairs of 3′ RNQ1 DNA) to target integration at the RNQ1 genomic locus. Plasmid pRNQ1–Ure2–GFP is a single-copy URA3-marked vector with the same URE2–GFP allele regulated by the RNQ1 promoter.
The L2888 [PSI+][RNQ+] and SY80 [PSI+][RNQ+] yeast strains were derived from the 74-D694 (MATa ade1-14UGAtrp1-289 his3-Δ200 ura3-52 leu2-3,112) (32). These strains have GFP-labeled Sup35 (NGMC) inserted into the SUP35 chromosomal locus.
Construction of Ure2–GFP plasmid
The Ure2–GFP was constructed by internally inserted the GFP fluorophore after residue 90. This was done by making three different PCR products and then annealing these products to obtain the GFP inserted at residue 90 of Ure2. The first PCR product started 500 base pairs into the promoter region of Ure2 and terminated at residue 90 of Ure2. The primers used for this reaction were gatcggatccgaaagaaagtcaagacc (forward) and gttcttctcctttacttgcctgttgttgttgtcgat (reverse). The second PCR product for insertion of the GFP between residues 90 and 91 of ure2 used the following primers: caacaacaacaggcaagtaaaggagaagaacttttcactgg (forward) and cgtgactcatatccgaaaatttgtatagttcatccatgccatg (reverse). The third PCR product started at residue 91 of Ure2 and extended 500 base pairs into the non-coding region used the following primers: gaactatacaaattttcggatatgagtcacgtggagtattcc (forward) and gatcggatcctcacatagtgtttgtaaac (reverse). The annealed PCR products were subcloned into pRS314 to make pURE2–Ure2–GFP. pRNQ1–Ure2–GFP was made using pRS316–Rnq1p–RNQ1 (pDCM137). Ure2–GFP was inserted into this plasmid after cutting out the RNQ1 gene. Plasmids were transformed into yeast as described previously (45). The centromeric Hsp42–mCherry plasmid used in this study was subcloned from the pAG415GPD–Hsp42–mCherry plasmid (O-2252) from the Alberti laboratory (29). The centromeric Cur1–RFP plasmid used in this study was subcloned from pBEE2 plasmid. All constructs were sequenced (Macrogen). A list of the plasmids used in this study is given in Table 1.
Table 1.
Plasmids
| Plasmids | Promoter | Copy number | Gene | Marker | Source |
|---|---|---|---|---|---|
| p1508 | GAL1 | Centromeric | Hsp42 | Leu2 | Ref. 17 |
| pHsp42–Cherry | GAL1 | Centromeric | Hsp42–Cherry | Leu2 | This study |
| pRU20 | GAL1 | Centromeric | Ydj1 | Leu2 | Ref. 9 |
| pRU14 | GAL1 | Centromeric | DnaJB6 | Leu2 | Ref. 9 |
| pBEE2 | GAL1 | Centromeric | Cur1 | Leu2 | This study |
| pCur1–RFP | GAL1 | Centromeric | Cur1–RFP | Trp1 | This study |
| pBEE1 | GAL1 | Centromeric | Btn2 | Leu2 | This study |
| pBtn2–RFP | GAL1 | High copy | Btn2–RFP | Ura3 | Ref. 6 |
| pRU21 | GAL1 | Centromeric | Ydj1 (D36N) | Leu2 | This study |
| pCur1–NLS–RFP | GAL1 | Centromeric | Cur1–NLS–RFP | Trp1 | This study |
| pSis1–GFP | ADH | High copy | Sis–GFP | Leu3 | Ref. 29 |
| pSis1 | GAL1 | Centromeric | Sis1 | Ura3 | Ref. 19 |
| pFL39–Hsp104 | GAL1 | Centromeric | Hsp104 | Trp1 | Ref. 3 |
Curing of yeast prions by overexpression of different proteins
Yeast grown in synthetic defined medium were washed with PBS and then grown overnight in raffinose medium. Galactose medium was then added, and the yeast growth was monitored by the absorbance at 600 nm. At the indicated times, yeast was plated on ½ YPD plates to measure the extent of curing. The presence of [URE3] was monitored y by use of an ADE2 allele regulated by the DAL5 promoter (PDAL5::ADE2), as described previously (19). When plating on limiting adenine (1/2 YPD plates), [URE3] and [ure-o] cells gave rise to white and red colonies, respectively. Curing of [URE3] was determined by the fraction of entirely red ([ure-o]) colonies after 3 days of incubation at 30 °C. The curing of [PSI+] was also monitored by red/white colony assay since the yeast strains have the ade1-14 nonsense mutation. Colonies that had any white were scored as [PSI+], whereas completely red colonies were scored as [psi−]. For each time point, we counted a minimum of 200 colonies. All curing curves were done a minimum of three times, and the data are plotted as the averages and standard deviation.
Microscopy
Imaging of the fluorescence was always done by Z-stack confocal imaging of live cells. As noted in the text, the different microscopes used to image were the Zeiss LSM 880 Airyscan confocal microscope equipped with a 63× 1.4 NA objective, the Nikon A1R confocal microscope equipped with a 60× 1.4 NA objective, and the Zeiss Live confocal microscope equipped with a 100× 1.4 NA objective. Yeast were imaged in 8-well, 25-mm, two-chambered coverslips (Lab-Tek, Rochester, NY).
Author contributions
X. Z., J. L., D. S., T. P., J. M. A., and E. D. S. performed the experiments and analyzed the data. E. E. B. constructed plasmids. E. E. provided critical advice regarding the design of the project and the revision of the manuscript. D. C. M. created yeast strains and contributed critical analysis to the project and the manuscript. L. E. G. supervised the project and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Reed Wickner (NIDDK, National Institutes of Health) and Dr. Simon Alberti (Max Planck Institute, Dresden, Germany) for plasmids and Dr. Susan Liebman (University of Nevada, Reno, Nevada) for the L2888 and SY80 yeast strains. We also thank Dr. Xufeng Wu for assistance and the NHLBI microscopy core and Antonio Barakat for data analysis and assisting with experiments.
Footnotes
This work was supported by National Institutes of Health Grant HL000516. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Liebman S. W., and Chernoff Y. O. (2012) Prions in yeast. Genetics 191, 1041–1072 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jung G., and Masison D. C. (2001) Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43, 7–10 10.1007/s002840010251 [DOI] [PubMed] [Google Scholar]
- 3. Wegrzyn R. D., Bapat K., Newnam G. P., Zink A. D., and Chernoff Y. O. (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell Biol. 21, 4656–4669 10.1128/MCB.21.14.4656-4669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eaglestone S. S., Ruddock L. W., Cox B. S., and Tuite M. F. (2000) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 240–244 10.1073/pnas.97.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newnam G. P., Birchmore J. L., and Chernoff Y. O. (2011) Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 408, 432–448 10.1016/j.jmb.2011.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kryndushkin D. S., Shewmaker F., and Wickner R. B. (2008) Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 27, 2725–2735 10.1038/emboj.2008.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ness F., Cox B. S., Wongwigkarn J., Naeimi W. R., and Tuite M. F. (2017) Over-expression of the molecular chaperone Hsp104 in Saccharomyces cerevisiae results in the malpartition of [PSI+] propagons. Mol. Microbiol. 104, 125–143 10.1111/mmi.13617 [DOI] [PubMed] [Google Scholar]
- 8. Park Y. N., Zhao X., Yim Y. I., Todor H., Ellerbrock R., Reidy M., Eisenberg E., Masison D. C., and Greene L. E. (2014) Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Eukaryot. Cell 13, 635–647 10.1128/EC.00300-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reidy M., Sharma R., Roberts B. L., and Masison D. C. (2016) Human J-protein DnaJB6b cures a subset of S. cerevisiae prions and selectively blocks assembly of structurally related amyloids. J. Biol. Chem. 291, 4035–4047 10.1074/jbc.M115.700393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patino M. M., Liu J. J., Glover J. R., and Lindquist S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622–626 10.1126/science.273.5275.622 [DOI] [PubMed] [Google Scholar]
- 11. Song Y., Wu Y. X., Jung G., Tutar Y., Eisenberg E., Greene L. E., and Masison D. C. (2005) Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4, 289–297 10.1128/EC.4.2.289-297.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Satpute-Krishnan P., and Serio T. R. (2005) Prion protein remodelling confers an immediate phenotypic switch. Nature 437, 262–265 10.1038/nature03981 [DOI] [PubMed] [Google Scholar]
- 13. Satpute-Krishnan P., Langseth S. X., and Serio T. R. (2007) Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLos Biol. 5, e24 10.1371/journal.pbio.0050024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derdowski A., Sindi S. S., Klaips C. L., DiSalvo S., and Serio T. R. (2010) A size threshold limits prion transmission and establishes phenotypic diversity. Science 330, 680–683 10.1126/science.1197785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park Y. N., Morales D., Rubinson E. H., Masison D., Eisenberg E., and Greene L. E. (2012) Differences in the curing of [PSI+] prion by various methods of Hsp104 inactivation. PLoS One 7, e37692 10.1371/journal.pone.0037692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edskes H. K., Gray V. T., and Wickner R. B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. U.S.A. 96, 1498–1503 10.1073/pnas.96.4.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wickner R. B., Bezsonov E., and Bateman D. A. (2014) Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. U.S.A. 111, E2711–E2720 10.1073/pnas.1409582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moriyama H., Edskes H. K., and Wickner R. B. (2000) [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell Biol. 20, 8916–8922 10.1128/MCB.20.23.8916-8922.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reidy M., Sharma R., Shastry S., Roberts B. L., Albino-Flores I., Wickner S., and Masison D. C. (2014) Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet. 10, e1004720 10.1371/journal.pgen.1004720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barbitoff Y. A., Matveenko A. G., Moskalenko S. E., Zemlyanko O. M., Newnam G. P., Patel A., Chernova T. A., Chernoff Y. O., and Zhouravleva G. A. (2017) To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 105, 242–257 10.1111/mmi.13697 [DOI] [PubMed] [Google Scholar]
- 21. Lian H. Y., Zhang H., Zhang Z. R., Loovers H. M., Jones G. W., Rowling P. J., Itzhaki L. S., Zhou J. M., and Perrett S. (2007) Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J. Biol. Chem. 282, 11931–11940 10.1074/jbc.M606856200 [DOI] [PubMed] [Google Scholar]
- 22. Månsson C., Kakkar V., Monsellier E., Sourigues Y., Härmark J., Kampinga H. H., Melki R., and Emanuelsson C. (2014) DNAJB6 is a peptide-binding chaperone which can suppress amyloid fibrillation of polyglutamine peptides at substoichiometric molar ratios. Cell Stress Chaperones 19, 227–239 10.1007/s12192-013-0448-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Månsson C., Arosio P., Hussein R., Kampinga H. H., Hashem R. M., Boelens W. C., Dobson C. M., Knowles T. P., Linse S., and Emanuelsson C. (2014) Interaction of the molecular chaperone DNAJB6 with growing amyloid-β42 (Aβ42) aggregates leads to sub-stoichiometric inhibition of amyloid formation. J. Biol. Chem. 289, 31066–31076 10.1074/jbc.M114.595124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillis J., Schipper-Krom S., Juenemann K., Gruber A., Coolen S., van den Nieuwendijk R., van Veen H., Overkleeft H., Goedhart J., Kampinga H. H., and Reits E. A. (2013) The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J. Biol. Chem. 288, 17225–17237 10.1074/jbc.M112.421685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brachmann A., Baxa U., and Wickner R. B. (2005) Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 10.1038/sj.emboj.7600772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kryndushkin D. S., Wickner R. B., and Tycko R. (2011) The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-β structure: evidence from solid-state NMR. J. Mol. Biol. 409, 263–277 10.1016/j.jmb.2011.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghaemmaghami S., Huh W.-K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 28. Wickner R. B., Kryndushkin D., Shewmaker F., McGlinchey R., and Edskes H. K. (2012) Study of amyloids using yeast. Methods Mol. Biol. 849, 321–346 10.1007/978-1-61779-551-0_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malinovska L., Kroschwald S., Munder M. C., Richter D., and Alberti S. (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 23, 3041–3056 10.1091/mbc.E12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma D., Stanley R. F., and Masison D. C. (2009) Curing of yeast [URE3] prion by the Hsp40 cochaperone Ydj1p is mediated by Hsp70. Genetics 181, 129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao X., Rodriguez R., Silberman R. E., Ahearn J. M., Saidha S., Cummins K. C., Eisenberg E., and Greene L. E. (2017) Heat shock protein 104 (Hsp104)-mediated curing of [PSI+] yeast prions depends on both [PSI+] conformation and the properties of the Hsp104 homologs. J. Biol. Chem. 292, 8630–8641 10.1074/jbc.M116.770719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathur V., Hong J. Y., and Liebman S. W. (2009) Ssa1 overexpression and [PIN+] variants cure [PSI+] by dilution of aggregates. J. Mol. Biol. 390, 155–167 10.1016/j.jmb.2009.04.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., and Liebman S. W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ripaud L., Maillet L., and Cullin C. (2003) The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. EMBO J. 22, 5251–5259 10.1093/emboj/cdg488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fernandez-Bellot E., Guillemet E., Ness F., Baudin-Baillieu A., Ripaud L., Tuite M., and Cullin C. (2002) The [URE3] phenotype: evidence for a soluble prion in yeast. EMBO Rep. 3, 76–81 10.1093/embo-reports/kvf011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Higurashi T., Hines J. K., Sahi C., Aron R., and Craig E. A. (2008) Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. U.S.A. 105, 16596–16601 10.1073/pnas.0808934105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aron R., Higurashi T., Sahi C., and Craig E. A. (2007) J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 26, 3794–3803 10.1038/sj.emboj.7601811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kushnirov V. V., Kryndushkin D. S., Boguta M., Smirnov V. N., and Ter-Avanesyan M. D. (2000) Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10, 1443–1446 10.1016/S0960-9822(00)00802-2 [DOI] [PubMed] [Google Scholar]
- 39. Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., and Liebman S. W. (2002) Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. U.S.A. 99, 16392–16399 10.1073/pnas.152330699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duennwald M. L., Echeverria A., and Shorter J. (2012) Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 10, e1001346 10.1371/journal.pbio.1001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reidy M., and Masison D. C. (2010) Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell Biol. 30, 3542–3552 10.1128/MCB.01292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moosavi B., Wongwigkarn J., and Tuite M. F. (2010) Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 27, 167–179 [DOI] [PubMed] [Google Scholar]
- 43. Specht S., Miller S. B., Mogk A., and Bukau B. (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 195, 617–629 10.1083/jcb.201106037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller S. B., Ho C. T., Winkler J., Khokhrina M., Neuner A., Mohamed M. Y., Guilbride D. L., Richter K., Lisby M., Schiebel E., Mogk A., and Bukau B. (2015) Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34, 778–797 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Escusa-Toret S., Vonk W. I., and Frydman J. (2013) Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15, 1231–1243 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma D., and Masison D. C. (2008) Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics 179, 1301–1311 10.1534/genetics.108.089458 [DOI] [PMC free article] [PubMed] [Google Scholar]