Abstract
The gut microbiome contributes to inflammatory bowel disease (IBD), in which bacteria can be present within the epithelium. Epithelial barrier function is decreased in IBD, and dysfunctional epithelial mitochondria and endoplasmic reticulum (ER) stress have been individually associated with IBD. We therefore hypothesized that the combination of ER and mitochondrial stresses significantly disrupt epithelial barrier function. Here, we treated human colonic biopsies, epithelial colonoids, and epithelial cells with an uncoupler of oxidative phosphorylation, dinitrophenol (DNP), with or without the ER stressor tunicamycin and assessed epithelial barrier function by monitoring internalization and translocation of commensal bacteria. We also examined barrier function and colitis in mice exposed to dextran sodium sulfate (DSS) or DNP and co-treated with DAPK6, an inhibitor of death-associated protein kinase 1 (DAPK1). Contrary to our hypothesis, induction of ER stress (i.e. the unfolded protein response) protected against decreased barrier function caused by the disruption of mitochondrial function. ER stress did not prevent DNP-driven uptake of bacteria; rather, specific mobilization of the ATF6 arm of ER stress and recruitment of DAPK1 resulted in enhanced autophagic killing (xenophagy) of bacteria. Of note, epithelia with a Crohn's disease–susceptibility mutation in the autophagy gene ATG16L1 exhibited less xenophagy. Systemic delivery of the DAPK1 inhibitor DAPK6 increased bacterial translocation in DSS- or DNP-treated mice. We conclude that promoting ER stress–ATF6–DAPK1 signaling in transporting enterocytes counters the transcellular passage of bacteria evoked by dysfunctional mitochondria, thereby reducing the potential for metabolic stress to reactivate or perpetuate inflammation.
Keywords: mitochondria, autophagy, endoplasmic reticulum stress (ER stress), bacteria, epithelial cell, DAPK1, intestinal barrier function, mitochondrial stress, bacterial translocation, IBD basic research, oxidative metabolism, inflammatory bowel disease, epithelial barrier;
Introduction
Bacteria within the epithelium are a pro-inflammatory threat. While observed in inflammatory bowel disease (IBD)3 (1–4), and putatively important in disease pathophysiology, the mechanism(s) underlying this barrier defect is unclear. Mutation in the organic cation transporter (OCTN2) gene is a susceptibility trait for IBD (5); this transporter facilitates carnitine uptake for fatty acid β-oxidation and ATP generation. Moreover, deranged mitochondrial ultrastructure, loss of mitochondrial proteins, lower levels of ATP, and increased susceptibility to uncouplers of oxidative phosphorylation occur in some patients with IBD (6). Similarly, suspected triggers of IBD reactivation can perturb mitochondrial function. For instance, non-steroidal anti-inflammatory drugs (NSAID) can cause relapses in IBD (7), and their use damages mitochondria contributing to decreased enteric barrier function (8, 9). Consequently, mitochondria are emerging as an organelle of interest in IBD and other enteropathies (10).
While idiopathic, the consensus is that uncontrolled immune reactivity against a component(s) of the gut microbiota in a genetically susceptible individual causes IBD (11). A defect in epithelial barrier function is implied in this hypothesis. With the exceptions of uptake by microfold cells or dendritic cell pseudopodia, material in the gut lumen enters the submucosa via the epithelial tight junctions (paracellular) or by crossing the enterocyte (transcellular) (12), and regulation of both pathways is energy-dependent. With that, it can be proposed that perturbed epithelial mitochondrial function would increase gut permeability to initiate disease or to precipitate relapses in IBD. Accordingly, mitochondrial damage induced by uncoupling oxidative phosphorylation with the H+ ionophore, dinitrophenol (DNP), to model the mitochondrial dysfunction observed in IBD resulted in increased internalization into and transcytosis of Escherichia coli across epithelial monolayers (13–15).
Mitochondria couple with the endoplasmic reticulum (ER), where reactive oxygen species (ROS) are important in protein folding (16). When improperly folded proteins accumulate in the ER, an ER stress program (or unfolded protein response (UPR)) occurs. Mice deficient in any of the three major arms of the ER stress response (inositol-requiring kinase/endonuclease 1 (IRE1), pancreatic ER kinase (PERK), and activating transcription factor 6 (ATF6)) are more susceptible to DSS-induced colitis (17, 18). Furthermore, mutation in the X-box–binding protein-1 (XBP1) gene (downstream of IRE1) is a susceptibility trait for Crohn's disease (17), and increased ER stress can occur in the epithelium of patients with IBD (17, 19, 20). We hypothesized that uncoupling oxidative phosphorylation (i.e. mitochondrial dysfunction) combined with ER stress would severely compromise epithelial integrity.
Results
ER stress protects epithelium transcellular permeability perturbed by mitochondrial stress
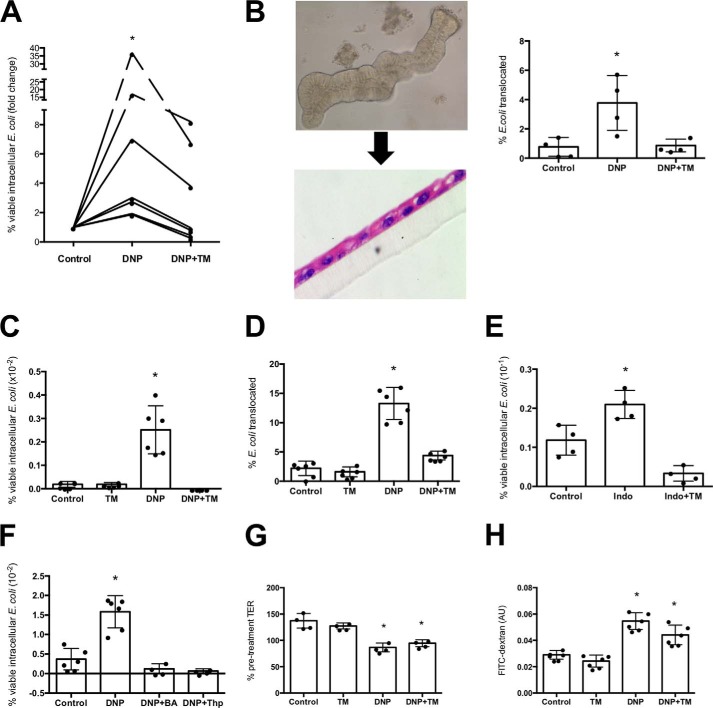
Human colonic biopsies were mounted in Ussing chambers; bacteria were added to the luminal buffer, and biopsies were treated with DNP ± the ER stressor tunicamycin (blocks protein glycosylation (21), which is often used to elicit ER stress in cell lines (19)). DNP promoted E. coli uptake; however, tunicamycin reduced this barrier defect (Fig. 1A). DNP treatment of colonic enteroid monolayers resulted in increased transcytosis of E. coli that was reduced by tunicamycin (Fig. 1B). Similarly, DNP-evoked increases in bacterial internalization into and translocation across the T84 (Fig. 1, C and D), Caco-2 (Fig. S1A), and HCT116 (Fig. S1B) human epithelial cell lines was significantly reduced by tunicamycin co-treatment. (Tunicamycin induction of ER stress was confirmed by GRP78 expression (Fig. S1C). Tunicamycin did not increase intracellular E. coli (Fig. S1D).) The NSAID indomethacin increased intracellularly viable E. coli in T84 cells (9), and this was reduced by tunicamycin (Fig. 1E). (HeLa cells provided similar data (Fig. S2).) Treatment with other ER stressors, brefeldin A (blocks Golgi transit) or thapsigargin (Ca2+-ATPase blocker that perturbs ER Ca2+ stores) (22), also reduced the number of viable intracellular bacteria after exposure to DNP (Fig. 1F). Epithelia treated with DNP display decreased TER (Fig. 1G) and increased FITC-dextran flux (Fig. 1H), and tunicamycin did not affect the increased paracellular permeability (Fig. 1, G and H). Respiratory rate (basal respiration, maximal respiration, and respiratory capacity) and ATP synthesis were reduced by DNP, and these bioenergetic parameters were not restored in tunicamycin co-treated cells (Fig. S3, A–C) (glycolysis was unaffected by DNP ± tunicamycin (Fig. S3D)).
Figure 1.
ER stress protects against epithelial barrier dysfunction evoked by metabolic stress. A, human colonic biopsies were mounted in Ussing chambers, live green fluorescent protein-labeled E. coli HS (108 cfu/ml) ± the metabolic stressor DNP (0.1 mm) or the ER stressor tunicamycin (TM, 10 μg/ml) added to the luminal side, and translocation into the serosal buffer assessed 3 h later (n = 7; *, p < 0.05 compared with control). B, mouse colonic enteroids were grown for 7 days, resuspended, and grown as monolayer, and then E. coli (strain HB101, 107 cfu) was added to the luminal surface ± DNP (0.1 mm) ± TM (10 μg/ml), and bacterial translocation into the basal compartment of the culture well was determined 16 h later (insets, phase contrast images of enteroid and subsequent filter-grown monolayer (10-μm section stained with hematoxylin and eosin)). C, monolayers of the human colon-derived T84 cells were grown to confluence (i.e. starting TER = 1000–1800 ohms·cm2) or to ∼70% confluence (by phase microscopy) on 12-well plates. Epithelia were treated with E. coli (strain HB101, 106 cfu) and DNP ± TM. Bacterial translocation from the apical surface of the epithelium into the basal compartment of the culture well (D) and bacterial internalization (as % of extracellular E. coli) were assessed 16 h later (one representative experiment of six experiments). E, increased epithelial (T84 cells) internalization of E. coli induced by the NSAID, indomethacin (indo; 1 μm; 16 h treatment), was reduced by TM co-treatment (n = 4 epithelial monolayers). F, shows the effect of two other ER stressors brefeldin-A (BA, 1 μm) and thapsigargin (Thp, 2 μm) on DNP-induced (16 h) T84 cell internalization of E. coli (n = 4–6). G and H show markers of paracellular permeability, TER, at 16 h post-treatment (normalized and presented as % pre-treatment value), and flux of 4-kDa FITC-dextran (100 μg/ml) over a 4-h period (after 16-h treatments) (1 representative experiment of two experiments) (mean ± S.D.; *, p < 0.05 compared with control).
Analysis of the 16 h post-DNP + E. coli ± tunicamycin exposure revealed no significant increase in T84 cell apoptosis; however, there was an ∼50% decrease in viability by 36 h post-DNP + E. coli treatment that was significantly reduced by tunicamycin (Fig. S4).
Tunicamycin promotes killing of internalized bacteria
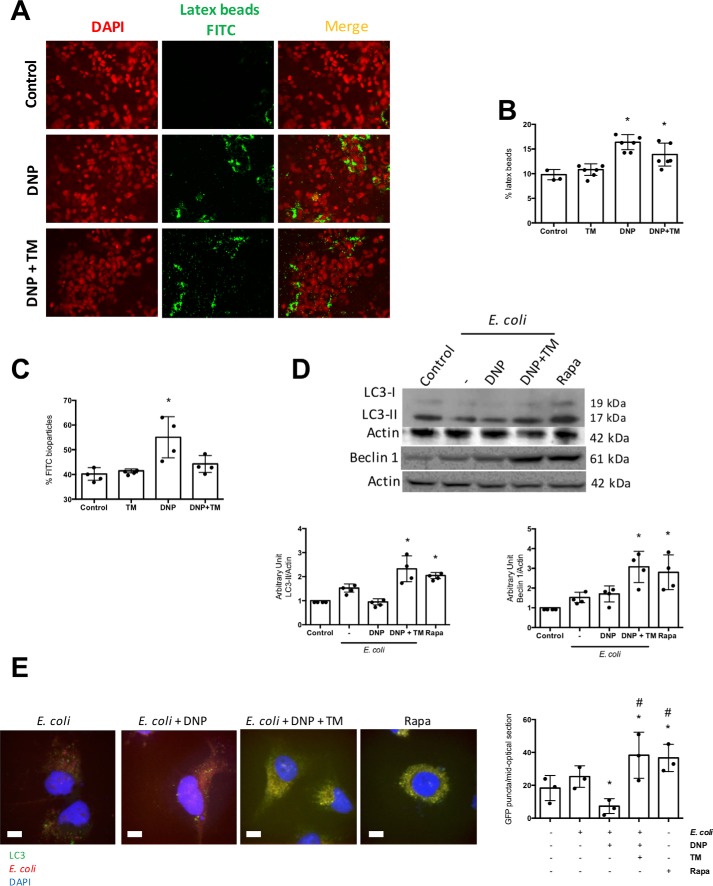
Reduced numbers of intracellular bacteria could reflect a reduction in extracellular bacteria; analysis at the end of each experiment revealed no significant differences in the extracellular bacteria numbers between the groups. The reduced number of viable intracellular E. coli in DNP + tunicamycin co-treated epithelia could be due to uptake inhibition. Immunolocalization (Fig. 2A) and flow cytometry (Fig. 2B) revealed tunicamycin did not affect the DNP-induced internalization of inert fluorescent beads, suggesting that inhibition of the rate of E. coli internalization is not the cause of the reduced viable bacteria in DNP + tunicamycin-treated T84 epithelia. However, the DNP-induced increase in inert E. coli (K-12) fluorescent bioparticles was abrogated by co-treatment with tunicamycin (Fig. 2C), suggestive of degradation and loss of fluorescence.
Figure 2.
Tunicamycin promotes killing of internalized bacteria and not suppression of uptake. T84 epithelial cells were incubated with fluorescent latex beads (108 1-μm diameter), DNP (0.1 mm), or DNP + TM (10 μg/ml) for 16 h, and monolayers were assessed by fluorescence microscopy (representative images, ×40 magnification) (A) and (B) flow cytometry (B). C, in parallel cultures, flow cytometry was used to assess intracellular levels of fluorescent dead E. coli F12 bioparticles (106/ml). D, T84 cells were treated with E. coli (HB101, 106 cfu) ± DNP or DNP + TM for 16 h, and autophagy was assessed by expression of LC3-II and beclin-1 on immunoblots that were quantified by densitometry and normalized to actin expression. Rapamycin (Rapa; 100 nm, 16 h) was a positive control for the induction of autophagy. E, LC3-GFP HeLa cells were treated as shown for 16 h and GFP+ puncta counted in randomly selected cells based on DAPI nuclear staining (representative images; scale bar, 65 μm). Co-localization of bacteria in phagosomes was obvious in the E. coli + DNP + TM-treated cells (mean ± S.D.; * and #, p < 0.05 compared with control and E. coli+ DNP-treated cells, respectively).
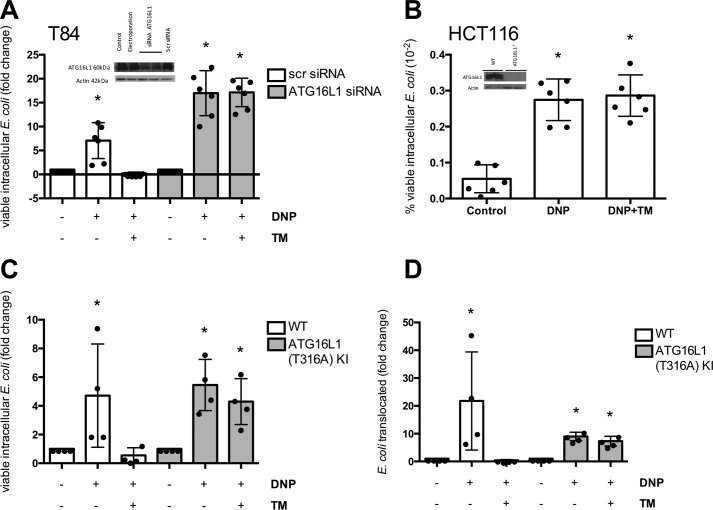
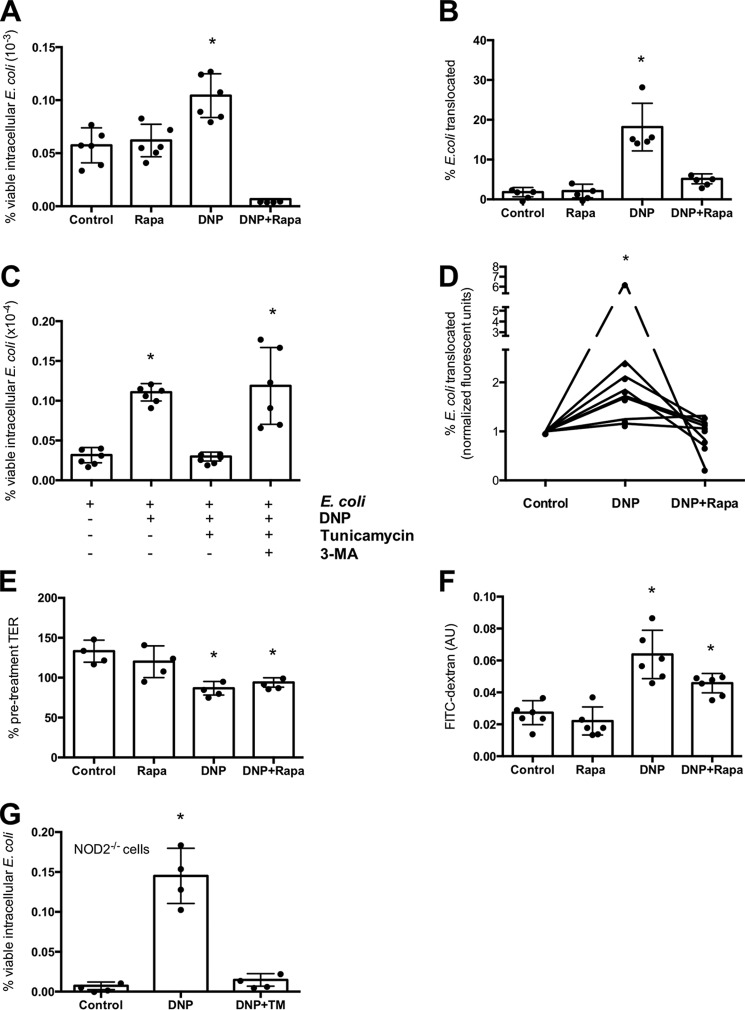
E. coli + DNP + tunicamycin-treated epithelia displayed enhanced autophagy as gauged by increased expression of LC3-II and beclin-1 and LC3 immunostaining (Fig. 2, D and E). Postulating that autophagy would target the internalized bacteria (i.e. xenophagy), co-localization of autophagosomes and internalized bacteria was observed in DNP + tunicamycin-treated cells (Fig. 2E). Furthermore, tunicamycin antagonism of DNP-induced E. coli internalization was lost in cells lacking the autophagy protein ATG16L1 (Fig. 3, A and B). Also, tunicamycin inhibition of the DNP-induced increased viable intracellular E. coli was not apparent in colonoids from mice expressing the knocked-in variant of Atg16l1T316A (homologous to the IBD susceptibility trait ATG16L1T300A) (Fig. 3, C and D). These findings suggest that ER stress ablated the DNP effect by mobilizing autophagy to kill the bacteria. In accordance, rapamycin (induces autophagy (23)) significantly reduced the number of viable intracellular bacteria and translocation across T84 cell monolayers (Fig. 4, A and B). Autophagy inhibition with 3-MA increased numbers of viable bacteria in tunicamycin-treated wildtype T84 cells (Fig. 4C). Moreover, rapamycin prevented the DNP-induced increase in live E. coli HS transcytosis across human colonic biopsies (Fig. 4D). Like tunicamycin, rapamycin did not affect the DNP-induced drop in TER or FITC-dextran flux across T84 epithelia (Fig. 4, E and F). Although the ability of tunicamycin to induce xenophagy was lost in ATG16L1 transgenic cells, tunicamycin reduced the number of viable bacteria in DNP-treated NOD2−/− T84 cells (Fig. 4G).
Figure 3.
Role of autophagy in ER stress protection of metabolic stress–induced barrier defect. The autophagy ATG16L1 gene was knocked down by siRNA (T84 cells) (A) or knocked out with CRISPR-Cas9 (HCT116 cells) (B). The cells were exposed to E. coli (106 cfu, strain HB101) and DNP (0.1 mm) ± TM (10 μg/ml) for 16 h, and the number of viable intracellular bacteria was assessed (one representative experiment of two experiments). Internalization (C) and translocation (D) of E. coli HB101 (107, cfu) across colonoids from mice with the knockin (KI) of ATG16L1T316A grown as monolayers on filter supports and treated with DNP ± TM for 16 h (mean ± S.D.; *, p < 0.05 compared with control).
Figure 4.
Induction of autophagy prevents DNP-induced barrier defect. The inducer of autophagy, rapamycin (Rapa, 100 nm, 16 h), blocked the effect of DNP on T84 epithelial cell internalization (A) and translocation (B) of bacteria across filter-grown monolayers. C, T84 cell treated with E. coli (strain HB101; 106 cfu) and DNP (0.1 mm) ± TM (10 μg/ml) ± the autophagy inhibitor 3-MA (5 μm) and bacterial internalization assessed 16 h later. D, increased mucosal-to-serosal flux of live green fluorescent protein-labeled E. coli HS across Ussing-chambered human colonic biopsies (DNP ± Rapa for 3 h). Rapamycin did not affect the DNP-induced reduction in TER (E) or the concomitant increased flux of 4-kDa FITC-dextran across T84 cell monolayers (F) (one representative experiment of two experiment s). G, T84 cells in which NOD2 was knocked out using shRNA also display increased DNP-induced internalization of E. coli that is inhibited by TM co-treatment (mean ± S.D.; *, p < 0.05 compared with control).
These data are in accordance with the supposition by Adolph et al. (24) that the ER stress observed in patients with IBD could compensate for defective/reduced autophagy, and, we add, a reaction to increased transcellular transport of commensal bacteria.
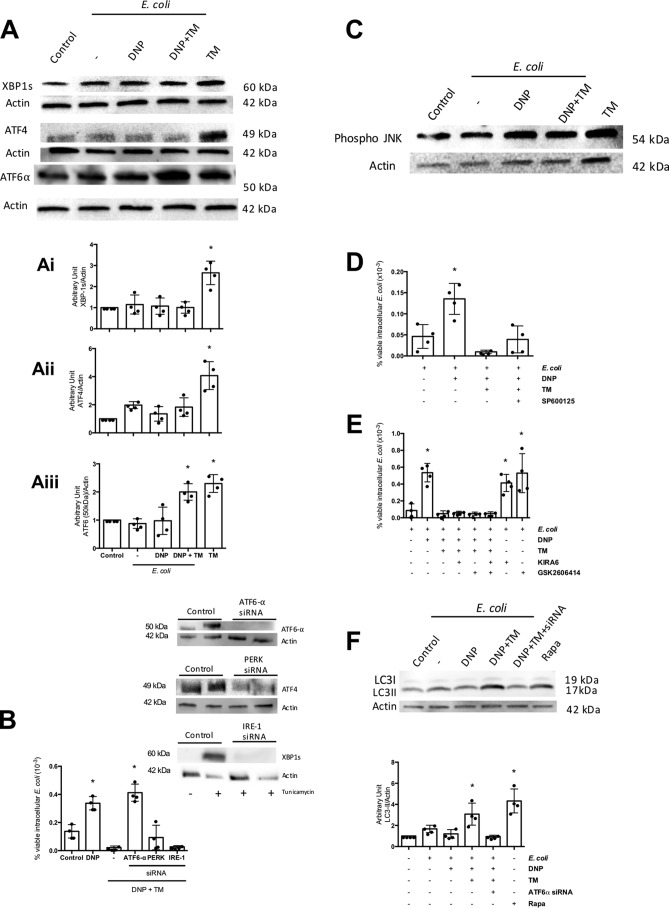
ATF-6 mediates inhibition of the DNP-effect
ER stress triggers three major pathways: IRE1 that activates the mitogen-activated protein kinase, JNK, and splicing of XBP1 transcription factor; PERK that leads to eIF2α phosphorylation, which acts as a transcription factor, and ATF4 mobilization, another transcription factor; and ATF6 that when truncated serves as a transcription factor (25). Tunicamycin resulted in subtle but significant increase in spliced XBP1, ATF4, and ATF6α in T84 cells (Fig. 5A). Transient siRNA knockdown of ATF6α, PERK, or IRE1 in T84 cells revealed that only the former blocked tunicamycin inhibition of DNP-evoked increases in viable intracellular E. coli (Fig. 5B). Selective pharmacological inhibitors of JNK (p-JNK increased in the cells) (Fig. 5C), IRE1, and PERK did not affect tunicamycin abrogation of the DNP effect (Fig. 5, D and E), focusing attention on ATF6. The increase in autophagy (i.e. LC3-II protein expression) evoked by tunicamycin was absent in ATF6α siRNA-treated T84 cells (Fig. 5F).
Figure 5.
ATF-6 mediates inhibition of the DNP effect. A, T84 cells treated with E. coli + DNP + tunicamycin show increased expression of ATF6 but not XBP1 or ATF4 (representative immunoblot; 16-h treatments; densitometry). B, treatment with ATF6α, but not ATF4 or XBP1, siRNA (inset, immunoblots showing siRNA knockdown of protein) prevents the ability of TM (10 μg/ml) to ablate the DNP (0.1 mm)-induced internalization of viable E. coli (106 cfu, strain HB101; 16 h) (one representative experiment of two experiments). C, immunoblotting of whole-cell extracts (representative of three blots) reveals increased phosphorylated JNK in TM + DNP-, but not E. coli only, treated T84 epithelial cells. D, co-treatment with the selective pharmacological inhibitor of JNK, SP600125 (40 nm), does not affect the TM suppression of DNP-evoked increases in viable intracellular E. coli in T84 epithelial cells (16 h). E, co-treatment with the selective IRE1 inhibitor (KIRA6, 600 nm) or PERK inhibitor (GSK2606414, 0.4 nm) did not affect DNP (0.1 mm, 16 h)-induced internalization of E. coli into T84 cells. F, increased autophagy as gauged by LC3-II protein expression observed in E. coli + DNP + TM-treated cells was not observed in T84 epithelia treated with siRNA against ATF6α. The inducer of autophagy, rapamycin (Rapa, 100 nm) is shown as a positive control (mean ± S.D.; *, p < 0.05 compared with control).
DAP kinase-1 is needed for tunicamycin inhibition of DNP-evoked decreased barrier function
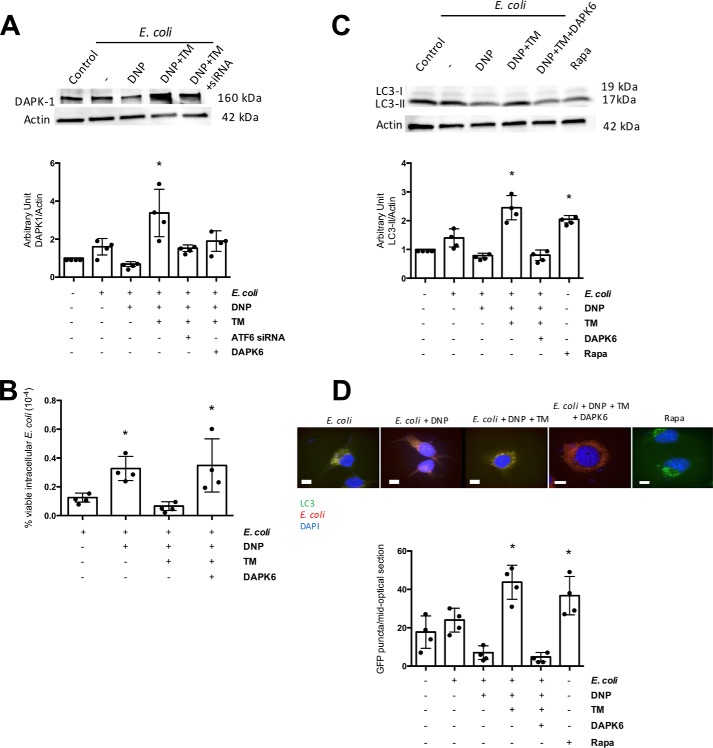
DAPK1 is mobilized downstream of ATF6 to regulate autophagy and apoptosis (26). T84 epithelia exposed to E. coli + DNP + tunicamycin displayed increased DAPK1 expression compared with cells treated with ATF6 siRNA (Fig. 6A) (PERK and IRE1 siRNA did not inhibit DAPK1 protein expression (Fig. S5)). Use of a selective DAPK1 inhibitor, designated DAPK6, blocked tunicamycin's ability to reduce DNP-evoked increases in viable intracellular E. coli by T84 cells (Fig. 6B) and the mobilization of autophagy (Fig. 6, C and D). Moreover, bacteria–autophagosome co-localization was completely lost in DAPK6 co-treated cells (Fig. 6D).
Figure 6.
DAP kinase-1 is needed for tunicamycin inhibition of DNP-evoked decreased barrier function. A, immunoblotting of whole-cell extracts of wildtype or ATF6 siRNA (2 nm)-treated T84 epithelial cells exposed to E. coli + DNP (0.1 mm) and TM (10 μg/ml) for 4 h shows an ATF6-dependent increase in DAPK1 expression. B, TM antagonism of DNP-induced increases in viable intracellular E. coli (106 cfu, 16 h) by T84 cells was not observed in cells co-treated with the selective inhibitor of DAPK1, DAPK6 (10 μm). C, DAPK6 suppressed autophagy evoked as a consequence of E. coli-DNP + TM treatment as assessed by immunoblotting for LC3-II in T84 cells; D, enumeration of LC3-GFP+ puncta formation in HeLa cells (mean ± S.D.; * and #, p < 0.05 compared with control and DNP, respectively).
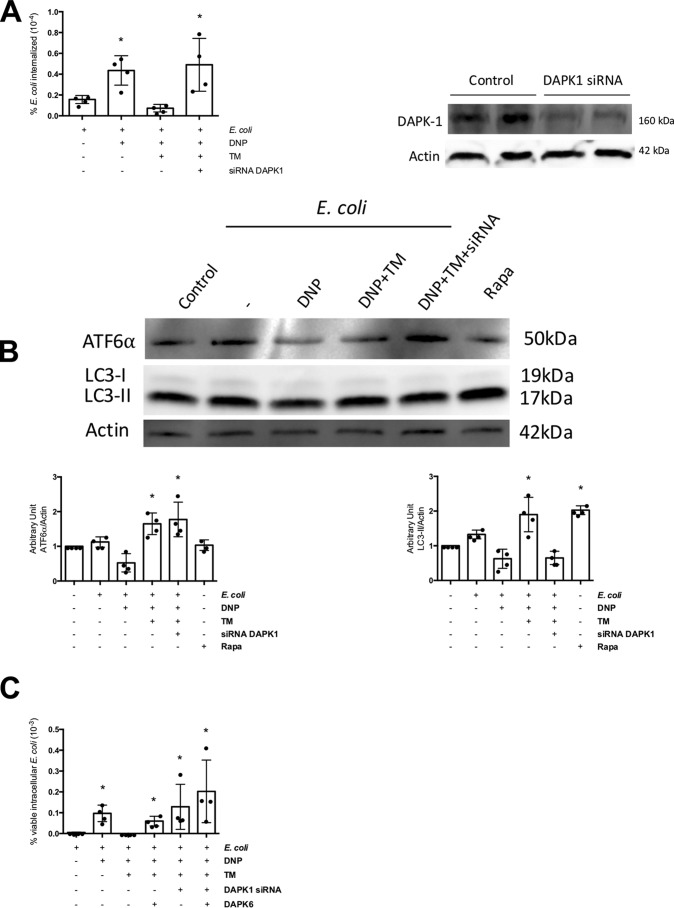
Likewise, knockdown of DAPK1 in T84 cells resulted in inhibition of the tunicamycin effect and increased viable intracellular E. coli numbers (Fig. 7A). Although the activation of ATF6 was not affected by DAPK1 siRNA in DNP + E. coli + tunicamycin-treated epithelia, the increased autophagy was lost, showing that the signal needed to up-regulate autophagy by ER stress was interrupted in the absence of DAPK1 (Fig. 7B). Co-treatment with DAPK1 siRNA and DAPK6 prevented the ability of tunicamycin to block the effects of DNP, but to a degree that was not statistically different from either DAPK6 or DAPK1 siRNA alone (Fig. 7C). These data also support the specificity of DAPK6 in targeting DAPK1, an important observation in the context of the murine in vivo experiments presented below.
Figure 7.
Knockdown of DAPK1 blocks tunicamycin inhibition of DNP-evoked decreased barrier function. A, DAPK1 knockdown with siRNA (40 pm) ablates the ability of the ER stressor TM (10 μg/ml) to reduce the number of viable intracellular E. coli (HB101, 106 CFU) in T84 epithelia evoked by exposure to DNP (0.1 mm; 16 h). B, expression of ATF6α and LC3-I and LC3-II as indicators of autophagy was analyzed by immunoblotting whole T84 cell extracts (± DAPK1 siRNA) 4 h after treatment with DNP, TM of DNP + TM. Rapamycin (Rapa; 100 nm) was used as a positive control to induce autophagy. C, co-treatment of T84 cells with DAPK1 siRNA and the inhibitor DAPK6 (10 μm) while blocking the effect of TM was not statistically significantly different from either agent alone (mean ± S.D.; *, p < 0.05 compared with control).
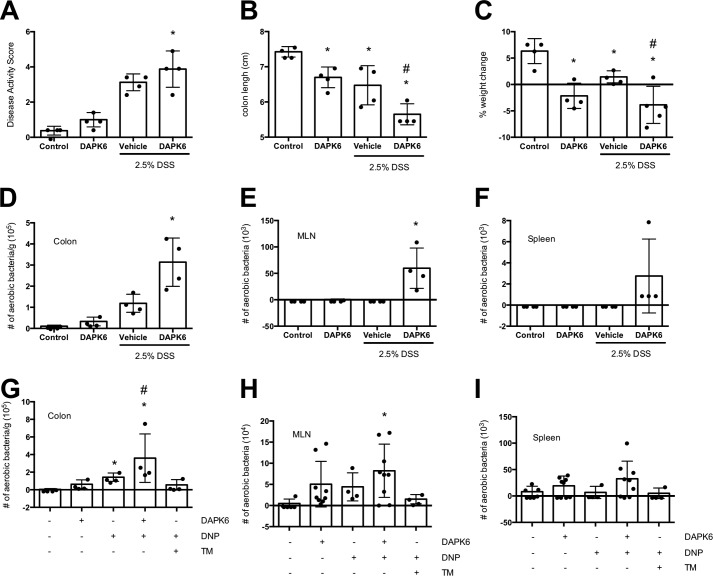
Mice treated with DAPK6 were more susceptible to DSS-induced colitis as gauged by disease activity score (Fig. 8A), colon length (Fig. 8B), and weight loss (Fig. 8C). DAPK6 treatment resulted in increased translocation of aerobic bacteria into the colonic mucosal, mesenteric lymph nodes (MLN), and the spleen (Fig. 8, D–F). Furthermore, intra-rectal delivery of DNP resulted in small increases in bacterial translocation that were aggravated by DAPK6 and alleviated by tunicamycin co-treatment (Fig. 8, G–I).
Figure 8.
Systemic delivery of DAPK1 enhances bacterial translocation in vivo. Male C57Bl/6 mice were treated with 2.5% (w/v) DSS (∼40 kDa) in their drinking water for 5 days (A–F). Additional mice received the DAPK1 inhibitor DAPK6 (20 μg/kg, i.p. daily). Mice were euthanized, and disease activity was assessed in the DSS-treated mice, and bacterial translocation into the colonic mucosa, MLN, and spleen was determined in all mice. G–I, male C57Bl/6 mice were intra-rectally administered DNP (3 mm) for 24 h. Additional mice received DAPK6 (20 μg/kg, i.p. daily) ± TM (20 μg/kg, i.p. daily). Mice were euthanized, and bacterial translocation into the colonic mucosa, MLN, and spleen was determined in all mice (mean ± S.D.; * and #, p < 0.05 compared with control and DSS or DNP, respectively).
Discussion
Increased numbers of bacteria, including E. coli, in the mucosa and gut epithelium can characterize IBD (1). Many lines of evidence point to dysregulated mitochondrial function in cohorts of patients with IBD (6, 10). By juxtaposing these two putative etiological factors in enteric inflammation, we showed increased E. coli internalization and transcytosis across epithelia treated with the hydrogen ionophore, DNP, that uncouples oxidative phosphorylation (15). Mitochondria do not function in isolation; they couple to the ER. Indeed, dysregulation of the UPR (or ER stress) can contribute to gut inflammation (27); data in support of this have accumulated mostly from analyses of Paneth (24) and goblet cells (28). The importance of mitochondria–ER interaction in the transporting enterocyte (the majority of the gut surface area) to host defense is poorly appreciated. The findings herein suggest that mobilization of the ATF6-arm of the UPR via DAPK1-dependent autophagy results in killing of bacteria taken into the cell because of disrupted mitochondrial function, effectively reducing the barrier defect elicited by metabolic stress. Support for this conclusion comes from the use of a variety of human-derived cell lines, human biopsies, primary mouse organoids, and murine models. Although each model has its pros and cons, the demonstration of the major phenomenon of this research in four model systems, adds, we believe, validity to the findings.
Many stimuli implicated in IBD reactivation can result in dysfunctional mitochondria (5). For instance, NSAID use can elicit disease relapse in some patients with IBD (29), and NSAID enteropathy is characterized by abnormal epithelial mitochondrial structure (8, 9). Modeling mitochondrial dysfunction with DNP treatment provided evidence that targeted loss of mitochondrial function resulted in ROS-dependent internalization of bacteria by enteric epithelia (15).
An excessive UPR can lead to colitis (28), and an inability to mount a normal UPR response increases susceptibility to colitis (17). Here, the hypothesis that a UPR-ER stress response would exaggerate an epithelial barrier defect triggered by increased oxidative stress proved to be incorrect; reduced translocation and fewer viable intracellular bacteria occurred in epithelia treated with ER stressors. Also, the apoptosis that occurred with longer DNP + E. coli exposure (36 h) was reduced by tunicamycin, which would preserve epithelial barrier function. ER stress occurs in response to pathogens: bacterial, viral, and fungi. The demonstration that provocation of an ER stress response to counter the effect of epithelial internalization of commensal bacteria caused by mitochondrial dysfunction underscores that this is an important aspect of innate immunity when challenged with intracellular microbes.
Autophagy is important in host defense, where, for example, protection against colitis by outer membrane vesicles from Bacteroides fragilis requires ATG16L1-competent dendritic cells (30). Tunicamycin did not affect internalization of inert beads into DNP-treated epithelia but enhanced autophagy markers suggesting that the reduction in viable intracellular bacteria was because of reduced survival. In accordance, the protective effect of tunicamycin was lost in epithelia lacking the autophagy protein, ATG16L1. The threonine-to-alanine (T300A) ATG16L1 variant that increases the risk for Crohn's disease results in degradation of the protein (31), reducing autophagic capacity and leaving the individual vulnerable to pathogenic bacteria (32, 33). The equivalent mutation in the mouse (T316A) also targets Atg16l1 for destruction (31). Monolayers from colonoids of the Atg16l1T316A knockin mouse not only had increased responses to DNP, as gauged by E. coli internalization but tunicamycin failed to reverse this event. These data support the view that the UPR via autophagy antagonizes the barrier defect evoked by perturbed mitochondrial function.
Given the significance of the colonic microbiota in IBD pathophysiology and that transporting enterocyte is the major constituent of the epithelium, we speculate that the impact of the ATG16L1T300A mutation would be compounded in the context of reduced mitochondrial activity due to NSAID use or other stimuli (6). Furthermore, a cytoprotective effect of ATG16L1 in epithelial cells promoted mitochondrial homeostasis and prevented necroptosis (34). Collectively, these data suggest that the development of drugs that drive autophagy (35) or an appropriate UPR (ER stress) pathway would be of value in treating enteropathies (e.g. IBD) characterized by loss of epithelial barrier function, i.e. uptake and passage of commensal bacteria, pathobionts, and pathogens, due to perturbed mitochondrial function. In this context, the anti-colitic pro-drug thioguanine promoted autophagy in human colon-derived epithelial cell lines, limiting Salmonella typhimurium replication (36).
The balance of this mitochondria–ER–autophagy cross-talk would be central to cell health. Up-regulation of autophagy would prevent the effects of excessive ER stress, and in the short-term, it has the benefit of targeting internalized bacteria as a consequence of mitochondrial dysfunction. Contrarily, prolonged ER stress and/or autophagy would be detrimental and potentially of pathophysiological significance (37, 38).
Essentially the UPR is a three-pronged pathway, in which any one arm can dominate (39), such as the mobilization of Ire1/Xbp1 signaling following bacterial infection or accumulation of Ire1α aggregates in Atg16l1ΔIEC mice that may contribute to Crohn's disease-like ileitis (37). Molecular knockout and use of PERK, IRE1, and JNK-selective inhibitors indicated that tunicamycin abrogation of the DNP-induced epithelial barrier defect was dependent on the ATF6 arm of the UPR. Other studies support a beneficial role for Atf6 in limiting enteric inflammation: mutation in a protease resulting in reduced Atf6 activation increased susceptibility to DSS-induced colitis (data on bacterial translocation were not presented) (40); Atf6−/− mice display increased susceptibility to bacteria correlating with decreased autophagy in macrophages (41). Furthermore, IL-10 can exert an anti-inflammatory effect via inhibition of TNFα-induced nuclear translocation of ATF6 (20).
ATF6 regulates DAPK1 synthesis (26), which can phosphorylate Beclin-1 to induce autophagy (42) and stimulate trafficking of Atg9-containing vesicles for autophagosome formation in response to starvation (43); dapk1−/− fibroblasts display less autophagy following tunicamycin treatment (44). Although preliminary, a polymorphism in the DAPK1 gene is reported in some patients with Crohn's disease (45). Noting that ATF6 siRNA-treated epithelia had reduced DAPK1 expression, it was determined that tunicamycin failed to ablate the DNP-evoked increase in viable intracellular E. coli in epithelia treated with a DAPK1-selective inhibitor, DAPK6, or DAPK1 siRNA, correlating with reduced autophagy. Moreover, treatment of mice with 2.5% DSS resulted in bacterial translocation into the colonic mucosa and MLNs that was significantly increased by DAPK6 co-treatment. Similarly, bacterial translocation (i.e. barrier defect) evoked by intra-rectal DNP (which was reduced by tunicamycin co-treatment) was enhanced by systemic delivery of DAPK6.
There is an elegance in assessing a single variable in an experimental system; however, the coordinated activity of intracellular signaling cascades regulates cell and tissue function. By defining an ATF6–to–DAPK1–to–autophagy (and apoptosis) pathway, we define how the UPR can preserve epithelial barrier function when faced with deranged mitochondrial function. The implications of this could be wide-ranging. In the context of IBD, the effect of reduced activity or the loss of ATG16L1 affects not only macrophages, goblet cells, and Paneth cells but the transporting enterocyte as well, negating the value of mobilizing ATF6 to counter the transcellular passage of microbes across metabolically-stressed epithelia. It will be important to address how the mitochondria–to–ATF6–DAPK1–autophagy interaction is impacted by other molecules that control epithelia–microbe interaction (e.g. NOD proteins, NFκβ, and the inflammasome (38, 46, 47)). For instance, cells devoid of NOD2 are more sensitive to DNP-induced barrier defects (48), and reduced epithelia autophagy due to knockout of the amino acid sensor GCN2 (a starvation model analogous to DNP exposure) was linked to ROS generation, inflammasome activation, and intestinal inflammation (47).
Epithelial–microbial interactions are at the nexus of IBD. Having defined a molecular pathway in the transporting enterocyte by which mobilization of the UPR abolishes the epithelial transcellular permeability defect (i.e. bacteria uptake and passage) in metabolically-stressed cells, we suggest that this is critical in gut homeostasis. Controlled ER stress, via autophagy, rescues the enterocyte from the loss of barrier function, as measured by bacterial transcytosis, caused by uncoupling oxidative phosphorylation. Reciprocally, the pathophysiological consequences of compromised autophagy or ER stress (both susceptibility traits for IBD) is compounded by perturbed mitochondrial function that drives the internalization of commensal bacteria by the epithelium, the enhanced survival of which could be a significant pro-inflammatory stimulus.
Experimental procedures
Human biopsies and murine colonoids
Human colonic biopsies were obtained from two healthy men and five women (mean age 78 years) at the Gastroenterology Department, University Hospital in Linköping. The Regional Committee of human ethics approved the study, and all subjects gave informed consent.
Mice with the Atg16l1T316A knockin gene (from M. van Lookeren Champagne, Genentech, San Francisco) were housed in standard conditions under protocol AC13-0015 (Animal Care Committee, University of Calgary). Experiments conformed to Canadian guidelines on animals in research. Colonic crypts were isolated (49), cultured in IntestiCultTM organoid growth medium (Stemcell, BC Canada), and monolayers grown on filter supports (50). E. coli HB101 was grown overnight in Luria-Bertani (LB) broth, and 107 colony-forming units (cfu) were added to the luminal aspect of filter-grown epithelia, and bacterial translocation was assessed by sampling medium from the basal compartment of the culture well.
Reagents
DNP, tunicamycin, rapamycin, 3-MA, brefeldin-A, thapsigargin, and FITC-4-kDa dextran were from Sigma. SP600125 (JNK inhibitor), DAPK6 (DAPK1 inhibitor), and GSK2606414 (PERK inhibitor) were all from Tocris Bioscience (Bristol, UK), and KIRA6 (IRE-1 inhibitor) was from EMD Millipore (Darmstadt, Germany). Green-yellow latex beads, E. coli F12 fluorescent BioParticles, and DAPI were from Molecular Probes (Eugene, OR). The E. coli strain HB101 was provided by Dr. Philip M. Sherman (Hospital for Sick Children, University of Toronto, Ontario, Canada).
Ussing chambers studies
Human colonic biopsies were mounted in Ussing chambers (1.8-mm2 surface area) (51). Live non-pathogenic GFP-labeled E. coli (HS strain; 108 cfu/ml) (52) were added to the mucosal side of the tissue with DNP (0.1 mm) ± tunicamycin (10 μg/ml) or rapamycin (100 nm). Additional biopsies from the same individual bathed in Krebs buffer only served as controls. All conditions were performed in duplicate biopsies. After 3 h, serosal buffers were collected, and fluorescence was measured at 488 nm in VICTORTM X3 multileader plate reader (PerkinElmer Life Sciences, Sweden). Other biopsies were homogenized in 200 μl of RIPA buffer, serially diluted, plated onto LB agar, cultured under aerobic conditions (∼16 h, 37 °C), and colony-forming units counted.
Cell culture, bacterial translocation, and intracellular viable bacteria
The T84, CaCo2, and HCT116 human colonic epithelial cell lines and the cervical epithelial HeLa cell line were maintained as described (15) (T84 NOD2−/− cells were used generated via shRNA (48)). Cells (106/ml) were seeded into 12-well plates and used at ∼70% confluence based on phase-contrast microscopy or onto 3.0-μm porous filter supports and cultured for 7 days until electrically confluent (T84 >1000 ohms·cm2; HT-29 >300 ohms·cm2) (15). E. coli HB101 was grown overnight in LB broth, and 106 cfu were added to the epithelium (luminal aspect of filter-grown polarized preparations). The intracellular viable bacteria number was assessed by killing extracellular bacteria with 200 μg/ml gentamicin (2 h), followed by enterocyte lysis (0.1% Triton X-100), and culturing serial dilutions of the cell extract. Bacterial translocation was assessed by sampling the medium from the basolateral compartment of filter-grown epithelia (15). All drugs were added simultaneously with the E. coli and were tested (at the doses used) for bacteriostatic and bactericidal effects.
TUNEL assay
Apoptotic cells were detected by TUNEL using ApopTag® in situ apoptosis detection kits (EDM Millipore).
Epithelial paracellular permeability
TER across filter-grown T84 cell monolayers was recorded at time 0 and 16 h later using a voltmeter and matched electrodes (Millicell-ERS, Millipore, Bedford, MA). Data are presented as a percentage of pretreatment TER value.
Following treatment, FITC-dextran (80 μg/ml, 4 kDa) in fresh culture medium was added to the apical surface of filter-grown T84 cell monolayers for 4 h at 37 °C. Samples were collected from the basolateral well of the chamber, and fluorescence was read at excitation (427 nm) and emission (536 nm) wavelengths.
Cell bioenergetics
The oxygen consumption rate and extracellular acidification rate were measured using a Seahorse XF-24 metabolic flux analyzer (Seahorse Bioscience, North Billerica, MA).
GFP-LC3 autophagy detection
HeLa cells expressing LC3-GFP (53) were used, and images were captured by confocal microscopy. ImageJ was used for unbiased LC3 puncta analysis in the mid-optical section of live cells. Cells were identified as a region with greater than 1 S.D. higher intensity MAP2 or HA stain than the mean intensity for that individual image. A punctum was identified as a region of LC3 stain that was more than 2 S.D. brighter than the individual image mean. All LC3 puncta larger than 0.1 μm within the MAP2 or HA stain were included in the analysis (smaller puncta are outside the size range of phagophores/autophagosomes at 0.2–10 μm).
siRNA
T84 cells in suspension (2 × 106/ml) ± ATG16L1 (20 nm), ATF6α (1000 nm), IRE1β (100 nm), PERK (1000 nm), or DAPK1 (40 nm) siRNA Silencer® pre-designed (Inventoried) siRNA (Thermo Fisher Scientific) were electroporated using the AMAXA electroporation device and then seeded into plates. Twenty four (ATF6α, IRE1β, and PERK) or 48 h (ATG16L1 and DAPK1) later, cells were rinsed and used in experiments (gene knockdown confirmed by immunoblotting).
ATG16L1 knockout
CRISPR-Cas9 methodology (54) was used to knockout ATG16L1 in the human HCT116 epithelial cell line. A gRNA target region (5′-ATGAGTATCCACATTGTCCT-3′) was identified using the crispr.mit.edu website (Zhang laboratory at MIT), cloned into pSpCas9(BB)-2A-Puro plasmid (Addgene plasmid 48139), and verified by sequencing. HCT116 cells were grown to 50% confluence in 100-mm tissue culture dishes and transfected with 2 μg of pSpCas9(gRNA)-2A-Puro plasmid using 20 μl of polyethyleneimine (Polysciences, Warrington, PA). Successfully transfected cells were selected 24 h later with 3 μg/ml puromycin. Limiting dilution was used to create clonal cell lines, of which the identity and knockout of ATG16L1 were verified by sequencing and immunoblotting for ATG16L1.
Latex bead phagocytosis
T84 cells plated in chamber slides (105/ml) at ∼70% confluence were treated with FluoSpheres® carboxylate (1 μm, yellow-green (505/515), 105/ml) or killed E. coli F12 fluorescent bioparticles (106/ml) (both from Thermo Fisher Scientific). Samples were analyzed by fluorescent microscopy and flow cytometry.
Protein immunodetection
Standard immunoblotting protocols were used to detect GRP78, ATF6α (Santa Cruz Biotechnology) (55), ATG16L1 (56), DAPK1 (57), LC3-II (58), sXBP1 (59), ATF4 (60), and p-JNK (61) (Cell Signaling Technology) in T84, CaCo2, and HCT116 epithelium using primary antibodies at 1:1000 dilution.
In vivo bacterial translocation
Data from mouse models have limitations when extrapolating to humans; indeed, there is no model of IBD, but there are useful models of colitis for pre-clinical investigation. Bearing this in mind, colitis was induced in 7–9-week-old male C57Bl/6 mice (Charles River Laboratories, Senneville, Quebec, Canada) by 2.5% (w/v) DSS (∼40 kDa) in drinking water for 5 days (an often used model (15)) ± daily DAPK6 (20 μg/kg; i.p.). At necropsy, a disease activity score based on body weight, colon length, and the animal's macroscopic appearance, colon, and stool was calculated. Two-cm pieces of colon, the mesenteric lymph nodes, and spleen were excised, rinsed in gentamicin (200 μg/ml), homogenized, serial dilution plated onto blood agar (37 °C, 24 h), and cfu counted.
Other mice received DNP (3 mm in 100 μl PBS) intra-rectally (13) ± DAPK6 or tunicamycin (20 μg/kg; i.p.), and 24 h later tissues were processed, and bacterial translocation was assessed.
Statistical analysis
Data are means ± S.D. Multiple group comparisons were performed by one-way analysis of variance, followed by pairwise post hoc tests, with p < 0.05 as a level of statistical significant difference.
Author contributions
F. L. and D. M. M. conceptualization; F L., A. S., J. L. R., N. M., A. A. R., A. W., C. B., M. D., Y. A., M. C., N. P., J. M. R., J. D. S., and D. M. M. data curation; F. L., Å. V. K., A. S., J. L. R., A. W., C. B., Y. A., M. C., W. M., H. J., D. J. P., and J. D. S. formal analysis; F. L. and D. M. M. validation; F. L., Å. V. K., N. M., A. A. R., A. W., Y. A., W. M., S. E. G., H. J., D. J. P., J. D. S., and D. M. M. investigation; F. L., Å. V. K., C. B., M. D., R. v. D., Y. A., M. C., W. M., H. J., and J. D. S. methodology; F. L. writing-original draft; A. W. and D. M. M. project administration; J. M. R., S. E. G., J. D. S., and D. M. M. funding acquisition; D. M. M. resources; D. M. M. supervision; D. M. M. writing-review and editing.
Supplementary Material
Acknowledgments
Facilities and expertise provided by the Live Cell Imaging Suite, Snyder Institute for Chronic Disease at the Cumming School of Medicine (University of Calgary), are gratefully acknowledged.
This work was supported in part by Canadian Institutes for Health Research Operating Grant MPO-126005 (to D. M. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- IBD
- inflammatory bowel disease
- DNP
- dinitrophenol
- ER
- endoplasmic reticulum
- DSS
- dextran sodium sulfate
- NSAID
- non-steroidal anti-inflammatory drug
- MLN
- mesenteric lymph node
- cfu
- colony-forming unit
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling
- TER
- transepithelial resistance
- ROS
- reactive oxygen species
- UPR
- unfolded protein response
- PERK
- pancreatic ER kinase
- TM
- tunicamycin
- 3-MA
- 3-methyladenine
- JNK
- c-Jun N-terminal kinase
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. Elliott T. R., Hudspith B. N., Wu G., Cooley M., Parkes G., Quiñones B., Randall L., Mandrell R. E., Fagerquist C. K., Brostoff J., Rayment N. B., Boussioutas A., Petrovska L., and Sanderson J. D. (2013) Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 19, 2326–2338 10.1097/MIB.0b013e3182a38a92 [DOI] [PubMed] [Google Scholar]
- 2. Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A. L., Barnich N., Bringer M. A., Swidsinski A., Beaugerie L., and Colombel J. F. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- 3. Keita A. V., Salim S. Y., Jiang T., Yang P. C., Franzén L., Söderkvist P., Magnusson K. E., and Söderholm J. D. (2008) Increased uptake of non-pathogenic E. coli via the follicle-associated epithelium in longstanding ileal Crohn's disease. J. Pathol. 215, 135–144 10.1002/path.2337 [DOI] [PubMed] [Google Scholar]
- 4. Moussata D., Goetz M., Gloeckner A., Kerner M., Campbell B., Hoffman A., Biesterfeld S., Flourie B., Saurin J. C., Galle P. R., Neurath M. F., Watson A. J., and Kiesslich R. (2011) Confocal laser endomicroscopy is a new imaging modality for recognition of intramucosal bacteria in inflammatory bowel disease in vivo. Gut 60, 26–33 10.1136/gut.2010.213264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Russell R. K., Drummond H. E., Nimmo E. R., Anderson N. H., Noble C. L., Wilson D. C., Gillett P. M., McGrogan P., Hassan K., Weaver L. T., Bisset W. M., Mahdi G., and Satsangi J. (2006) Analysis of the influence of OCTN1/2 variants within the IBD5 locus on disease susceptibility and growth indices in early onset inflammatory bowel disease. Gut 55, 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schoultz I., Söderholm J. D., and McKay D. M. (2011) Is metabolic stress a common denominator in inflammatory bowel disease? Inflamm. Bowel Dis. 17, 2008–2018 10.1002/ibd.21556 [DOI] [PubMed] [Google Scholar]
- 7. Klein A., and Eliakim R. (2010) Non-steroidal anti-inflammatory drugs and inflammatory bowel disease. Pharmaceuticals 3, 1084–1092 10.3390/ph3041084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Somasundaram S., Sigthorsson G., Simpson R. J., Watts J., Jacob M., Tavares I. A., Rafi S., Roseth A., Foster R., Price A. B., Wrigglesworth J. M., and Bjarnason I. (2000) Uncoupling of intestinal mitochondrial oxidative phosphorylation and inhibition of cyclooxygenase are required for the development of NSAID-enteropathy in the rat. Aliment. Pharmacol. Ther. 14, 639–650 10.1046/j.1365-2036.2000.00723.x [DOI] [PubMed] [Google Scholar]
- 9. Schoultz I., McKay C. M., Graepel R., Phan V. C., Wang A., Söderholm J., and McKay D. M. (2012) Indomethacin-induced translocation of bacteria across enteric epithelia is reactive oxygen species-dependent and reduced by vitamin C. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G536–G545 10.1152/ajpgi.00125.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rath E., and Haller D. (2012) Mitochondria at the interface between danger signaling and metabolism: role of unfolded protein responses in chronic inflammation. Inflamm. Bowel Dis. 18, 1364–1377 10.1002/ibd.21944 [DOI] [PubMed] [Google Scholar]
- 11. Khor B., Gardet A., and Xavier R. J. (2011) Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 10.1038/nature10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odenwald M. A., and Turner J. R. (2017) The intestinal epithelial barrier: a therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 14, 9–21 10.1038/nrgastro.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nazli A., Yang P. C., Jury J., Howe K., Watson J. L., Söderholm J. D., Sherman P. M., Perdue M. H., and McKay D. M. (2004) Epithelia under metabolic stress perceive commensal bacteria as a threat. Am. J. Pathol. 164, 947–957 10.1016/S0002-9440(10)63182-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewis K., Lutgendorff F., Phan V., Söderholm J. D., Sherman P. M., and McKay D. M. (2010) Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm. Bowel Dis. 16, 1138–1148 10.1002/ibd.21177 [DOI] [PubMed] [Google Scholar]
- 15. Wang A., Keita Å. V., Phan V., McKay C. M., Schoultz I., Lee J., Murphy M. P., Fernando M., Ronaghan N., Balce D., Yates R., Dicay M., Beck P. L., MacNaughton W. K., Söderholm J. D., and McKay D. M. (2014) Targeting mitochondria-derived reactive oxygen species to reduce epithelial barrier dysfunction and colitis. Am. J. Pathol. 184, 2516–2527 10.1016/j.ajpath.2014.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marchi S., Patergnani S., and Pinton P. (2014) The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim. Biophys. Acta 1837, 461–469 10.1016/j.bbabio.2013.10.015 [DOI] [PubMed] [Google Scholar]
- 17. Kaser A., Lee A. H., Franke A., Glickman J. N., Zeissig S., Tilg H., Nieuwenhuis E. E., Higgins D. E., Schreiber S., Glimcher L. H., and Blumberg R. S. (2008) XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H. S., Chen Y., Fan L., Xi Q. L., Wu G. H., Li X. X., Yuan T. L., He S. Q., Yu Y., Shao M. L., Liu Y., Bai C. G., Ling Z. Q., Li M., Liu Y., and Fang J. (2015) The endoplasmic reticulum stress sensor IRE1α in intestinal epithelial cells is essential for protecting against colitis. J. Biol. Chem. 290, 15327–15336 10.1074/jbc.M114.633560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao S. S., Zimmermann E. M., Chuang B. M., Song B., Nwokoye A., Wilkinson J. E., Eaton K. A., and Kaufman R. J. (2013) The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology 144, 989–1000 10.1053/j.gastro.2013.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shkoda A., Ruiz P. A., Daniel H., Kim S. C., Rogler G., Sartor R. B., and Haller D. (2007) Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology 132, 190–207 10.1053/j.gastro.2006.10.030 [DOI] [PubMed] [Google Scholar]
- 21. Ling Y. H., Li T., Perez-Soler R., and Haigentz M. Jr. (2009) Activation of ER stress and inhibition of EGFR N-glycosylation by tunicamycin enhances susceptibility of human non-small cell lung cancer cells to erlotinib. Cancer Chemother. Pharmacol. 64, 539–548 10.1007/s00280-008-0902-8 [DOI] [PubMed] [Google Scholar]
- 22. Shen T., Li Y., Chen Z., Liang S., Guo Z., Wang P., Wu Q., Ba G., and Fu Q. (2017) CHOP negatively regulates Polo-like kinase 2 expression via recruiting C/EBPα to the upstream-promoter in human osteosarcoma cell line during ER stress. Int. J. Biochem. Cell Biol. 89, 207–215 10.1016/j.biocel.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 23. Drion C. M., Borm L. E., Kooijman L., Aronica E., Wadman W. J., Hartog A. F., van Vliet E. A., and Gorter J. A. (2016) Effects of rapamycin and curcumin treatment on the development of epilepsy after electrically induced status epilepticus in rats. Epilepsia 57, 688–697 10.1111/epi.13345 [DOI] [PubMed] [Google Scholar]
- 24. Adolph T. E., Tomczak M. F., Niederreiter L., Ko H. J., Böck J., Martinez-Naves E., Glickman J. N., Tschurtschenthaler M., Hartwig J., Hosomi S., Flak M. B., Cusick J. L., Kohno K., Iwawaki T., Billmann-Born S., et al. (2013) Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 10.1038/nature12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grootjans J., Kaser A., Kaufman R. J., and Blumberg R. S. (2016) The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 16, 469–484 10.1038/nri.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gade P., Kimball A. S., DiNardo A. C., Gangwal P., Ross D. D., Boswell H. S., Keay S. K., and Kalvakolanu D. V. (2016) Death-associated protein kinase-1 expression and autophagy in chronic lymphocytic leukemia are dependent on activating transcription factor-6 and CCAAT/enhancer-binding protein-β. J. Biol. Chem. 291, 22030–22042 10.1074/jbc.M116.725796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bogaert S., De Vos M., Olievier K., Peeters H., Elewaut D., Lambrecht B., Pouliot P., and Laukens D. (2011) Involvement of endoplasmic reticulum stress in inflammatory bowel disease: a different implication for colonic and ileal disease? PLoS One 6, e25589 10.1371/journal.pone.0025589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heazlewood C. K., Cook M. C., Eri R., Price G. R., Tauro S. B., Taupin D., Thornton D. J., Png C. W., Crockford T. L., Cornall R. J., Adams R., Kato M., Nelms K. A., Hong N. A., Florin T. H., Goodnow C. C., and McGuckin M. A. (2008) Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 5, e54 10.1371/journal.pmed.0050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kvasnovsky C. L., Aujla U., and Bjarnason I. (2015) Nonsteroidal anti-inflammatory drugs and exacerbations of inflammatory bowel disease. Scand. J. Gastroenterol. 50, 255–263 10.3109/00365521.2014.966753 [DOI] [PubMed] [Google Scholar]
- 30. Chu H., Khosravi A., Kusumawardhani I. P., Kwon A. H., Vasconcelos A. C., Cunha L. D., Mayer A. E., Shen Y., Wu W. L., Kambal A., Targan S. R., Xavier R. J., Ernst P. B., Green D. R., McGovern D. P., Virgin H. W., and Mazmanian S. K. (2016) Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 10.1126/science.aad9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murthy A., Li Y., Peng I., Reichelt M., Katakam A. K., Noubade R., Roose-Girma M., DeVoss J., Diehl L., Graham R. R., and van Lookeren Campagne M. (2014) A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- 32. Lassen K. G., Kuballa P., Conway K. L., Patel K. K., Becker C. E., Peloquin J. M., Villablanca E. J., Norman J. M., Liu T. C., Heath R. J., Becker M. L., Fagbami L., Horn H., Mercer J., Yilmaz O. H., et al. (2014) Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl. Acad. Sci. U.S.A. 111, 7741–7746 10.1073/pnas.1407001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sadaghian Sadabad M., Regeling A., de Goffau M. C., Blokzijl T., Weersma R. K., Penders J., Faber K. N., Harmsen H. J., and Dijkstra G. (2015) The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut 64, 1546–1552 10.1136/gutjnl-2014-307289 [DOI] [PubMed] [Google Scholar]
- 34. Matsuzawa-Ishimoto Y., Shono Y., Gomez L. E., Hubbard-Lucey V. M., Cammer M., Neil J., Dewan M. Z., Lieberman S. R., Lazrak A., Marinis J. M., Beal A., Harris P. A., Bertin J., Liu C., Ding Y., van den Brink M. R. M., and Cadwell K. (2017) Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214, 3687–3705 10.1084/jem.20170558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuo S. Y., Castoreno A. B., Aldrich L. N., Lassen K. G., Goel G., Dančík V., Kuballa P., Latorre I., Conway K. L., Sarkar S., Maetzel D., Jaenisch R., Clemons P. A., Schreiber S. L., Shamji A. F., and Xavier R. J. (2015) Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl. Acad. Sci. U.S.A. 112, E4281–E4287 10.1073/pnas.1512289112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oancea I., Movva R., Das I., Aguirre de Cárcer D., Schreiber V., Yang Y., Purdon A., Harrington B., Proctor M., Wang R., Sheng Y., Lobb M., Lourie R., Ó Cuív P., Duley J. A., Begun J., and Florin T. H. (2017) Colonic microbiota can promote rapid local improvement of murine colitis by thioguanine independently of T lymphocytes and host metabolism. Gut 66, 59–69 10.1136/gutjnl-2015-310874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tschurtschenthaler M., Adolph T. E., Ashcroft J. W., Niederreiter L., Bharti R., Saveljeva S., Bhattacharyya J., Flak M. B., Shih D. Q., Fuhler G. M., Parkes M., Kohno K., Iwawaki T., Janneke van der Woude C., Harding H. P., et al. (2017) Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis. J. Exp. Med. 214, 401–422 10.1084/jem.20160791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diamanti M. A., Gupta J., Bennecke M., De Oliveira T., Ramakrishnan M., Braczynski A. K., Richter B., Beli P., Hu Y., Saleh M., Mittelbronn M., Dikic I., and Greten F. R. (2017) IKKα controls ATG16L1 degradation to prevent ER stress during inflammation. J. Exp. Med. 214, 423–437 10.1084/jem.20161867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamamoto K., Yoshida H., Kokame K., Kaufman R. J., and Mori K. (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 136, 343–350 10.1093/jb/mvh122 [DOI] [PubMed] [Google Scholar]
- 40. Brandl K., Rutschmann S., Li X., Du X., Xiao N., Schnabl B., Brenner D. A., and Beutler B. (2009) Enhanced sensitivity to DSS colitis caused by a hypomorphic Mbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 106, 3300–3305 10.1073/pnas.0813036106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gade P., Ramachandran G., Maachani U. B., Rizzo M. A., Okada T., Prywes R., Cross A. S., Mori K., and Kalvakolanu D. V. (2012) An IFN-γ-stimulated ATF6-C/EBP-β-signaling pathway critical for the expression of death associated protein kinase 1 and induction of autophagy. Proc. Natl. Acad. Sci. U.S.A. 109, 10316–10321 10.1073/pnas.1119273109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zalckvar E., Berissi H., Eisenstein M., and Kimchi A. (2009) Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 5, 720–722 10.4161/auto.5.5.8625 [DOI] [PubMed] [Google Scholar]
- 43. Tang H. W., Liao H. M., Peng W. H., Lin H. R., Chen C. H., and Chen G. C. (2013) Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress–induced JNK activation and autophagy induction. Dev. Cell 27, 489–503 10.1016/j.devcel.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 44. Gozuacik D., Bialik S., Raveh T., Mitou G., Shohat G., Sabanay H., Mizushima N., Yoshimori T., and Kimchi A. (2008) DAP-kinase is a mediator of endoplasmic reticulum stress–induced caspase activation and autophagic cell death. Cell Death Differ. 15, 1875–1886 10.1038/cdd.2008.121 [DOI] [PubMed] [Google Scholar]
- 45. Kaur M., Panikkath D., Yan X., Liu Z., Berel D., Li D., Vasiliauskas E. A., Ippoliti A., Dubinsky M., Shih D. Q., Melmed G. Y., Haritunians T., Fleshner P., Targan S. R., and McGovern D. P. (2016) Perianal Crohn's disease is associated with distal colonic disease, stricturing disease behavior, IBD-associated serologies and genetic variation in the JAK-STAT pathway. Inflamm. Bowel Dis. 22, 862–869 10.1097/MIB.0000000000000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keestra-Gounder A. M., Byndloss M. X., Seyffert N., Young B. M., Chávez-Arroyo A., Tsai A. Y., Cevallos S. A., Winter M. G., Pham O. H., Tiffany C. R., de Jong M. F., Kerrinnes T., Ravindran R., Luciw P. A., McSorley S. J., Bäumler A. J., and Tsolis R. M. (2016) NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397 10.1038/nature17631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ravindran R., Loebbermann J., Nakaya H. I., Khan N., Ma H., Gama L., Machiah D. K., Lawson B., Hakimpour P., Wang Y. C., Li S., Sharma P., Kaufman R. J., Martinez J., and Pulendran B. (2016) The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527 10.1038/nature17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saxena A., Lopes F., Poon K. K. H., and McKay D. M. (2017) Absence of the NOD2 protein renders epithelia more susceptible to barrier dysfunction due to mitochondrial dysfunction. Am. J. Physiol. Gastrointest. Liver Physiol. 313, G26–G38 10.1152/ajpgi.00070.2017 [DOI] [PubMed] [Google Scholar]
- 49. Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K., Clevers H., and Watanabe M. (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat. Med. 18, 618–623 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- 50. Fernando E. H., Dicay M., Stahl M., Gordon M. H., Vegso A., Baggio C., Alston L., Lopes F., Baker K., Hirota S., McKay D. M., Vallance B., and MacNaughton W. K. (2017) A simple, cost-effective method for generating murine colonic 3D enteroids and 2D monolayers for studies of primary epithelial cell function. Am. J. Physiol. Gastrointest. Liver Physiol. 313, G467–G475 10.1152/ajpgi.00152.2017 [DOI] [PubMed] [Google Scholar]
- 51. Wallon C., Braaf Y., Wolving M., Olaison G., and Söderholm J. D. (2005) Endoscopic biopsies in Ussing chambers evaluated for studies of macromolecular permeability in the human colon. Scand. J. Gastroenterol. 40, 586–595 10.1080/00365520510012235 [DOI] [PubMed] [Google Scholar]
- 52. Keita A. V., Gullberg E., Ericson A. C., Salim S. Y., Wallon C., Kald A., Artursson P., and Söderholm J. D. (2006) Characterization of antigen and bacterial transport in the follicle-associated epithelium of human ileum. Lab. Invest. 86, 504–516 10.1038/labinvest.3700397 [DOI] [PubMed] [Google Scholar]
- 53. Begun J., Lassen K. G., Jijon H. B., Baxt L. A., Goel G., Heath R. J., Ng A., Tam J. M., Kuo S. Y., Villablanca E. J., Fagbami L., Oosting M., Kumar V., Schenone M., Carr S. A., et al. (2015) Integrated genomics of Crohn's disease risk variant identifies a role for CLEC12A in antibacterial autophagy. Cell Rep. 11, 1905–1918 10.1016/j.celrep.2015.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C., Tang Y., Li Y., Xie L., Zhuang W., Liu J., and Gong J. (2017) Unfolded protein response plays a critical role in heart damage after myocardial ischemia/reperfusion in rats. PLoS One 12, e0179042 10.1371/journal.pone.0179042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Merrill N. M., Schipper J. L., Karnes J. B., Kauffman A. L., Martin K. R., and MacKeigan J. P. (2017) PI3K-C2α knockdown decreases autophagy and maturation of endocytic vesicles. PLoS One 12, e0184909 10.1371/journal.pone.0184909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mitsui Y., Chang I., Fukuhara S., Hiraki M., Arichi N., Yasumoto H., Hirata H., Yamamura S., Shahryari V., Deng G., Wong D. K., Majid S., Shiina H., Dahiya R., and Tanaka Y. (2015) CYP1B1 promotes tumorigenesis via altered expression of CDC20 and DAPK1 genes in renal cell carcinoma. BMC Cancer 15, 942 10.1186/s12885-015-1951-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Z., Bu J., Yao X., Liu C., Shen H., Li X., Li H., and Chen G. (2017) Phosphorylation at S153 as a functional switch of phosphatidylethanolamine binding protein 1 in cerebral ischemia-reperfusion injury in rats. Front. Mol. Neurosci. 10, 358 10.3389/fnmol.2017.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van de Beek M. C., Ofman R., Dijkstra I., Wijburg F., Engelen M., Wanders R., and Kemp S. (2017) Lipid-induced endoplasmic reticulum stress in X-linked adrenoleukodystrophy. Biochim. Biophys. Acta 1863, 2255–2265 10.1016/j.bbadis.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 60. Inoue Y., Kawachi S., Ohkubo T., Nagasaka M., Ito S., Fukuura K., Itoh Y., Ohoka N., Morishita D., and Hayashi H. (2017) The CDK inhibitor p21 is a novel target gene of ATF4 and contributes to cell survival under ER stress. FEBS Lett. 591, 3682–3691 10.1002/1873-3468.12869 [DOI] [PubMed] [Google Scholar]
- 61. Zhang Q., Sun J., Wang Y., He W., Wang L., Zheng Y., Wu J., Zhang Y., and Jiang X. (2017) Antimycobacterial and anti-inflammatory mechanisms of baicalin via induced autophagy in macrophages infected with mycobacterium tuberculosis. Front. Microbiol. 8, 2142 10.3389/fmicb.2017.02142 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.