Abstract
The stringent response is a global reprogramming of bacterial physiology that renders cells more tolerant to antibiotics and induces virulence gene expression in pathogens in response to stress. This process is driven by accumulation of the intracellular alarmone guanosine-5′-di(tri)phosphate-3′-diphosphate ((p)ppGpp), which is produced by enzymes of the RelA SpoT homologue (RSH) family. The Gram-positive Firmicute pathogen, Staphylococcus aureus, encodes three RSH enzymes: a multidomain RSH (Rel) that senses amino acid starvation on the ribosome and two small alarmone synthetase (SAS) enzymes, RelQ (SAS1) and RelP (SAS2). In Bacillus subtilis, RelQ (SAS1) was shown to form a tetramer that is activated by pppGpp and inhibited by single-stranded RNA, but the structural and functional regulation of RelP (SAS2) is unexplored. Here, we present crystal structures of S. aureus RelP in two major functional states, pre-catalytic (bound to GTP and the non-hydrolyzable ATP analogue, adenosine 5′-(α,β-methylene)triphosphate (AMP-CPP)), and post-catalytic (bound to pppGpp). We observed that RelP also forms a tetramer, but unlike RelQ (SAS1), it is strongly inhibited by both pppGpp and ppGpp and is insensitive to inhibition by RNA. We also identified putative metal ion–binding sites at the subunit interfaces that were consistent with the observed activation of the enzyme by Zn2+ ions. The structures reported here reveal the details of the catalytic mechanism of SAS enzymes and provide a molecular basis for understanding differential regulation of SAS enzymes in Firmicute bacteria.
Keywords: stress response, Staphylococcus aureus (S. aureus), X-ray crystallography, enzyme mechanism, bacterial pathogenesis, (p)ppGpp, small alarmone synthetase (SAS), stringent reponse
Introduction
The bacterial stringent response is a wide-ranging transcriptional and metabolic reprogramming that is induced in response to a range of stress conditions such as amino acid starvation and heat shock (1, 2). Activation of the stringent response causes a complete transcriptional reprogramming and is accompanied by inhibition of ribosome assembly, protein translation, and replication, and consequently induces a halt in cell division until growth conditions improve (3). In addition, the stringent response is of potential medical importance because it regulates virulence gene expression in some bacterial species and can render bacteria tolerant to antibiotic treatment (4, 5).
At the molecular level, the stringent response is mediated by two alarmone nucleotides, guanosine-5′-(di)phosphate-3′-diphosphate (ppGpp)2 and guanosine-5′-(tri)phosphate-3′-diphosphate (pppGpp), which are 3′ hyper-phosphorylated versions of GDP and GTP, respectively (6). Synthesis and degradation of the two alarmones, collectively referred to as (p)ppGpp, are mediated by enzymes belonging to the RelA-SpoT homologue (RSH) family. This family comprises both the large multidomain enzymes (the “long” RSH's), RelA and SpoT, as well as two classes of single-domain RSH's, the small alarmone synthetases (SASs) and small alarmone hydrolases that synthesize and hydrolyze (p)ppGpp, respectively (7). In the long RSHs, the N-terminal half contains both (p)ppGpp synthesis (SYNTH) and hydrolysis (HD) domains, whereas the C-terminal domains are purely regulatory (7–10). The crystal structure of a bifunctional catalytic fragment of Streptococcus equisimilis Rel (SeRel) revealed two reciprocal conformations, one in complex with the substrate GDP (hydrolase-OFF, synthetase-ON) and another bound to both GDP and an unusual cyclic (p)ppGpp derivative, guanosine 5′-diphosphate 2′-3′ cyclic monophosphate (hydrolase-ON, synthetase-OFF) (11). The (p)ppGpp synthesis activities of the ribosome-associated Rel and RelA enzymes are induced by binding of uncharged tRNA to the A-site during amino acid starvation (12, 13). Structural work has demonstrated that upon ribosome binding, the C-terminal domains are removed from the synthetase domain, thus activating the enzyme in the ribosome-bound form (8–10).
SAS enzymes lack both the hydrolase and C-terminal regulatory domains of the long RSHs and are thus minimalist (p)ppGpp synthetase enzymes (7). Two such enzymes, RelQ (SAS1, YjbM) and RelP (SAS2, YwaC), were first identified in the Firmicute, Streptococcus mutans (14). Both alkaline shock and cell-wall stress (such as exposure to cell-wall active antibiotics) induce expression of the SAS enzymes, cause accumulation of (p)ppGpp and thus improve the fitness of the bacterial cells (15, 16). The human pathogen, Staphylococcus aureus, also encodes both the RelP (SAS2) and RelQ (SAS1) enzymes (17). Clinically, S. aureus gives rise to a diverse array of diseases ranging from mild skin infections to life threatening conditions such as endocarditis and osteomyelitis that can be difficult to treat with antibiotics (18). Similar to S. mutants, expression of the SAS enzymes in S. aureus is strongly induced by cell-wall active antibiotics such as ampicillin and vancomycin, and mutants lacking the relP and relQ genes show a significantly reduced survival rate upon exposure to these antibiotics (17). A study of Bacillus subtilis RelQ (BsRelQ) revealed that the protein forms a homotetramer with four active sites and structures were determined of the enzyme in complex with either ATP (one of the two substrates) or pppGpp (product) (16). This study further revealed two allosteric nucleotide-binding sites at the subunit interface and provided evidence that binding of pppGpp (but not ppGpp) to these sites dramatically increases catalytic activity of the enzyme. Based on the BsRelQ structure it was also proposed that RelP (SAS2) would have a similar structure and catalytic mechanism but be differently regulated. Finally, RelQ from Enterococcus faecalis (EfRelQ) is negatively regulated by single-stranded RNA, likely through binding to the same allosteric site as the effect is mutually exclusive with regulation by pppGpp (19).
Here, we present crystal structures of S. aureus RelP (SAS2, SaRelP) in the complete pre-catalytic state (i.e. bound to both GTP and the non-hydrolysable ATP analogue, AMP-CPP) as well as in the post-catalytic state (bound to pppGpp). Together, these structures complete the view of the reaction cycle of the bacterial SAS enzymes and allow us to propose a detailed reaction mechanism. We also demonstrate that the allosteric pppGpp-binding site is not present in RelP (SAS2). This is consistent with functional analysis showing that neither ppGpp nor pppGpp induce the enzymatic activity of SaRelP and instead inhibit the enzyme, likely through active site binding. In contrast to EfRelQ, SaRelP does not bind and is not inhibited by single-stranded RNA, underscoring the difference in regulatory logic between the two classes of SAS enzymes. Finally, we identify two putative divalent metal ion–binding sites at the subunit interfaces and show that Zn2+ is a potent activator of SaRelP.
Results
Structure determination and overall structure of SaRelP
To understand why some Gram-positive bacteria require two very similar SAS enzymes and delineate the molecular basis for the proposed differences in regulation, we decided to determine the crystal structure of SaRelP by X-ray crystallography. The protein has a monomer mass of approximately 25 kDa but elutes with an apparent molecular mass of ∼100 kDa during size exclusion chromatography, suggesting that it forms a tetramer similar to BsRelQ (Fig. S1) (16). The purified protein was concentrated and incubated with equimolar amounts of ATP and GTP prior to crystallization. Large, single crystals belonging to the centered tetragonal space group I4122 and diffracting to 2.8-Å were obtained and the structure was determined by molecular replacement using the structure of BsRelQ bound to pppGpp (PDB code 5DED) (16) as a search model (see Table 1 for crystallographic data collection and refinement statistics). There are two SaRelP molecules per crystallographic asymmetric unit, which are arranged as a dimer, and the final structure covers 194 residues of a total of 230. The poorly conserved N terminus (residues 1–29) and the loop region 194–201 are disordered and not visible in the electron density. The final structure has Rwork = 21.8% (Rfree = 26.3%) with good overall geometry (Table 1).
Table 1.
Crystallographic data collection and refinement statistics
| SaRelP–AMP-CPP:GTP | SaRelP–pppGpp | Below Fe3+ edge | Above Fe3+ edge | Above Zn2+ edge | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 0.979 | 0.979 | 1.79 (6.900 keV) | 1.71 (7.200 keV) | 1.26 (9.800 keV) |
| Resolution range | 63.1–2.24 (2.32–2.24)a | 53.7–2.78 (2.88–2.78) | 69.0–3.07 (3.18–3.07) | 54.3–3.09 (3.20–3.09) | 54.4–2.98 (3.09–2.98) |
| Space group | I4122 | I4122 | I4122 | I4122 | I4122 |
| Unit cell dimensions | |||||
| a, b, c, (Å) | 125.1, 125.1, 219.1 | 124.3, 124.3, 213.2 | 126.4, 126.4, 217.0 | 126.4, 126.4, 217.3 | 125.9, 125.9, 216.1 |
| α, β, γ (o) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Total reflections | 556,165 (53,841) | 133,328 (10,390) | 122,971 (11,057) | 126,224 (13,018) | 119,309 (11887) |
| Unique reflections | 42,049 (4098) | 21,284 (2055) | 16,809 (1621) | 16,498 (1622) | 18,140 (1738) |
| Multiplicity | 13.2 (13.1) | 6.3 (5.1) | 7.3 (7.0) | 7.7 (8.0) | 6.6 (6.5) |
| Completeness (%) | 99.9 (99.9) | 99.3 (96.9) | 99.9 (99.2) | 99.8 (99.8) | 99.9 (99.2) |
| Rmerge (%) | 5.5 (181) | 7.3 (105) | 10.1 (135) | 5.9 (113) | 13.5 (172) |
| I/σ(I) | 24.8 (1.4) | 12.6 (1.4) | 8.6 (1.2) | 21.5 (1.7) | 8.0 (1.3) |
| CC1/2 | 1.00 (0.66) | 1.00 (0.53) | 0.99 (0.51) | 0.99 (0.62) | 0.99 ( 0.53) |
| Refinement | |||||
| Average B-factor (Å2) | 83.0 | 97.5 | |||
| No. of reflections | 42,042 (4098) | 21,297 (2,055) | |||
| No. of reflections (free) | 2,100 (227) | 1,049 (90) | |||
| Rwork (%) | 19.3 (29.8) | 22.4 (34.5) | |||
| Rfree (%) | 23.2 (35.7) | 26.4 (43.6) | |||
| No. of atoms | |||||
| Protein | 3,386 | 3,246 | |||
| Solvent | 99 | 25 | |||
| Ligand | 129 | 81 | |||
| Root mean square deviation bonds (Å) | 0.015 | 0.010 | |||
| Root mean square deviation angles (o) | 1.17 | 1.12 | |||
| Ramachandran statistics | |||||
| Favored (%) | 96.2 | 96.4 | |||
| Allowed (%) | 3.3 | 2.6 | |||
| Outliers (%) | 0.5 | 1.0 | |||
a Numbers in parentheses refer to the outermost resolution shell.
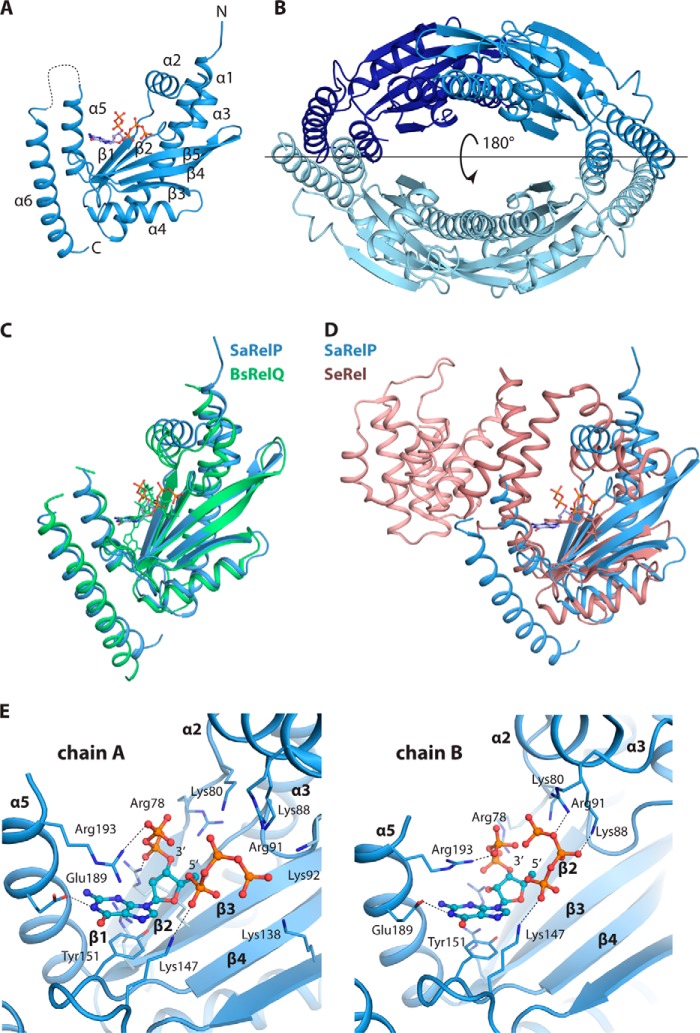
The SaRelP monomer displays a Rossmann-like fold consisting of a five-stranded anti-parallel β-sheet sandwiched between six α-helices (Fig. 1A). In accordance with the behavior of the protein in solution and as previously observed for BsRelQ, four SaRelP molecules come together to form a ring-shaped tetramer with a large cavity at the center (Fig. 1B). In the I4122 space group, this tetramer is generated across a crystallographic 2-fold axis (parallel to the horizontal line in Fig. 1B), so that the two dimers are perfectly identical. This is in contrast to the BsRelQ structures, which have one or two full tetramers in the crystallographic asymmetric unit and thus allow for asymmetry. Nevertheless, the overall structure of SaRelP is very similar to both BsRelQ (Fig. 1C) as well as the catalytic domain of SeRel (Fig. 1D).
Figure 1.
Overall structure of SaRelP in the post-catalytic state. A, crystal structure of the pppGpp-bound SaRelP monomer shown in schematic with N and C termini and secondary structure elements indicated and pppGpp in ball and stick. The disordered loop 194–201 is shown with a dashed line. B, overview of the SaRelP homotetramer with the four chains in various shades of blue. The horizontal line indicates the location of the 2-fold crystallographic axis that generates the tetramer. C, structural alignment of SaRelP (blue) and BsRelQ (PDB code 5DED, green) (16). The pppGpp molecule bound at the active site in both structures is shown in ball and stick in matching colors. D, structural alignment of SaRelP (blue) with the catalytic domain of SeRel (PDB code 1VJ7, brown) (11). The GDP molecule in the SeRel active site is shown in ball and stick. The hydrolase domain of SeRel is highlighted with a darker shade of brown. E, details of the interactions between pppGpp and SaRelP in chains A and B of the structure. The pppGpp molecule is shown in ball and stick with relevant interacting residues and secondary structure elements labeled.
pppGpp is found in two different conformations inside the active site
Inspection of the active site cavities of the two SaRelP monomers revealed clear density compatible with neither of the added ligands (ATP and GTP) but only pppGpp (Fig. S2A). This suggested that the reaction took place prior to crystallization and left the product in the active site similarly to what was observed for BsRelQ (16). The active site pocket is formed mainly by the β-strands and the loops β1/α2, α3/β2, and β3/β4 in addition to two small helices, α2 and α5 (Fig. 1A). pppGpp adopts different conformations inside the two active sites (chains A and B) of the dimer (Fig. 1E and Fig. S2B). Both of these conformations are different from the one observed in BsRelQ, where the two phosphate arms (5′ and 3′) display an open conformation and point in opposite directions. In contrast, in SaRelP, the phosphate arms either point in the same direction (chain A, Fig. 1E, left, and Fig. S2B) or toward each other (chain B, Fig. 1E, right). In chain A where the 5′ γ-phosphate groups face away from the 3′-phosphate arm, α2 is shifted by ∼4.6 Å compared with BsRelQ, apparently to adapt to the more distant position of the 5′ γ-phosphate (Fig. S2C, left). This shift is less (3.7 Å) but still significant in chain B, where the two phosphate arms face each other (Fig. S2C, right). Together, these data suggest that the product nucleotide is loosely bound within the enzyme post-catalysis, possibly to promote its dissociation.
The interaction between pppGpp and SaRelP is stabilized by the universally conserved Tyr-151, which stacks on the guanine base, whereas Glu-189 forms a hydrogen bond with the N1 atom. The combined negative charge of the phosphates is balanced by a large number of basic residues in the vicinity, including Arg-78, Lys-80, Lys-88, Arg-91, Lys-92, Lys-138, Lys-147, and Arg-193 (Fig. 1E). Not surprisingly, several of the residues involved in the interaction between SaRelP and pppGpp (Arg-78, Lys-80, Lys-88, and Lys-147) are among the most conserved in SAS/Rel enzymes (Fig. S3). Active site differences between SaRelP and BsRelQ include Lys-138, which interacts with the outermost 5′ γ-phosphate in chain A and Arg-193, which interacts both with the guanine base and the 3′-phosphate moieties of pppGpp in SaRelP (chain A) but are not conserved in BsRelQ.
Structure of SaRelP in the pre-catalytic state
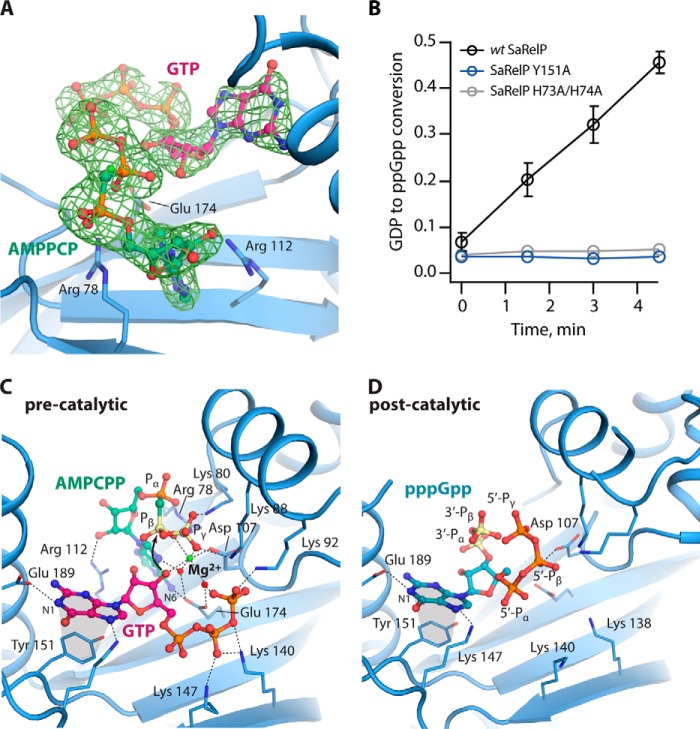
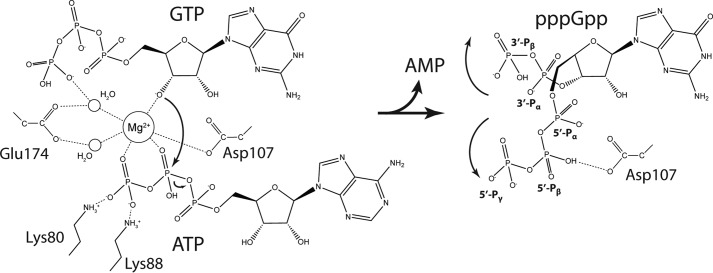
SAS enzymes catalyze transfer of a pyrophosphate moiety from ATP to the 3′-OH group of either GDP or GTP forming ppGpp or pppGpp, respectively. However, no structure of the complete pre-catalytic state of a SAS enzyme, i.e. bound to both GTP and ATP, has so far been determined. To understand the molecular details of the catalytic mechanism, we therefore determined the crystal structure of SaRelP in the pre-catalytic state by incubating the protein with GTP and the non-hydrolysable ATP analogue, AMP-CPP, prior to crystallization. Complete data were collected to 2.2 Å and the structure was determined by molecular replacement using the pppGpp-bound SaRelP structure (Table 1). In these crystals, which belong to the same space group as those for the post-catalytic state, we observe clear electron density for both AMP-CPP and GTP in both active sites (Fig. 2A). Moreover, the overall B factors are lower and we were able to build the flexible loop region 194–201, which was disordered in the pppGpp-bound state, suggesting the enzyme became stabilized. The conformation of the nucleotides is identical in chains A and B with GTP most closely resembling the pppGpp conformation found in chain B of the post-catalytic state. The 3′-OH of GTP is positioned in close proximity (3.2 Å) to the β-phosphate of AMP-CPP, and thus poised for the nucleophilic attack (Fig. 2C). The adenine base of AMP-CPP is sandwiched through π-stacking between Arg-78 and Arg-112, and Glu-174 further stabilizes the base through interaction with its N6 group (Fig. 2, A and C). Glu-174 also coordinates two water molecules that organize a single magnesium ion, which is further coordinated by oxygen atoms from the β- and γ-phosphate groups of AMP-CPP as well as the 3′-OH of GTP (Fig. 2C). The 3′-OH of GTP does not make any direct interactions with SaRelP but interacts with Glu-174 and Asp-107 indirectly through the magnesium ion (Fig. 2C).
Figure 2.
Structures of SaRelP in the pre- and post-catalytic states. A, binding of AMP-CPP and GTP in the active site of SaRelP in the pre-catalytic state. The nucleotides are shown in ball and stick with interacting residues as labeled sticks. The OMIT difference map (calculated before inclusion of nucleotides in the model), is shown contoured at 2.0 σ. B, the fraction of GDP converted to ppGpp by wt SaRelP and the Y151A and H73A/H74A mutants at various time points. Error bars represent S.D. of three independent measurements. C, overview of the pre-catalytic state of SaRelP bound to GTP and AMP-CPP. Interacting residues are shown in labeled sticks along with the magnesium ion (green sphere) and relevant water molecules (red spheres). GTP is shown in a purple, AMP-CPP in green, and the two phosphate groups of AMP-CPP (Pβ and Pγ) that get transferred during the reaction are colored yellow. Stacking interactions are shown with gray areas. D, overview of the post-catalytic state of SaRelP (chain B) bound to pppGpp. Residues interacting with pppGpp are shown with labeled sticks and the phosphate groups that were transferred (3′-Pα and 3′-Pβ) are colored yellow.
The orientation of both the ribose and base of GTP is very similar to pppGpp in the post-catalytic state (Fig. 2D), suggesting that no major rearrangements of these groups take place during the reaction. In contrast, the phosphate arms move significantly so that the 5′ γ-phosphate group of pppGpp assumes the position of the γ-phosphate group of ATP prior to the reaction. Moreover, helix α2 is ∼1.5 Å closer to the guanosine nucleotide in the pre-catalytic state compared with the post-catalytic state. The guanine base stacks on Tyr-151 in the pre-catalytic state similar to what was observed for SeRel (PDB code 1VJ7) in complex with GDP, where it corresponds to Tyr-308 (Fig. S3) (11). Consistent with the central role of this residue, mutation of Tyr-151 to alanine renders SaRelP completely inactive (Fig. 2B) without affecting the tetrameric state of the enzyme (Fig. S4A). Recognition of GDP in SeRel also involves Lys-304, Asn-306, Asp-264, Glu-323, and His-312, which are all conserved in SaRelP and correspond to Lys-147, Asn-149, Asp-107, Glu-174, and His-155, respectively. Together, the pre-catalytic and post-catalytic structures presented here thus complete our picture of the reaction taking place in the SAS enzymes and allow for a detailed analysis of how these enzymes specifically transfer a pyrophosphate from ATP to the 3′-OH position of guanosine nucleotides.
SaRelP lacks the cleft allosteric pppGpp-binding site
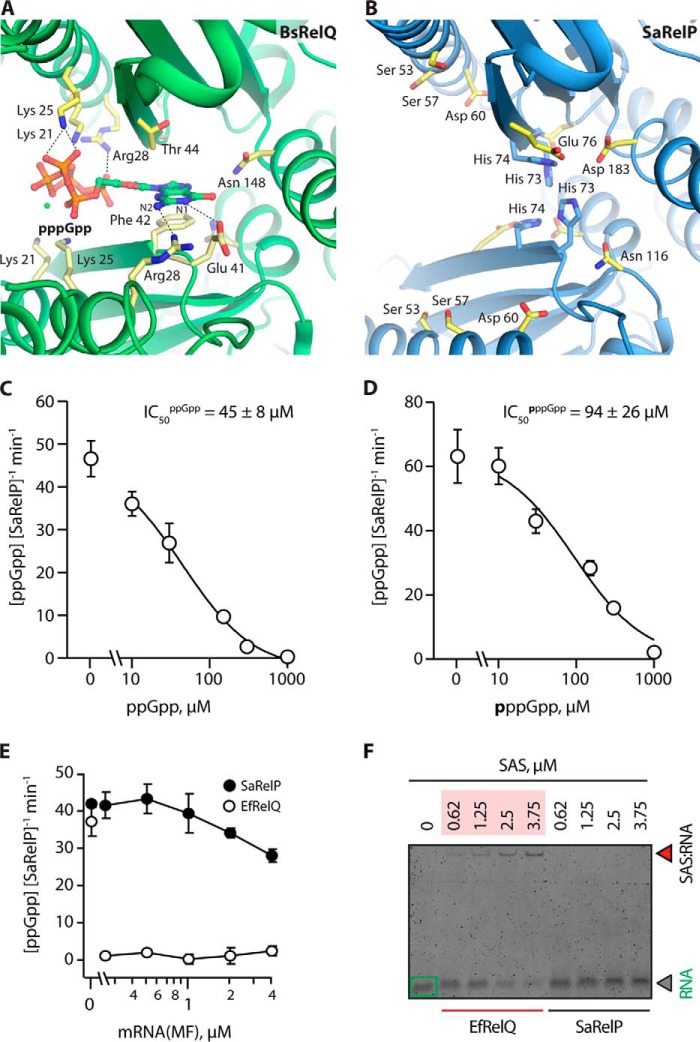
The structure of BsRelQ revealed an unexpected allosteric binding site at the subunit interface between two dimers that was shown to positively affect activity upon binding of pppGpp but not ppGpp (16). In this site, the negative charges of pppGpp are balanced by pairs of Lys-21, Lys-25, and Arg-28 residues originating from neighboring chains on either side of the subunit cleft (Fig. 3A). Phe-42 stacks with the guanine base, one Arg-28 residue interacts with the N2 atom, whereas Glu-41 forms a hydrogen bond with the N1 position ensuring specificity for guanosine (16). Interestingly, none of the residues that interact specifically with pppGpp at the BsRelQ allosteric site are conserved in SaRelP (Fig. 3B and Fig. S3). Lys-21 and Lys-25 have both been replaced by serine residues (Ser-53 and Ser-57, respectively), Arg-28 has been replaced by Asp-50 and thus has an inverted charge, whereas Phe-42 is replaced by His-73. Moreover, Ser-38, which allows Arg-28 to contact the guanine base in BsRelQ, is replaced by the larger Asn-70 in SaRelP. Altogether, a clear pattern of differences between RelP (YwaC) and RelQ (YjbM) enzymes in the region around α1 and β1 emerges that indicate that they bind different regulatory molecules (Fig. S3). In summary, structural and sequence data strongly indicate that SaRelP does not bind pppGpp at the cleft allosteric binding site and consequently that it may be regulated in a different way.
Figure 3.
SaRelP is negatively regulated by (p)ppGpp-binding to the active site. A, overview of the allosteric pppGpp-binding site in BsRelQ (PDB entry 5DED) with relevant interacting side chains labeled (21). B, the same site in SaRelP with equivalent residues (as per the alignment) labeled. The positively charged Lys-21, Lys-25, and Arg-28 have been replaced by Ser-53, Ser-57, and Asp-60, respectively, thus changing the electrostatic behavior of the binding site. His-73 and His-74, which are unique to SaRelP, are shown in blue. C, activity of SaRelP (measured as production of ppGpp from GDP and ATP per enzyme/min) in the presence of increasing amounts of ppGpp. The calculated IC50 value is shown. D, same titration as in C but using pppGpp. Error bars represent S.D. of the turnover estimates determined by linear regression. Each experiment was performed at least three times. E, effect of model single-stranded mRNA (MF) on the enzymatic activity of EfRelQ (empty circles) and SaRelP (filled circles) measured as production of ppGpp from GDP and ATP per enzyme/min. Enzymatic assays were performed with 250 nm (62.5 nm tetramer) enzyme, 200 μm [3H]GDP, and 1 mm ATP. Error bars represent S.D. of the turnover estimates determined by linear regression; each experiment was performed at least three times. F, EMSA analysis of mRNA (MF) complex formation using increasing concentrations of EfRelQ and SaRelP. EMSAs were performed using 0.19 μm mRNA (MF) and increasing concentrations of SaRelP or EfRelQ as indicated on the figure.
SaRelP is inhibited by (p)ppGpp but not single-stranded RNA
Because the cleft allosteric pppGpp-binding site appears to be lacking in SaRelP, we next wanted to understand if the product nucleotides pppGpp and ppGpp have any effect on the activity of the enzyme. For this analysis, we employed the approach previously used for EfRelQ, where the synthesis of ppGpp is followed by thin layer chromatography after incubation of purified enzyme with 3H-labeled GDP and ATP as substrates (19). For SaRelP, upon addition of increasing amounts of either ppGpp (Fig. 3C) or pppGpp (Fig. 3D) to this reaction, we observe gradual inhibition of ppGpp synthesis activity, with a complete loss of activity occurring at 1 mm concentration of either product nucleotide. We note that ppGpp appears to be a slightly more potent inhibitor (IC50ppGpp = 45 ± 8 μm) than pppGpp (IC50pppGpp = 94 ± 26 μm).
For EfRelQ, allosteric activation by pppGpp (but not ppGpp) was found to be mutually exclusive with inhibition through binding of single-stranded RNA suggesting that RNA competes for binding at the cleft allosteric site (19). Using EfRelQ as a positive control, we next tested if SaRelP is also affected by single-stranded RNA. However, unlike EfRelQ, SaRelP is neither inhibited by model mRNA (Fig. 3E) nor forms a protein–RNA complex as judged by electrophoretic mobility shift assay (EMSA) (Fig. 3F).
SaRelP contains an iron-binding site at the dimer interface
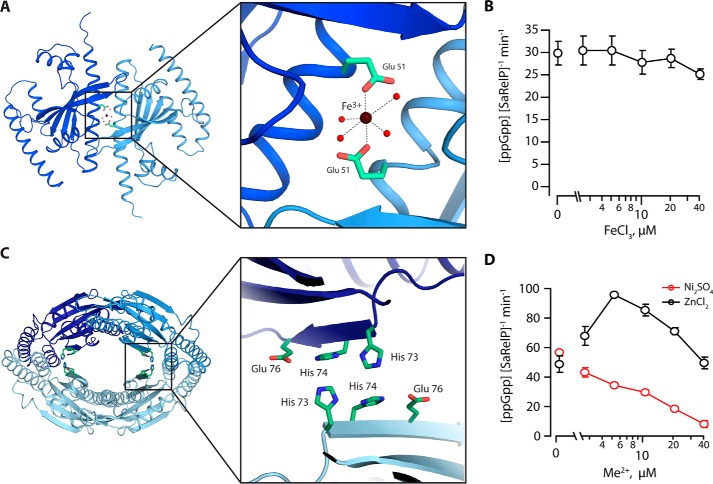
Inspection of the 2mFo − DFc density at the SaRelP dimer interface revealed a significant, spherical blob between the side chains of two equivalent Glu-51 residues from adjacent molecules, i.e. at the non-crystallographic 2-fold symmetry axis between monomers (Fig. S5A). To understand if this peak represents a metal ion–binding site, we performed broad wavelength range fluorescence scans on crystals of SaRelP in the pppGpp-bound form, which indicated that both Zn2+ and Fe3+ were present (data not shown). To check whether one of these metals could correspond to the site identified between the Glu-51 side chains, complete anomalous data sets were collected both above and below the K-edge absorption edges for Zn2+ and Fe3+ and used for calculation of anomalous difference maps (Table 1 and Fig. S5B). For the data collected above the Fe3+ K-edge at 7.200 keV, a significant anomalous peak (3.2σ) was observed between the two Glu-51 side chains, whereas there was no signal in the anomalous map below the absorption edge (6.900 keV) nor in the map calculated from data collected above the Zn2+ K-edge at 9.800 keV.
Based on these data and the presence of a peak in the 2 DFc − mFc refined maps of both SaRelP structures (pre-catalytic and post-catalytic), we conclude that iron (Fe2+ or Fe3+) binds at the dimerization interface. Because no Fe3+ was present in the buffers used for purification and crystallization, this ion must have been present in the protein during expression and carried along during purification. The ion displays a clear octahedral coordination geometry involving carboxylate oxygen atoms from the two Glu-51 side chains and four surrounding water molecules (Fig. 4A). In this context, we note that octahedral configuration is the preferred coordination for Fe3+, whereas the zinc ion (Zn2+) prefers tetragonal coordination and is very rarely octahedral (20). Glu-51 is highly conserved in the SAS enzyme family, but is, intriguingly, replaced by alanine in BsRelP (YwaC) suggesting that the iron-binding site is not present in this protein (Fig. S3). To check for a possible regulatory effect on enzyme activity, we repeated the activity assay in the presence of increasing concentrations of Fe3+ (Fig. 4B), which revealed no strong effect. Because Fe2+ and Fe3+ are readily interchangeable in aqueous solutions, Mn2+ is often used as a surrogate (21). We therefore also tested the effect of Mn2+ on the catalytic activity of SaRelP and consistently found no strong effect (Fig. S4B). We therefore conclude that SaRelP binds Fe2+ or Fe3+ at the dimer interface and that this might represent a structural ion-binding site.
Figure 4.
Metal ion ligand–binding sites in SaRelP. A, a top view of the SaRelP dimer shown with Fe3+ bound at the subunit–subunit interface of the dimer and coordinated by the two Glu-51 residues originating from neighboring chains. The inset displays a close-up view with the octahedral coordination of Fe3+ indicated. Fe3+ is shown as a brown sphere, relevant water molecules with red spheres, and the glutamate residues with green sticks. B, enzymatic activity of SaRelP (measured as production of ppGpp from GDP and ATP per enzyme per minute) as a function of increasing FeCl3 concentrations. Assays were performed using 250 nm SaRelP, 200 μm [3H]GDP, and 1 mm ATP. C, a putative second metal-binding site is found at the dimer–dimer interface of the tetramer. The inset shows a close-up view of the region with residues forming the site shown in green sticks. D, enzymatic activity of SaRelP as a function of increasing Ni2SO4 and ZnCl2 concentrations. Error bars represent S.D. of the turnover estimates determined by linear regression. Each experiment was performed at least three times.
SaRelP contains a putative Zn2+-binding site
Binding of pppGpp at the cleft allosteric site in BsRelP requires Arg-28, Glu-41, and Phe-42, which interact with the guanine N1/N2 atoms and stack with the base, respectively (Fig. 3A) (16). In SaRelP, these residues have been replaced by two histidines (His-73 and His-74) suggestive of another putative metal (possibly zinc)-binding site. The two histidine residues are located across from the equivalent residues in the other half of the tetramer and thus form a four-histidine site (Fig. 4C). Intriguingly, this unique combination of residues appears to be conserved among certain RelP/SAS2 enzymes, such as Staphylococcus schleiferi RelP (Fig. S3), whereas B. subtilis RelP (YwaC) contains one residue (Glu) from the BsRelQ motif (EF) and a histidine from the SaRelP motif (HH). To understand if Zn2+ could bind to this site, crystals of SaRelP in the pppGpp-bound state were soaked in 5 mm ZnCl2 before data collection. However, no difference in electron density peaks were observed following refinement (data not shown) and it is therefore presently uncertain if this site is able to bind a metal ion. Surprisingly, activity tests carried out in the presence of increasing amounts of Zn2+ demonstrated that the ion acts as an activator at lower concentrations (peaking at ∼5 μm), and as an inhibitor at higher concentrations (Fig. 4D). Ni2+, used here as a specificity control, had only the inhibitory effect, nearly completely abolishing the enzymatic activity at 40 μm (Fig. 4D). To test if this effect is mediated by the HH motif, we finally mutated the two histidine residues (His-73 and His-74) to alanine. Surprisingly, this mutation completely abolished the activity of the enzyme (Fig. 2B) but did not affect the tetrameric state (Fig. S4A). Thus, despite our inability to demonstrate structurally that Zn2+ binds at the His site, our data suggest that the site is indeed functionally important, and also that divalent metal ions can affect the enzyme activity. In summary, RelP/SAS2 (as opposed to RelQ/SAS1) enzymes appear to be regulated by metal ions and future investigations should therefore focus on revealing the mechanism by which this occurs.
Discussion
In this paper, we present crystal structures of S. aureus RelP/SAS2 in two key functional states, pre-catalytic and post-catalytic. Together, these structures provide a clear picture of the catalytic mechanism of the single-domain SAS enzymes (Fig. 5). The complete pre-catalytic state presented here demonstrates that Glu-174 plays a key role in organizing two water molecules that form part of the interaction sphere of a Mg2+ ion in the active site (Figs. 2C and 5). This residue corresponds to Glu-139 in BsRelQ, which was proposed to function as a general base and deprotonate the 3′-OH of GTP directly during the reaction (16). This appears not to be the case in SaRelP, because the Mg2+ ion is the only group that directly contacts the 3′-OH group of GTP in the pre-catalytic state. It is therefore rather the electrophilic Mg2+ ion that promotes deprotonation of the 3′-OH group and subsequently the nucleophilic attack of the resulting alkoxide (R-O−) on the β-phosphate of ATP (Fig. 5, left, curved arrow). The pentavalent transition state is stabilized by a large number of positively charged residues in the vicinity, one of which likely also serves as a general acid to protonate the phosphate leaving group on AMP. Interestingly, the conformation of the 5′- and 3′-phosphate arms in the product nucleotide (pppGpp) deviate from the position of the corresponding groups in the pre-catalytic state suggesting that some conformational change takes place during the reaction (Fig. 5).
Figure 5.
Mechanism of (p)ppGpp synthesis by SAS enzymes. Detailed catalytic mechanism for (p)ppGpp synthesis by SAS enzymes based on the structures of the pre-catalytic (left) and post-catalytic (right) state structures of SaRelP. In the pre-catalytic state (left), a Mg2+ ion is held firmly in place by tight octahedral coordination facilitated by the β- and λ-phosphate oxygen atoms of ATP, two water molecules (which again are constrained by both carboxylate oxygen atoms of Glu-174), one carboxylate oxygen from Asp-107, and the 3′-OH group of GTP. The electrophilic metal ion promotes deprotonation of the 3′-OH group on GTP and subsequently a nucleophilic attack (large, curved arrow) of the resulting alkoxide ion onto the β-phosphate group of ATP causing hydrolysis of the phosphate ester between the α- and β-phosphate groups (small, curved arrow). The developing negative charge of the pentavalent transition state is stabilized by the combined positive charge of two arginine residues and six lysine residues in the neighborhood (not all shown). In the post-catalytic state (right), the 5′ β- and λ-phosphate groups of pppGpp flip over and the λ-phosphate group assumes the position of the λ-phosphate group of ATP in the pre-catalytic state, stabilized through hydrogen bonding with Asp-107. Likewise, the new 3′ α- and β-phosphate groups shift position to generate the U-shaped conformation of pppGpp found in chain A of the post-catalytic state.
Although the reaction mechanisms of RelP and RelQ enzymes are very likely the same, our data also highlight the functional and structural differences between these homologous enzymes in Firmicute bacteria. We demonstrate that SaRelP, unlike BsRelQ, is not stimulated by pppGpp binding, in fact, we observe that the activity drops in the presence of both ppGpp and pppGpp. This, together with the observation that the allosteric pppGpp-binding site is absent from SaRelP, suggests that the effect we observe is orthosteric, i.e. arising from simple competitive product binding at the active site. In this scenario, one would predict that the relative level of inhibition would increase as the substrate (GDP) concentration is lowered due to competition for the same binding site. However, we do not observe this effect, which suggests that GDP and (p)ppGpp do not compete for the same binding sites directly (data not shown). It is, however, possible, that the effect is related to the tetrameric state of the enzyme and thus, cross-talk between active sites. For example, binding of a product nucleotide to one active site could negatively affect the other active sites in the tetramer without directly competing with binding of GDP.
If not regulated by (p)ppGpp, the question remains: what, if anything, regulates RelP/SAS2 in Firmicute bacteria, and why are there two homologous SAS enzymes? We present evidence for the existence of an iron-binding site located between monomers of a dimer and identify a second metal-binding site at the dimer–dimer interface. Surprisingly, we were not able to observe any significant effect on the catalytic rate upon addition of Fe3+, whereas Zn2+, which could not be located at the second binding site, did have an effect. Moreover, we note that the in vitro activity of SaRelP in the presence of Zn2+ shows a biphasic response curve: up to 4 μm the ion acts as an activator, whereas at higher concentrations, inhibition occurs (Fig. 4D). The location of two histidine residues adjacent to two equivalent histidines from the other half of the tetramer is unusual and highly suggestive of a zinc-binding site. But we have not been able to identify Zn2+ at this site, not even when the crystals were soaked in millimolar concentrations of Zn2+. Nevertheless, we speculate that the three unique combinations of residues (HH, EH, and EF) observed among homologous SAS sequences at this position (Fig. S3, green boxes) represent different specificities for ligands yet to be uncovered. The four histidine residues located very close to each other in SaRelP thus represent a different regulatory site than the one that has been observed in BsRelQ, which has Glu–Phe at this position. Although we have not been able to identify what binds to this site in SaRelP, we note the presence of a significant and continuous blob of electron density at the interface between dimers in the structure of the pre-catalytic state that is not compatible with a bound nucleotide (Fig. S5C). This density is found adjacent to the putative Zn2+ site, close to Arg-77 and the backbone of residues 73–77 and was only observed for the structure of the pre-catalytic state determined in the presence of AMP-CPP and GTP.
Altogether, our results suggest that Firmicute bacteria contain two separate SAS enzymes because they need to respond to more environmental signals than can be handled by a single allosteric site. Most likely, RelP/SAS2 arose by gene duplication and diversification eventually led to two enzymes catalyzing the same reaction but responding to different signals. Although the details of this diverse regulation are not yet clear, RelP/SAS2 appears to respond to metal ions to a greater extent than RelQ/SAS1, which likely responds only to nucleotides. In this context, it is interesting to note that the presence of Zn2+ ions in the medium has been observed to induce a stringent response and (p)ppGpp production in B. subtilis and that YwaC (SAS2/RelP) is one of several proteins that are up-regulated as a response (22). A working hypothesis could therefore be that RelP evolved to respond to oxidative stress induced by chelation of low amounts of metal ions present in the environment. This could also explain why we only see activation of SaRelP at low Zn2+ concentrations. At higher concentrations, it is possible that the presence of Zn2+ negatively affects the enzyme in a non-physiological way. Future research in the field should now be focused on understanding where metal ions interact with RelP/SAS2 and how this affects activity. Second, it remains possible that the enzyme is regulated by additional (larger) compounds, as indicated by our unexplained density at the tetramer interface. Careful purification and MS analysis of protein produced under stress conditions in S. aureus might allow identification of such metabolite(s).
Experimental procedures
Cloning of SaRelP
The complete S. aureus relP ORF (NWMN_2405) was amplified by PCR (Phusion Hot Start II DNA Polymerase, ThermoFisher Scientific) using S. aureus strain Newman chromosomal DNA as template and the primer pair: 5′-GATACATCTAGATTAAGAAGGAGATATACCATGTATGTAGATCGAAAACCATC-3′, 5′-CGCGGATCCTTAGTGATGGTGATGGTGATGACCCTCTGTTATTTCAGAATGAATTTG-3′, which contains an upstream ribosomal-binding site (rbs) and the coding sequence for a C-terminal His6 tag. The PCR fragment was cloned into pET28a using the XbaI/BamHI restriction sites and verified by sequencing. The Y151A and H73A/H74A mutants were constructed using the Agilent QuikChange Multi Lightning Kit, according to the instructions by the manufacturer.
Protein expression and purification
Escherichia coli BL21(DE3) competent cells carrying the pET28a-SaRelP-His expression plasmid were grown overnight in lysogeny broth (LB) medium supplemented with 50 μg/ml of kanamycin at 37 °C, then used to inoculate 4 liters of LB medium supplemented with kanamycin and grown at 37 °C until the A600 reached ∼0.6–0.7. The culture was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside and protein expression was allowed to proceed for 3 h at 30 °C in a shaking incubator. Cells were harvested (∼7,000 × g for 15 min), resuspended in a lysis buffer containing 50 mm Tris-HCl, pH 8.5, 500 mm NaCl, 5 mm MgCl2, 10 mm imidazole, 3 mm β-mercaptoethanol (β-ME), and 100 mm phenylmethylsulfonyl fluoride, and disrupted by sonication. Cell debris was pelleted by a fast centrifugation (∼23,000 × g for 45 min) and the supernatant applied to a 1-ml HisTrap HP Ni-affinity column (GE Healthcare) equilibrated with lysis buffer. After washing with 5–10 column volumes, the protein was eluted with a 5-step gradient using 30, 50, 75, 150, and 300 mm imidazole over 10 column volumes. At this stage, protein for biochemistry was concentrated and exchanged into 30 mm Tris, pH 8, 300 mm NaCl, 5 mm MgCl2, 5 mm β-ME, and 5% glycerol and used directly. Protein for crystallography was further purified on a 1-ml Source15Q (GE Healthcare) anion exchange column equilibrated in 50 mm Tris-HCl, pH 8.5, 100 mm NaCl, and 5 mm β-ME and eluted using a linear gradient into 1 m NaCl. The eluate was concentrated by spin filtration (VivaSpin 30,000 MWCO, Sartorius) and applied to a Superdex 200 Increase 10/300 size exclusion column (GE Healthcare) equilibrated in 30 mm Tris-HCl, pH 8.5, 300 mm NaCl, 5 mm MgCl2, and 5 mm β-ME. Peak fractions were pooled and concentrated to 10–15 mg/ml and the final protein purity was assessed by SDS-PAGE. Protein concentrations were measured spectroscopically using A280.
Crystallization
All crystallization experiments were carried out using the sitting drop vapor diffusion method, where the protein sample was mixed 1:1 with reservoir buffer in 96-well Swissci trays (Molecular Dimensions) using a Mosquito® liquid handler (TTP Labtech). For the pppGpp-bound structure, purified SaRelP (10 mg/ml) was incubated with 1 mm ATP and 1 mm GTP at 30 °C for 5 min, then kept on ice. Initial crystal hits were obtained after 2 days and optimized to a final condition containing 30–40% (v/v) pentaerythritol propoxylate (5/4 PO/OH), 0.2 m sodium thiocyanate, and 0.1 m Hepes-NaOH, pH 7.0, at 19 °C. For the AMP-CPP:GTP-bound structure, SaRelP (11 mg/ml) was incubated with 1 mm AMP-CPP and 1 mm GTP at 30 °C for 5 min, then kept on ice. Crystals were obtained at 19 °C in a buffer containing 25% (w/v) PEG 3350 and 0.1 m Tris-HCl, pH 8.5.
Data collection and structure determination
Crystals of SaRelP on the pppGpp-bound form grown in 32% (v/v) pentaerythritol propoxylate (5/4 PO/OH), 0.2 m sodium thiocyanate, and 0.1 m Hepes-NaOH, pH 7.0 were cryo-protected in the mother liquor by addition of 20% glycerol, whereas crystals on the AMP-CPP:GTP-bound form were cryo-protected by addition of 30% glycerol. Complete native diffraction data were collected at 100 K at the European Synchrotron Radiation Facility (ESRF) beamline ID29, Grenoble, France (pppGpp-bound form), and PETRA III, beamline P13, Hamburg, Germany (AMP-CPP:GTP-bound form and anomalous data). All data were processed using the xia2 automated processing system (23) using the XDS pipeline (24). Data quality was assessed using Phenix.xtriage (25). The structures were determined by molecular replacement in Phenix.phaser (26) using the BsRelQ structure (PDB code 5DED) as search model (16). The structures were built in Coot (27) and iteratively refined in Phenix.refine (25) with manual rebuilding. Figures were created using PyMOL (28). Anomalous data were collected from a crystal of SaRelP on the pppGpp-bound form above the Zn2+ K absorption edge at 1.26 Å (9.800 keV), and both below (1.79Å, 6.900 keV) and above (1.71Å, 7.200 keV) the Fe3+ K-edge. The structure was refined briefly against each of these data sets before anomalous difference maps were calculated in Phenix.
Enzymatic assays
Experiments were performed at 37 °C in Hepes:Polymix buffer with 5 mm Mg2+ (29, 30) supplemented with 1 mm β-ME (see supporting Experimental procedures for details). Reaction mixtures containing 250 nm SaRelP or EfRelQ and 200 μm [3H]GDP were preincubated for 2 min at 37 °C in the presence of increasing concentrations of different effectors; nucleotides, metal ions (Ni2SO4, ZnCl2, or FeCl3), or model mRNA coding for a Met-Phe (MF) dipeptide, 5′-GGCAAGGAGGUAAAAAUGUUCAAA-3′ (19). Reactions were initiated by addition of 1 mm ATP after which 5-μl aliquots were taken throughout the time course of the reaction. Aliquots were quenched with 4 μl of 70% formic acid supplemented with a cold nucleotide standard (10 mm GDP + 10 mm GTP) and analyzed by UV-shadowing after separation on polyethyleneimine-TLC plates (Macherey-Nagel). TLC analysis was performed as described by Mechold and colleagues (31) with modifications. Nucleotides were resolved in 0.5 m KH2PO4, pH 3.5, buffer, and plates were dried and cut into sections as guided by UV-shadowing. 3H radioactivity was quantified by scintillation counting using Optisafe-3 (Fisher) scintillation mixture. Conversion of substrate to product was quantified as described previously (32).
Electrophoretic mobility shift assay
Reactions were carried out in 8 μl total in Hepes:Polymix buffer as described above. Prior to assembling the reaction mixtures, stock RNA was incubated for 2 min at 65 °C to melt potential secondary structure. Reaction mixtures were assembled by adding increasing concentrations of SaRelP or EfRelQ to the RNA (0.19 μm final concentration), followed by addition of 4 units/μl of RiboLock RNase Inhibitor (Thermo Scientific). After incubation for 10 min at 37 °C, 5 μl of loading dye, 40% (w/v) sucrose supplemented with bromphenol blue, was added per 8 μl (i.e. per 1.5 pmol of mRNA) and the samples resolved on 12–15% Tris:borate:EDTA gels run at 4 °C (120–140 V) for 1.5–2 h. Gels were stained with SYBR Gold nucleic acid stain (Life Technologies) for 20 min prior to visualization using a Typhoon Trio variable mode imager (Amersham Biosciences).
Author contributions
M. C. M., M. S. B., T. T., H. I., V. H., and D. E. B. conceptualization; M. C. M., J. B., and M. S. B. data curation; M. C. M., J. B., H. I., V. H., and D. E. B. formal analysis; M. C. M., J. B., V. H., and D. E. B. validation; M. C. M., J. B., M. S. B., T. T., H. I., V. H., and D. E. B. investigation; M. C. M., J. B., V. H., and D. E. B. visualization; M. C. M., J. B., M. S. B., T. T., H. I., V. H., and D. E. B. writing-original draft; M. C. M., J. B., M. S. B., T. T., H. I., V. H., and D. E. B. writing-review and editing; M. S. B., T. T., H. I., V. H., and D. E. B. resources; T. T., H. I., V. H., and D. E. B. supervision; T. T., H. I., V. H., and D. E. B. funding acquisition; T. T., H. I., V. H., and D. E. B. project administration.
Supplementary Material
Acknowledgments
We thank Rasmus Koch Flygaard and the beamline staff at ESRF and EMBL PETRA beamlines ID29 and P13 for help during data collection.
This work was supported by Danish National Research Foundation Grant DNRF120 (to D. E. B.), Estonian Research Council Grant IUT2-22 (to T. T.), the European Regional Development Fund through the Centre of Excellence for Molecular Cell Technology (to V. H. and T. T.), Swedish Research Council Vetenskapsrådet Grant 2013–4680 (to V. H.), and the Ragnar Söderberg foundation (to V. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains ”Experimental procedures” and Figs. S1–S5.
The atomic coordinates and structure factors (codes 6EX0 and 6EWZ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ppGpp
- guanosine-5′-(di)phosphate-3′-diphosphate
- pppGpp
- guanosine-5′-(tri)phosphate-3′-diphosphate
- RSH
- RelA-SpoT homologue
- SAS
- small alarmone synthetase
- AMP-CPP
- adenosine 5′-(α,β-methylene)triphosphate
- EMSA
- electrophoretic mobility shift assay
- PDB
- Protein Data Bank
- β-ME
- β-mercapthoethanol.
References
- 1. Cashel M., and Kalbacher B. (1970) The control of ribonucleic acid synthesis in Escherichia coli: V. characterization of a nucleotide associated with the stringent response. J. Biol. Chem. 245, 2309–2318 [PubMed] [Google Scholar]
- 2. Pao C. C., and Dyess B. T. (1981) Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim. Biophys. Acta 677, 358–362 10.1016/0304-4165(81)90247-6 [DOI] [PubMed] [Google Scholar]
- 3. Potrykus K., and Cashel M. (2008) (p) ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 4. Dalebroux Z. D., Svensson S. L., Gaynor E. C., and Swanson M. S. (2010) ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199 10.1128/MMBR.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harms A., Maisonneuve E., and Gerdes K. (2016) Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, aaf4268 10.1126/science.aaf4268 [DOI] [PubMed] [Google Scholar]
- 6. Cashel M., and Gallant J. (1969) Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841 10.1038/221838a0 [DOI] [PubMed] [Google Scholar]
- 7. Atkinson G. C., Tenson T., and Hauryliuk V. (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6, e23479 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arenz S., Abdelshahid M., Sohmen D., Payoe R., Starosta A. L., Berninghausen O., Hauryliuk V., Beckmann R., and Wilson D. N. (2016) The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 44, 6471–6481 10.1093/nar/gkw470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown A., Fernández I. S., Gordiyenko Y., and Ramakrishnan V. (2016) Ribosome-dependent activation of stringent control. Nature 534, 277–280 10.1038/nature17675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loveland A. B., Bah E., Madireddy R., Zhang Y., Brilot A. F., Grigorieff N., and Korostelev A. A. (2016) Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hogg T., Mechold U., Malke H., Cashel M., and Hilgenfeld R. (2004) Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates(p) ppGpp metabolism during the stringent response (corrected). Cell 117, 57–68 10.1016/S0092-8674(04)00260-0 [DOI] [PubMed] [Google Scholar]
- 12. Avarbock D., Avarbock A., and Rubin H. (2000) Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA·ribosome·mRNA·RelMtb complex. Biochemistry 39, 11640–11648 10.1021/bi001256k [DOI] [PubMed] [Google Scholar]
- 13. Haseltine W. A., and Block R. (1973) Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 70, 1564–1568 10.1073/pnas.70.5.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemos J. A., Lin V. K., Nascimento M. M., Abranches J., and Burne R. A. (2007) Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581 10.1111/j.1365-2958.2007.05897.x [DOI] [PubMed] [Google Scholar]
- 15. Nanamiya H., Kasai K., Nozawa A., Yun C. S., Narisawa T., Murakami K., Natori Y., Kawamura F., and Tozawa Y. (2008) Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304 [DOI] [PubMed] [Google Scholar]
- 16. Steinchen W., Schuhmacher J. S., Altegoer F., Fage C. D., Srinivasan V., Linne U., Marahiel M. A., and Bange G. (2015) Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl. Acad. Sci. U.S.A. 112, 13348–13353 10.1073/pnas.1505271112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geiger T., Kästle B., Gratani F. L., Goerke C., and Wolz C. (2014) Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 196, 894–902 10.1128/JB.01201-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boucher H., Miller L. G., and Razonable R. R. (2010) Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 51, S183–197 10.1086/653519 [DOI] [PubMed] [Google Scholar]
- 19. Beljantseva J., Kudrin P., Andresen L., Shingler V., Atkinson G. C., Tenson T., and Hauryliuk V. (2017) Negative allosteric regulation of Enterococcus faecalis small alarmone synthetase RelQ by single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 114, 3726–3731 10.1073/pnas.1617868114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laitaoja M., Valjakka J., and Jänis J. (2013) Zinc coordination spheres in protein structures. Inorg. Chem. 52, 10983–10991 10.1021/ic401072d [DOI] [PubMed] [Google Scholar]
- 21. Friedman Y. E., and O'Brian M. R. (2004) The ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum is an iron-responsive transcriptional repressor in vitro. J. Biol. Chem. 279, 32100–32105 10.1074/jbc.M404924200 [DOI] [PubMed] [Google Scholar]
- 22. Luche S., Eymard-Vernain E., Diemer H., Van Dorsselaer A., Rabilloud T., and Lelong C. (2016) Zinc oxide induces the stringent response and major reorientations in the central metabolism of Bacillus subtilis. J. Proteomics 135, 170–180 10.1016/j.jprot.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 23. Winter G. (2010) xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 [Google Scholar]
- 24. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 28. Schrodinger L. L. C. (2015) The PyMOL Molecular Graphics System, version 1.8, Schroedinger, LLC, New York [Google Scholar]
- 29. Antoun A., Pavlov M. Y., Tenson T., and Ehrenberg M. (2004) Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol. Proced. Online 6, 35–54 10.1251/bpo71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jelenc P. C., and Kurland C. G. (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. U.S.A. 76, 3174–3178 10.1073/pnas.76.7.3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mechold U., Murphy H., Brown L., and Cashel M. (2002) Intramolecular regulation of the opposing (p) ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888 10.1128/JB.184.11.2878-2888.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shyp V., Tankov S., Ermakov A., Kudrin P., English B. P., Ehrenberg M., Tenson T., Elf J., and Hauryliuk V. (2012) Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 13, 835–839 10.1038/embor.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.