Abstract
Mitochondrial DNA mutations have been reported to be associated with bipolar disorder (BD). In this study, we performed genome-wide analyses to assess mitochondrial single-nucleotide polymorphism (mtSNP) effects on BD risk and early-onset BD (EOBD) among BD patients, focusing on interaction effects between nuclear SNPs (nSNPs) and mtSNPs. Common nSNP and mtSNP data from European American BD cases (n = 1,001) and controls (n = 1,034) from the Genetic Association Information Network BD study were analyzed to assess the joint effect of nSNP and nSNP-mtSNP interaction on the risk of BD and EOBD. The effect of nSNP-mtSNP interactions was also assessed. For BD risk, the strongest evidence of an association was obtained for nSNP rs1880924 in MGAM and mtSNP rs3088309 in CytB (pjoint = 8.2 × 10−8, pint = 1.4 × 10−4). Our results also suggest that the minor allele of the nSNP rs583990 in CTNNA2 increases the risk of EOBD among carriers of the mtSNP rs3088309 minor allele, while the nSNP has no effect among those carrying the mtSNP major allele (OR = 4.53 vs. 1.05, pjoint = 2.1 × 10−7, pint = 1.16 × 10−6). While our results are not statistically significant after multiple testing correction and a large-sample replication is required, our exploratory study demonstrates the potential importance of considering the mitochondrial genome for identifying genetic factors associated with BD.
Keywords: Mitochondrial genome, Bipolar disorder, Genome-wide association study, Interaction, Single-nucleotide polymorphism
Introduction
Bipolar disorder (BD) is a serious and persistent mental illness characterized by recurrent episodes of mania/hypomania and depression [1,2]. Among all mental and neurological disorders, BD was ranked fourth as a cause of global burden based on disability-adjusted life years [3]. Many genetic studies, including both candidate gene studies and genome-wide association studies (GWAS), have been performed to identify BD susceptibility genes. By searching the nuclear genome, GWAS of large samples have identified multiple loci associated with the risk of BD, including CACNA1C, ANK3, ODZ4, SYNE1, and TRANK1 [4,5]. However, genetic variants from these loci only explain a small portion of BD heritability [6].
Mitochondrial dysfunction has been implicated in BD [7,8,9,10]. The energy dysregulation has been characterized by increased reactive oxygen species production, decreased mitochondrial complex subunits in the brain, ATP-dependent proteasome degradation, and an increase in lactate with a corresponding decreased intracellular pH [11,12,13,14,15,16]. Because mitochondrial DNA (mtDNA) encodes a number of mitochondrial proteins, it has been hypothesized that inherited variation in the mitochondrial genome may affect mitochondrial dysfunction and thus BD risk. Consistent with this hypothesis, clinical studies have found that subjects with maternal relatives with BD have a higher disease risk than those with paternal relatives with BD, supporting a potential maternal mtDNA transmission of risk [17]. In addition, a recent study based on induced pluripotent stem cell lines showed that mtDNA genes were differently expressed in young BD hippocampal neurons compared to normal neurons and in lithium-responsive BD neurons with and without lithium treatment [18]. Research on the role of the mitochondrial genome in BD risk has revealed candidate associations with mitochondrial point mutations, deletions, haplogroups (subjects sharing the same maternal ancestral haplotypes), and copy number variations [19,20,21,22]. For instance, the rare mtDNA mutation 3644T>C was found to be associated with BD [7,19]. In addition, various mitochondrial single-nucleotide polymorphisms (mtSNPs) have been proposed to be associated with BD [19,22,23,24]. For example, the 10398A variant was significantly associated with the risk of BD and a better response to lithium, as well as impaired prefrontal glucose utilizations [23,25,26]. MtDNA deletion (4,977-bp deletion), known as “the common deletion,” was shown to be overrepresented in BD brain tissue compared to controls, in particular in the dorsolateral prefrontal cortex [21]. With regard to mitochondrial haplogroups, overrepresentation of N9a in BD patients has been reported [22]. In addition, some haplogroups (U, K, and Uk) showed significantly lower cerebellar pH, leading to speculation that pH variation in the brain could be inherited through mtDNA and constitute a risk factor for BD. A decrease in mtDNA copy numbers has also been observed in brains of BD subjects [27]. These studies suggest that mitochondrial genome variation plays a role in BD; however, they are mainly small studies without adequate multiple testing correction and no replication, and thus further research is needed to determine the impact of mtDNA variation on the risk of BD and related phenotypes.
It is also important to note that mitochondrial proteins are encoded by both nuclear and mtDNA, with the vast majority of genes encoding mitochondrial proteins being products of nuclear DNA (nDNA). In fact, MitoCarta, which is an inventory of genes encoding proteins with strong support of mitochondrial localization, includes more than 1,000 genes encoded by the human nuclear genome [28]. The products of these nuclear and mitochondrial genes work together to control transcription and translation of mitochondrial genes and to form the complexes of the respiratory chain that carry out oxidative phosphorylation and energy production. Therefore, genetic variations from both mtDNA and nDNA may affect BD risk by influencing mitochondrial function. While further investigation of the role of mitochondrion-related genes in BD is needed, variation in certain mitochondrion-related nDNA genes has been reported to be associated with BD. For example, DISC-1, which encodes a protein involved in mitochondrial dynamics and trafficking, was identified as a potential susceptibility gene for schizophrenia and BD in a large Scottish pedigree [29,30]. Studies have also suggested that genetic variation in the promoter region of NDUFV2, a mitochondrial complex I gene, is associated with BD [31,32]. A potential role of this gene in BD was also supported by the observed downregulation of NDUFV2 in lymphoblastoid cell lines from patients with BD type I but not BD type II [33]. While the association of BD with nDNA-encoded mitochondrial genes is not yet well established, it warrants further investigation.
Many commonly used genome-wide genotyping platforms include mtSNP content and thus many existing GWAS datasets contain mtSNP data. However, mtSNPs have typically been excluded from complex-trait genetic risk discovery efforts, and studies incorporating both nuclear and mitochondrial genome data are extremely rare [34]. Exceptions include a study of outcomes following traumatic brain injury (interaction between nuclear gene APOE and mitochondrial haplogroup K), a study of late-onset Alzheimer's disease (interaction between nuclear variant APOE4+ status and mitochondrial subhaplogroup H5), and an ulcerative colitis study (mtSNP A10550G being an independent risk factor from nucleus-encoded susceptibility loci) [34,35,36].
In BD, a recent GWAS using the Genetic Association Information Network (GAIN) BD dataset reported that 2 mtSNPs in ND1 (rs28357968) and CytB (rs28357375) showed a nominal association (without correcting for multiple testing) with the risk of BD [37]. However, no prior studies in BD have investigated potential interaction effects of genetic variations from both genomes despite the fact that mitochondrial proteins are encoded by both genomes.
In this study, we performed genome-wide analyses to assess mtSNP effects on BD risk, focusing on interaction effects between nuclear SNPs (nSNPs) and mtSNPs. We hypothesized that the impact of nuclear genetic variation on BD risk could be modified by the mtDNA that a person carries and perhaps more nuclear BD risk loci would be uncovered by incorporating mitochondrial interactions into the risk model. In addition, we also investigated a potential role of mtSNP-nSNP interactions for early-onset BD (EOBD) compared to later-onset cases. Given previous studies showing the significance of age of onset of BD as a predictor of familial risk (relatives of EOBD patients had significantly greater risks of BD than those of later-onset BD patients) [38], we hypothesized that by considering both the nuclear and the mitochondrial genomes additional genetic risk loci could be uncovered that influence age at BD onset.
Methods and Materials
Study Description
This study utilized publically available GWAS data from the GAIN BD study, which included 2,035 European American subjects (i.e., 1,001 BD cases [EOBD, n = 419] and 1,034 controls) [39]. All BD cases met criteria for DSM-IV-defined bipolar I disorder, and controls were matched on age, sex, and ethnicity [39,40]. The age at BD onset (in years) was obtained from diagnostic interviews for the BD cases; EOBD was defined as a BD onset age of 19 years or younger [41]. Genotyping was performed using the Affymetrix 6.0 array that included nSNPs and mtSNPs. Basic quality control criteria were applied to exclude SNPs with a low minor allele frequency (MAF; <10%) and low call rates (<95%), resulting in 544,209 nSNPs and 15 mtSNPs [42]. We chose a higher cutoff for MAF (i.e., 10% rather than a standard cutoff of 1-5%), because this study focused on nSNP-mtSNP interaction effects, and the power to detect interactions declines more rapidly with decreasing MAF than does the power to detect main effects. A list of the 15 mtSNPs included in our analyses with detailed annotation information is presented in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000464444).
Statistical Analysis
For the nuclear genome, the effective number of independent SNPs was calculated to be used for Bonferroni multiple testing correction [43]. For the mitochondrial genome, the number of principal components (PCs) explaining at least 95% of the variability in mitochondrial genotypes was calculated. The total number of tests to be corrected for this study was then set to the effective number of independent nSNPs (n = 273,063) multiplied by the number of PCs from the mitochondrial genome (5 PCs). Therefore, association results with p < 3.7 × 10−8 (=273,063 × 5) were considered statistically significant after multiple testing correction.
For each outcome (BD risk and EOBD among BD cases), logistic regression models were used to assess the joint effects of the nSNP main effect and the interaction effect between nSNP and mtSNP, for each pair of mtSNP and nSNP, using 2df likelihood ratio tests. The 2df test jointly evaluates the effect of the last 2 terms (i.e., the nSNP effect and the nSNP × mtSNP interaction effect) from the following logistic regression model:
| Logit (p) = a₀ + a1 × mtSNP + a2 × nSNP + a3 × nSNP × mtSNP, |
where p is the probability of the outcome (e.g., BD vs. control), and a₀/a1/a2/a3 are regression parameters for terms included in the model. The rationale for this analysis is to jointly assess the effect of the nSNP main effect and/or the nSNP × mtSNP interaction term (i.e., testing if both a2 = 0 and a3 = 0), thereby testing the nSNP effect while allowing for modification of the effect by the mtSNP genotype. In addition, 1df likelihood ratio tests of the nSNP × mtSNP interaction terms were also performed (i.e., test assessing whether a3 = 0).
All models were further adjusted for the first PC from the nuclear genome (nPC1), and the interaction between nPC1 and mtSNP to prevent potential confounding by population stratification. For mtSNPs showing the strongest associations based on the joint tests, genome-wide analyses were repeated using imputed SNP data to generate Manhattan plots and locus zoom plots. Genome-wide imputation for the GAIN BD data was conducted using the 1,000 Genome Project cosmopolitan reference panel, and the detailed procedures have been previously described [44].
In addition to SNP level analysis, gene set analyses were conducted using a competitive method implemented in MAGENTA [45] to test whether nSNPs in mitochondrion-related genes were more enriched for interactions with individual mtSNPs compared to those not related to mitochondria. A gene set analysis was performed for each outcome (risk of BD and EOBD), comparing the mtSNP interaction results for nSNPs in 967 autosomal mitochondrial genes described in MitoCarta [28], with mtSNP interaction results for autosomal nSNPs outside of this set of 967 genes. Since both outcomes are considered highly polygenic, a gene set enrichment analysis (GSEA) cutoff using the top 25% of gene level statistics, recommended by the authors of MAGENTA, was used for comparison between the 967 genes in MitoCarta and those not in the MitoCarta list. Gene boundaries were based on the provided build 37 locations in the MAGENTA software with a 40-kb downstream and 110-kb upstream buffer for each gene. The HLA genes were removed as suggested by the authors of MAGENTA. GSEA p values were computed via permutation, with a minimum of 10,000 and a maximum of 1,000,000 permutations.
Results
None of the association results from the 2df tests or the 1df interaction tests passed the stringent significance threshold corrected for multiple testing (p < 3.7 × 10−8). Tables 1 and 2 present the top 3 association signals based on the 2df test for the analyses of BD risk and EOBD, respectively; Tables 1 and 2 also include 2 subsequent association results for each of the 3 top mtSNPs. In Tables 1 and 2 we report top findings based on the joint effect (2df test), while not including any results with p > 0.001 for the nSNP × mtSNP interaction effect. We only present joint effect test results for pairs of SNP with at least “marginal” evidence of an interaction effect (defined as pint ≤ 0.001), because the association results for nSNP-mtSNP pairs with little evidence for an interaction effect (pint > 0.001) are mainly driven by the nSNP main effects that have been extensively tested in prior GWAS studies and are not the focus of this study.
Table 1.
Top 3 association results based on 2df tests for risk of bipolar disorder, and 2 subsequent association results for each of the top mtSNPs
| mtSNP |
nSNP |
Association results |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | BP | gene | MA | MAF | SNP | Chr: BP | closest gene | MA | MAF | 2df p value | interaction p value | OR1 | OR2 |
| rs3088309 | 15,452 | CytB | T | 0.21 | rs1880924 | 7: 141717225 | MGAM | A | 0.14 | 8.2×10−8 | 1.4×10−4 | 1.27 | 3.42 |
| rs12733666 | 1: 77927820 | AK5 | C | 0.18 | 2.9×10−6 | 7.1×10−7 | 1.14 | 0.38 | |||||
| rs7782502 | 7: 105695500 | CDHR3 | G | 0.40 | 6.6×10−6 | 3.6×10−5 | 0.72 | 1.45 | |||||
| rs3915952 | 11,251 | ND4 | C | 0.22 | rs1880924 | 7: 141717225 | MGAM | A | 0.14 | 4.1×10−7 | 6.9×10−4 | 1.29 | 3.10 |
| rs7782502 | 7: 105695500 | CDHR3 | G | 0.40 | 2.6×10−6 | 1.2×10−5 | 0.71 | 1.48 | |||||
| rs10948994 | 6: 56184046 | COL21A1 | G | 0.35 | 2.6×10−6 | 4.7×10−4 | 1.50 | 0.93 | |||||
| rs28358279 | 10,463 | tRNA | G | 0.11 | rs1124376 | 3: 20108546 | PCAF/KAT2B | A | 0.22 | 2.8×10−6 | 5.0×10−5 | 0.87 | 0.26 |
| rs10000984 | 4: 175644564 | GLRA3 | G | 0.44 | 2.8×10−6 | 9.8×10−7 | 0.97 | 3.06 | |||||
| rs11764581 | 7: 141712467 | MGAM | C | 0.13 | 7.0×10−6 | 6.7×10−4 | 1.25 | 3.85 | |||||
MtSNP-nSNP interaction test results are also shown for the same SNP pairs. mtSNP, mitochondrial single-nucleotide polymorphism; nSNP, nuclear single-nucleotide polymorphism; SNP, single-nucleotide polymorphism; BP, base pair; MA, minor allele; MAF, minor allele frequency; Chr, chromosome.
In the presence of a major mtSNP allele, OR for adding 1 copy of an nSNP minor allele based on the interaction analysis.
In the presence of a minor mtSNP allele, OR for adding 1 copy of an nSNP minor allele based on the interaction analysis.
Table 2.
Top 3 association results based on 2df tests for risk of early-onset bipolar disorder, and 2 subsequent association results for each of the top mtSNPs
| mtSNP |
nSNP |
Association results |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | BP | gene | MA | MAF | SNP | Chr: BP | closest gene | MA | MAF | 2df p value | interaction p value | OR1 | OR2 |
| rs3088309 | 15,452 | CytB | T | 0.21 | rs583990 | 2: 80213652 | CTNNA2 | A | 0.25 | 2.1×10−7 | 1.6×10−6 | 1.05 | 4.53 |
| rs9933834 | 16: 70683178 | IL34 | C | 0.34 | 1.1×10−6 | 4.7×10−5 | 1.69 | 0.60 | |||||
| rs11122534 | 1: 230688466 | LOC729257 | C | 0.17 | 1.4×10−5 | 2.7×10−4 | 0.82 | 0.19 | |||||
| rs3915952 | 11,251 | ND4 | C | 0.22 | rs9933834 | 16: 70683178 | IL34 | C | 0.34 | 2.9×10−7 | 1.1×10−5 | 1.74 | 0.58 |
| rs556003 | 2: 80216399 | CTNNA2 | G | 0.38 | 3.1×10−7 | 3.2×10−7 | 0.95 | 3.75 | |||||
| rs1975145 | 18: 55931179 | NEDD4L | T | 0.21 | 5.4×10−6 | 1.4×10−6 | 1.49 | 0.32 | |||||
| rs3928306 | 3,010 | RNR2 | A | 0.24 | rs2420932 | 10: 123118218 | FGFR2 | G | 0.18 | 4.5×10−7 | 9.4×10−8 | 1.29 | 0.20 |
| rs6597183 | 6: 6042429 | NRN1 | T | 0.22 | 1.4×10−6 | 2.1×10−5 | 1.07 | 1.07 | |||||
| rs7987059 | 13: 27541625 | LOC100129306 | T | 0.19 | 1.2×10−5 | 2.5×10−6 | 0.66 | 0.65 | |||||
MtSNP-nSNP interaction test results are also shown for the same SNP pairs. mtSNP, mitochondrial single-nucleotide polymorphism; nSNP, nuclear single-nucleotide polymorphism; SNP, single-nucleotide polymorphism; BP, base pair; MA, minor allele; MAF, minor allele frequency; Chr, chromosome.
In the presence of a major mtSNP allele, OR for adding 1 copy of an nSNP minor allele.
In the presence of a minor mtSNP allele, OR for adding 1 copy of an nSNP minor allele.
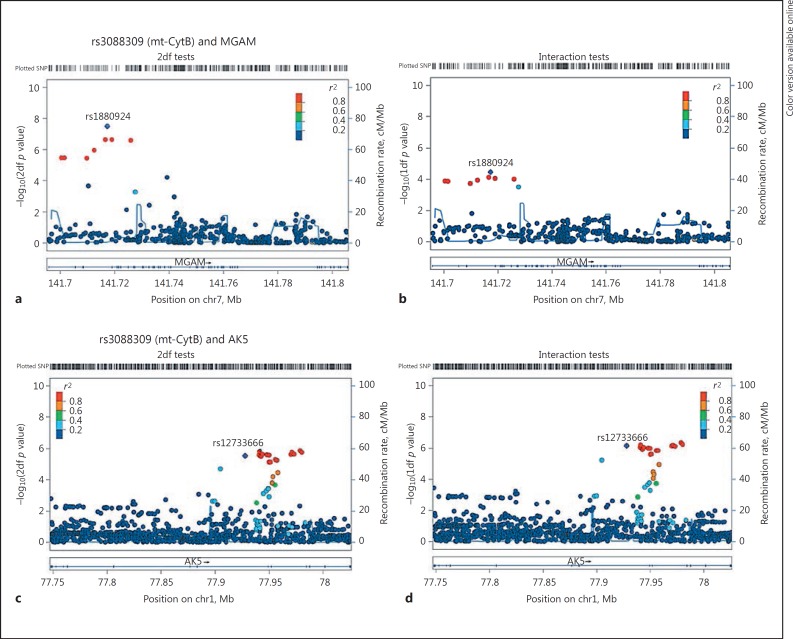
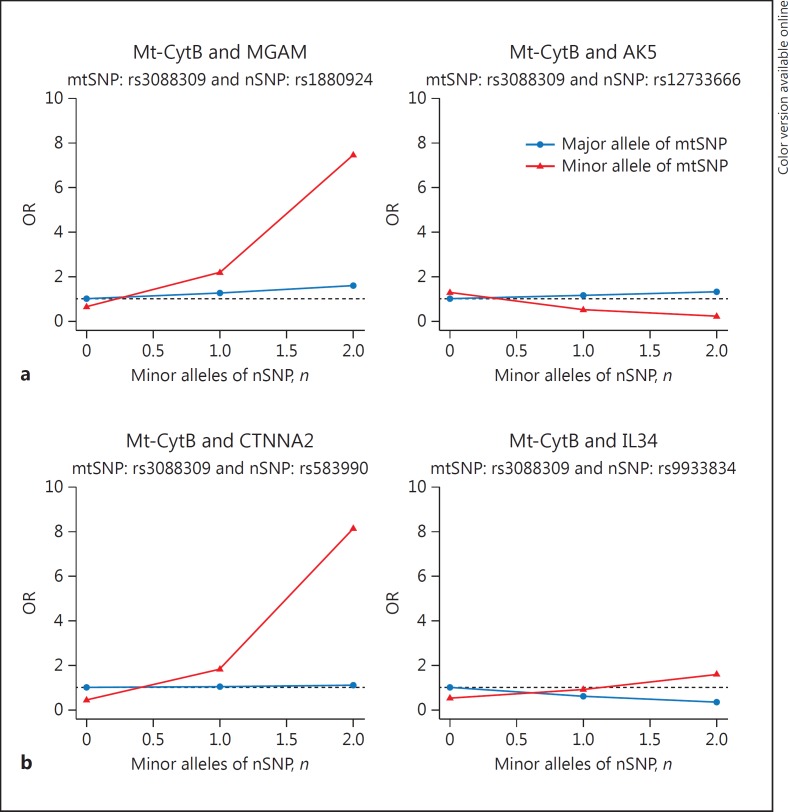
The strongest evidence of an association with BD risk was observed for mtSNP rs3088309 (MAF = 0.21 for allele T) located in the mitochondrial encoded cytochrome B (CytB) gene and the nSNP rs1880924 (MAF = 0.14 for allele A) located in the maltase-glucoamylase (MGAM) on chromosome 7, a gene involved in glycosaminoglycan metabolism pathways (p2df = 8.2 × 10−8; Table 1; Fig. 1; online suppl. Fig. 1). The interaction between these variants showed a moderate level of association with BD risk (Table 1; pint = 1.4 × 10−4), implying the joint test signal is partially due to the nSNP main effect and partially due to the nSNP-mtSNP interaction effect supporting the epistatic interaction between the nuclear and mitochondrial genes. The odds ratio (OR) for one copy of the minor nSNP allele was 3.42 among carriers of the minor allele A of the mtSNP and 1.27 among carriers of the common allele G of the mtSNP (Fig. 3a). Among other top association results involving the same mtSNP (rs3088309), the joint effect with rs12733666 located in adenylate kinase 5 (AK5) showed a trend for significance (p2df = 2.9 × 10−6), with this joint test result being entirely driven by the nSNP-mtSNP interaction (pint = 7.1 × 10−7). The second most significant association was observed with rs3915952 located in Mt-ND4 and nSNP rs1880924, the same MGAM SNP from the top association result discussed above. Rs3915952 is highly correlated with the top mtSNP rs3088309 (r2 = 0.95), implying that the second association signal is not independent of the top hit. In fact, both mtSNPs rs3088309 and rs3915952 are part of mitochondrial haplogroup JT defining SNP.
Fig. 1.
Regional association plots for the top 2 gene regions (MGAM and AK5) with rs3088309 (Mt-CytB) for risk of bipolar disorder. a, cp values from 2df tests. b, d Interaction test p values. chr7, chromosome 7; chr1, chromosome 1; SNP, single-nucleotide polymorphism.
Fig. 3.
OR for each combination of mtSNP rs3088309 (0/1 for the minor allele) and nSNP (0/1/2 for the number of minor alleles) genotypes for top 2 association results from 2df tests for the risk of bipolar disorder (a) and early-onset bipolar disorder (b). The pair (mtSNP = 0 and nSNP = 0) was used as the reference category (i.e., OR = 1). mtSNP, mitochondrial single-nucleotide polymorphism; nSNP, nuclear single-nucleotide polymorphism.
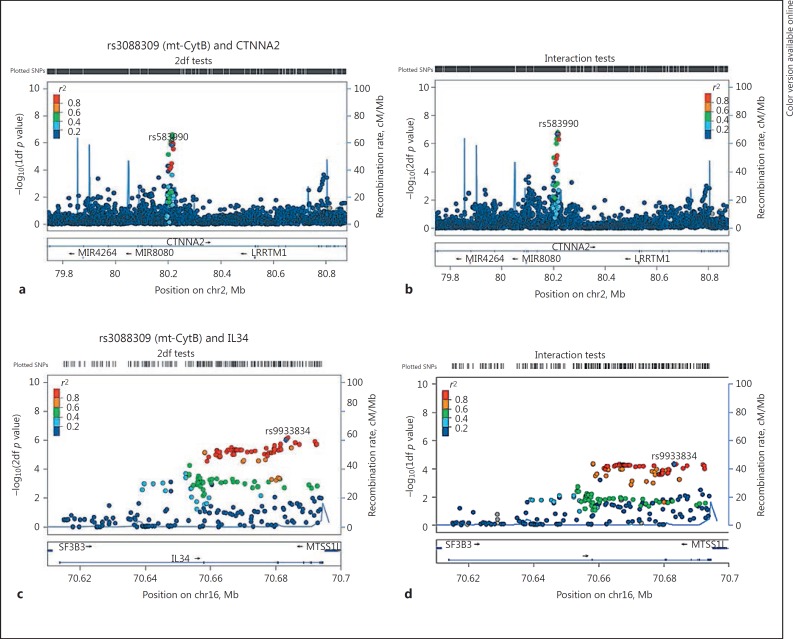
For EOBD risk among BD patients, the top association signal was with the mtSNP rs3088309 in CytB (the same mtSNP that is involved in the top BD risk association), and the nSNP rs583990 (MAF = 0.25 for allele A) located in the Cadherin-associated protein, Alpha2 (CTNNA2) on chromosome 2 (p2df = 2.1 × 10−7 and pint = 1.6 × 10−6). The results suggest that among BD patients the minor allele A of nSNP rs583990 is associated with a greater risk of EOBD (i.e., a higher risk of developing BD by the age of 19 years), and the effect of the nSNP is greater for carriers of the minor mtSNP rs3088309 T allele, compared to those with the major mtSNP allele (OR = 4.53 vs. 1.05; Fig. 3b). The mtSNP rs3088309 also has suggestive evidence for interacting with nSNP rs9933834 located in interleukin 34 (IL34; pjoint = 1.1 × 10−6; interaction pint = 1.6 × 10−5) for the risk of EOBD (Table 2; Fig. 2b, 3b). The second highest association signal involved the mtSNP rs3915952 in ND4 and rs9933834 in IL34 on chromosome 16 (Table 2; Fig. 2; online suppl. Fig. 2).
Fig. 2.
Regional association plots for top 2 gene regions (CTNNA2 and IL34) with rs3088309 (Mt-CytB) for the risk of early-onset bipolar disorder. a, cp values from 2df tests. b, d Interaction test p values. chr2, chromosome 2; chr16, chromosome 16; SNP, single-nucleotide polymorphism.
When testing for enrichment of nSNP-mtSNP interaction association, no significant enrichment was found for the 967 mitochondrial-related genes (online suppl. Table 2). The highest evidence of enrichment was observed for mtSNP rs28359178 located in ND5 for risk of BD (240 for the observed number of genes above the cutoff compared to 217 expected genes; GSEA p = 0.05), and with rs3899498 (located in ND5; GSEA p = 0.05) and rs28357682 (located in CytB; GSEA p = 0.05) for risk of EOBD. These gene set analysis results are not significant after correction for multiple testing incurred by performing an enrichment analysis for each of 15 mtSNPs with an MAF >0.10.
Discussion
To our knowledge, this is the first GWAS to investigate mtSNP contribution in BD, focusing on nSNP-mtSNP epistatic interaction and the risk of BD as well as EOBD (a BD subphenotype). Although no association results were genome-wide significant, some highly notable associations were detected when accounting for mtSNP interaction effects, including nuclear genes that were previously reported BD susceptibility genes or have been implicated in neuropathology. This proof-of-concept study suggests the potential importance of investigating both nuclear and mitochondrial genomes in studying the complex genetic predisposition for BD as well as the age of disease onset. By modeling the modifying effects of mtSNPs on the contribution of nSNPs to BD risk and age of disease onset, additional genetic factors may be identified.
Mt-CytB SNP rs3088309 (and the highly correlated mtSNP rs3915952 in ND4) variants were the top mtSNP with suggestive evidence of interaction with nSNPs in contributing to BD risk and EOBD. The Mt-CytB gene is responsible for making cytochrome B, which is a key protein for transferring the electrons (carrier), and cytochrome B is the only component of complex III that is produced by mtDNA [46]. The mtSNP rs3088309 (merged into rs527236209) located in the Mt-CytB gene, is a missense variant that causes an amino acid change from leucine (major allele) to isoleucine (minor allele) at position 236. Given the relevance of the Mt-CytB gene to the electron transport chain, any mutation in this gene potentially contributes reactive oxygen species, which has been shown to be elevated in BD [14,15]. Being essential for electron transport chain, the mutations in CytB rs3088309 and ND4 rs3915952 might leave neurons more vulnerable to genetic and environmental risk factors. In addition, mitochondrial haplogroup JT (the top mtSNPs rs3088309 and rs3915952 are part of the haplogroup-defining SNP) has been implicated in early-onset schizophrenia, though the mechanism underlying this association is still unknown [47].
The results of this study suggest that for the risk of BD the effects of nuclear genes MGAM and AK5 are modulated by variants in mitochondrial genes. AK5 is mainly expressed in the brain and it has been implicated in temporal lobe epilepsy, autoimmune limbic encephalitis, and Parkinson's and Alzheimer's diseases [48,49,50,51]. Our results suggest that carrying the minor allele of AK5 rs12733666 may have a protective effect for BD in those carrying the minor mtSNP allele. The MGAM gene, related to starch metabolism, has been shown to be downregulated by ZNF804A overexpression (a gene implicated in susceptibility to schizophrenia) [52]. In addition, subjects with autism have dysregulated mRNA expression of MGAM [53]. Considering its central role in starch metabolism [54], it is plausible that genetic variation in MGAM coupled with particular mutations in the mitochondrial genome may result in mitochondrial dysfunction, which may have significant implications for BD pathophysiology.
Our results also suggest that nuclear genes CTNNA2 and IL34 may interact with mitochondrial variants in modulating the risk of EOBD. CTNNA2 is highly expressed in the brain and encodes an α-catenin important for synaptic contact and neuronal plasticity. A CTNNA2 polymorphism has been implicated in a highly heritable excitement-seeking trait, a common trait in BD [55]. CTNNA2 knockout mice show axon migration defects as well as hippocampal and cerebellar lamination defects that are associated with impairments in startle responses and fear conditioning [56,57]. Furthermore, a CTNNA2 SNP was associated with BD in a GWAS study [58]. IL34 is a cytokine that is implicated in immune response and might play an important role in inflammatory mechanisms in mood disorders [59,60]. For instance, IL34 cytokine is important for the survival of microglia, the macrophages of the brain that determine the levels of inflammation in the cellular environments [61]. It has been suggested that mitochondrial dysfunction may be induced by excessive reactive oxygen species produced by activated microglia [62].
This work demonstrates a novel approach to studying genetic effects on BD; however, the results must be interpreted in light of the limitations of this study. Detection of interaction effects requires a significantly larger sample size compared to standard main-effect genetic analysis. This study was based on a small sample, which limited the power to detect genome-wide significant associations; to partially overcome challenges related to power in this study we limited the analyses to common SNPs with an MAF >0.1. While none of the mtSNP-nSNP interactions were significant after multiple testing correction, replication of the top interaction signals would strengthen the presented findings. A further limitation is that this study used mtSNP genotypes from blood samples, not from brain samples. However, although mtDNA accumulates different mutations in different tissues leading to heteroplasmy differences between tissues, previous research has demonstrated perfect concordance between homoplasmic variants (such as mtSNPs used in this study) in 11 brain regions and blood, suggesting that blood is suitable for the study of homoplasmic mtDNA variation [37].
Although none of the associations were statistically significant after multiple testing correction, this study is the first GWAS investigating the effects of mitochondrial genome variation on the risk of BD and a BD subphenotype (early onset vs. adult onset), while considering the combined effects of nuclear and mitochondrial genetic variants. Our novel approach of investigating mtSNP modulation of the nSNP contribution to the risk of BD found suggestive evidence of the joint effect of mtSNPs and several candidate genes with biological relevance to BD and its onset. This concept of analyzing nSNP effects while accounting for interactions with mtSNPs warrants further investigation in larger samples.
In conclusion, our study demonstrated the potential importance of considering the mitochondrial genome for uncovering additional genetic factors in BD, especially for nuclear genes that do not have a strong signal alone but whose effects depend on mitochondrial genetic variants.
Disclosure Statement
The authors have nothing to disclose.
Supplementary Material
Supplementary data
Acknowledgements
This study was funded by the Marriot Foundation, the Mayo Clinic Center for Individualized Medicine, and a small grant award supported by the Department of Health Sciences Research, Mayo Clinic.
References
- 1.Frye MA. Clinical practice: bipolar disorder – a focus on depression. N Engl J Med. 2011;364:51–59. doi: 10.1056/NEJMcp1000402. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association . ed 5. Arlington: APA; 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 3.Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, Anderson W, Dhansay MA, Phillips A, Shurin S, et al. Grand challenges in global mental health. Nature. 2011;475:27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinozaki G, Potash JB. New developments in the genetics of bipolar disorder. Curr Psychiatry Rep. 2014;16:493. doi: 10.1007/s11920-014-0493-5. [DOI] [PubMed] [Google Scholar]
- 5.Ou X, Crane DE, MacIntosh BJ, Young LT, Arnold P, Ameis S, Goldstein BI. CACNA1C rs1006737 genotype and bipolar disorder: focus on intermediate phenotypes and cardiovascular comorbidity. Neurosci Biobehav Rev. 2015;55:198–210. doi: 10.1016/j.neubiorev.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Gershon ES, Alliey-Rodriguez N, Liu C. After GWAS: searching for genetic risk for schizophrenia and bipolar disorder. Am J Psychiatry. 2011;168:253–256. doi: 10.1176/appi.ajp.2010.10091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa RT, Machado-Vieira R, Zarate CA, Jr, Manji HK. Targeting mitochondrially mediated plasticity to develop improved therapeutics for bipolar disorder. Expert Opin Ther Targets. 2014;18:1131–1147. doi: 10.1517/14728222.2014.940893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011;29:311–324. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sequeira A, Rollins B, Magnan C, van Oven M, Baldi P, Myers RM, Barchas JD, Schatzberg AF, Watson SJ, Akil H, et al. Mitochondrial mutations in subjects with psychiatric disorders. PLoS One. 2015;10:e0127280. doi: 10.1371/journal.pone.0127280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreazza AC, Young LT. The neurobiology of bipolar disorder: identifying targets for specific agents and synergies for combination treatment. Int J Neuropsychopharmacol. 2014;17:1039–1052. doi: 10.1017/S1461145713000096. [DOI] [PubMed] [Google Scholar]
- 11.Andreazza AC, Frey BN, Erdtmann B, Salvador M, Rombaldi F, Santin A, Goncalves CA, Kapczinski F. DNA damage in bipolar disorder. Psychiatry Res. 2007;153:27–32. doi: 10.1016/j.psychres.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Scola G, Kim HK, Young LT, Andreazza AC. A fresh look at complex I in microarray data: clues to understanding disease-specific mitochondrial alterations in bipolar disorder. Biol Psychiatry. 2013;73:e4–e5. doi: 10.1016/j.biopsych.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- 14.Andreazza AC, Wang JF, Salmasi F, Shao L, Young LT. Specific subcellular changes in oxidative stress in prefrontal cortex from patients with bipolar disorder. J Neurochem. 2013;127:552–561. doi: 10.1111/jnc.12316. [DOI] [PubMed] [Google Scholar]
- 15.Andreazza AC, Shao L, Wang JF, Young LT. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch Gen Psychiatry. 2010;67:360–368. doi: 10.1001/archgenpsychiatry.2010.22. [DOI] [PubMed] [Google Scholar]
- 16.Kim HK, Mendonca KM, Howson PA, Brotchie JM, Andreazza AC. The link between mitochondrial complex I and brain-derived neurotrophic factor in SH-SY5Y cells – the potential of JNX1001 as a therapeutic agent. Eur J Pharmacol. 2015;764:379–384. doi: 10.1016/j.ejphar.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Anglin RE, Garside SL, Tarnopolsky MA, Mazurek MF, Rosebush PI. The psychiatric manifestations of mitochondrial disorders: a case and review of the literature. J Clin Psychiatry. 2012;73:506–512. doi: 10.4088/JCP.11r07237. [DOI] [PubMed] [Google Scholar]
- 18.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, Zheng Y, Diffenderfer KE, Zhang J, Soltani S, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munakata K, Tanaka M, Mori K, Washizuka S, Yoneda M, Tajima O, Akiyama T, Nanko S, Kunugi H, Tadokoro K, et al. Mitochondrial DNA 3644T->C mutation associated with bipolar disorder. Genomics. 2004;84:1041–1050. doi: 10.1016/j.ygeno.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Chang CC, Jou SH, Lin TT, Liu CS. Mitochondrial DNA variation and increased oxidative damage in euthymic patients with bipolar disorder. Psychiatry Clin Neurosci. 2014;68:551–557. doi: 10.1111/pcn.12163. [DOI] [PubMed] [Google Scholar]
- 21.Kato T, Takahashi Y. Deletion of leukocyte mitochondrial DNA in bipolar disorder. J Affect Disord. 1996;37:67–73. doi: 10.1016/0165-0327(95)00050-x. [DOI] [PubMed] [Google Scholar]
- 22.Kazuno AA, Munakata K, Mori K, Nanko S, Kunugi H, Nakamura K, Mori N, Yamada K, Yoshikawa T, Kato N, et al. Mitochondrial DNA haplogroup analysis in patients with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:243–247. doi: 10.1002/ajmg.b.30804. [DOI] [PubMed] [Google Scholar]
- 23.Washizuka S, Ikeda A, Kato N, Kato T. Possible relationship between mitochondrial DNA polymorphisms and lithium response in bipolar disorder. Int J Neuropsychopharmacol. 2003;6:421–424. doi: 10.1017/S1461145703003778. [DOI] [PubMed] [Google Scholar]
- 24.Kato T, Kunugi H, Nanko S, Kato N. Association of bipolar disorder with the 5178 polymorphism in mitochondrial DNA. Am J Med Genet. 2000;96:182–186. [PubMed] [Google Scholar]
- 25.Li CT, Bai YM, Hsieh JC, Lee HC, Yang BH, Chen MH, Lin WC, Tsai CF, Tu PC, Wang SJ, et al. Peripheral and central glucose utilizations modulated by mitochondrial DNA 10398A in bipolar disorder. Psychoneuroendocrinology. 2015;55:72–80. doi: 10.1016/j.psyneuen.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Kato T. The other, forgotten genome: mitochondrial DNA and mental disorders. Mol Psychiatry. 2001;6:625–633. doi: 10.1038/sj.mp.4000926. [DOI] [PubMed] [Google Scholar]
- 27.Kakiuchi C, Ishiwata M, Kametani M, Nelson C, Iwamoto K, Kato T. Quantitative analysis of mitochondrial DNA deletions in the brains of patients with bipolar disorder and schizophrenia. Int J Neuropsychopharmacol. 2005;8:515–522. doi: 10.1017/S1461145705005213. [DOI] [PubMed] [Google Scholar]
- 28.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatr. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, Revilla-Sanchez R, Kelly MP, Dunlop AJ, Murdoch H, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatr. 2011;16:1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu C, Li PP, Kennedy JL, Green M, Hughes B, Cooke RG, Parikh SV, Warsh JJ. Further support for association of the mitochondrial complex I subunit gene NDUFV2 with bipolar disorder. Bipolar Disord. 2008;10:105–110. doi: 10.1111/j.1399-5618.2008.00535.x. [DOI] [PubMed] [Google Scholar]
- 32.Doyle GA, Dahl JP, Bloch PJ, Weller AE, Lohoff FW, Ferraro TN, Berrettini WH. Association study of polymorphisms in the autosomal mitochondrial complex I subunit gene, NADH dehydrogenase (ubiquinone) flavoprotein 2, and bipolar disorder. Psychiatr Genet. 2011;21:51–52. doi: 10.1097/YPG.0b013e328341333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washizuka S, Iwamoto K, Kakiuchi C, Bundo M, Kato T. Expression of mitochondrial complex I subunit gene NDUFV2 in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. Neurosci Res. 2009;63:199–204. doi: 10.1016/j.neures.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Rosa A, Abrantes P, Sousa I, Francisco V, Santos P, Francisco D, Xavier JM, Oliveira SA. Ulcerative colitis is under dual (mitochondrial and nuclear) genetic control. Inflamm Bowel Dis. 2016;22:774–781. doi: 10.1097/MIB.0000000000000694. [DOI] [PubMed] [Google Scholar]
- 35.Bulstrode H, Nicoll JA, Hudson G, Chinnery PF, Di Pietro V, Belli A. Mitochondrial DNA and traumatic brain injury. Ann Neurol. 2014;75:186–195. doi: 10.1002/ana.24116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruszak A, Safranow K, Branicki W, Gaweda-Walerych K, Pospiech E, Gabryelewicz T, Canter JA, Barcikowska M, Zekanowski C. The impact of mitochondrial and nuclear DNA variants on late-onset Alzheimer's disease risk. J Alzheimers Dis. 2011;27:197–210. doi: 10.3233/JAD-2011-110710. [DOI] [PubMed] [Google Scholar]
- 37.Sequeira A, Martin MV, Rollins B, Moon EA, Bunney WE, Macciardi F, Lupoli S, Smith EN, Kelsoe J, Magnan CN, et al. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somanath CP, Jain S, Reddy YC. A family study of early-onset bipolar I disorder. J Affect Disord. 2002;70:91–94. doi: 10.1016/s0165-0327(00)00372-4. [DOI] [PubMed] [Google Scholar]
- 39.Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatr. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winham SJ, Cuellar-Barboza AB, Oliveros A, McElroy SL, Crow S, Colby C, Choi DS, Chauhan M, Frye M, Biernacka JM. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol Psychiatr. 2014;19:1010–1016. doi: 10.1038/mp.2013.159. [DOI] [PubMed] [Google Scholar]
- 41.Mahon PB, Pirooznia M, Goes FS, Seifuddin F, Steele J, Lee PH, Huang J, Hamshere ML, DePaulo JR, Kelsoe JR, et al. Genome-wide association analysis of age at onset and psychotic symptoms in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:370–378. doi: 10.1002/ajmg.b.31172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsen KK, Nievergelt CM, Zayats T, Greenwood TA, Anttila V, Akiskal HS, Haavik J, Fasmer OB, Kelsoe JR, Johansson S, et al. Genome wide association study identifies variants in NBEA associated with migraine in bipolar disorder. J Affect Disord. 2015;172:453–461. doi: 10.1016/j.jad.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendricks AE, Dupuis J, Logue MW, Myers RH, Lunetta KL. Correction for multiple testing in a gene region. Eur J Hum Genet. 2014;22:414–418. doi: 10.1038/ejhg.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winham SJ, Cuellar-Barboza AB, McElroy SL, Oliveros A, Crow S, Colby CL, Choi DS, Chauhan M, Frye MA, Biernacka JM. Bipolar disorder with comorbid binge eating history: a genome-wide association study implicates APOB. J Affect Disord. 2014;165:151–158. doi: 10.1016/j.jad.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D, DIAGRAM Consortium MAGIC Investigators Common inherited variation in mitochondrial ghenes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magri C, Gardella R, Barlati SD, Valsecchi P, Sacchetti E, Barlati S. Mitochondrial DNA haplogroups and age at onset of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:496–501. doi: 10.1002/ajmg.b.30496. [DOI] [PubMed] [Google Scholar]
- 48.Lai Y, Hu X, Chen G, Wang X, Zhu B. Down-regulation of adenylate kinase 5 in temporal lobe epilepsy patients and rat model. J Neurol Sci. 2016;366:20–26. doi: 10.1016/j.jns.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 49.Ng AS, Kramer J, Centurion A, Dalmau J, Huang E, Cotter JA, Geschwind MD. Clinico-pathological correlation in adenylate kinase 5 autoimmune limbic encephalitis. J Neuroimmunol. 2015;287:31–35. doi: 10.1016/j.jneuroim.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Esparcia P, Hernandez-Ortega K, Ansoleaga B, Carmona M, Ferrer I. Purine metabolism gene deregulation in Parkinson's disease. Neuropathol Appl Neurobiol. 2015;41:926–940. doi: 10.1111/nan.12221. [DOI] [PubMed] [Google Scholar]
- 51.Ansoleaga B, Jove M, Schluter A, Garcia-Esparcia P, Moreno J, Pujol A, Pamplona R, Portero-Otin M, Ferrer I. Deregulation of purine metabolism in Alzheimer's disease. Neurobiol Aging. 2015;36:68–80. doi: 10.1016/j.neurobiolaging.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Umeda-Yano S, Hashimoto R, Yamamori H, Okada T, Yasuda Y, Ohi K, Fukumoto M, Ito A, Takeda M. The regulation of gene expression involved in TGF-beta signaling by ZNF804A, a risk gene for schizophrenia. Schizophr Res. 2013;146:273–278. doi: 10.1016/j.schres.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 53.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, Bennett A, Jabado O, Hirschberg DL, Lipkin WI. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang R, Oddy VH, Archibald AL, Vercoe PE, Dalrymple BP. Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. Peer J. 2016;4:e1762. doi: 10.7717/peerj.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terracciano A, Esko T, Sutin AR, de Moor MH, Meirelles O, Zhu G, Tanaka T, Giegling I, Nutile T, Realo A, et al. Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Transl Psychiatry. 2011;1:e49. doi: 10.1038/tp.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding alpha N-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–284. doi: 10.1038/ng908. [DOI] [PubMed] [Google Scholar]
- 57.Uemura M, Takeichi M. Alpha N-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Dev Dyn. 2006;235:2559–2566. doi: 10.1002/dvdy.20841. [DOI] [PubMed] [Google Scholar]
- 58.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 60.Wohleb ES, Franklin T, Iwata M, Duman RS. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci. 2016;17:497–511. doi: 10.1038/nrn.2016.69. [DOI] [PubMed] [Google Scholar]
- 61.Hodes GE, Kana V, Menard C, Merad M, Russo SJ. Neuroimmune mechanisms of depression. Nat Neurosci. 2015;18:1386–1393. doi: 10.1038/nn.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Witte ME, Mahad DJ, Lassmann H, van Horssen J. Mitochondrial dysfunction contributes to neurodegeneration in multiple sclerosis. Trends Mol Med. 2014;20:179–187. doi: 10.1016/j.molmed.2013.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data