Abstract
Mutations in the MED13L gene, which encodes a subunit of a transcriptional regulatory complex, result in a complex phenotype entailing physical and cognitive anomalies. Deep language impairment has been reported in affected individuals, mostly in patients with copy number variations. We report on a child with a nonsynonymous p.Cys63Arg change in MED13L (chr12:116675396A>G, GRCh37) who exhibits profound language impairment in the expressive domain, cognitive delay, behavioral disturbances, and an autism-like phenotype. Because of the brain areas in which MED13L is expressed and because of the functional links between MED13L and the products of selected candidate genes for cognitive disorders involving language deficits, the proband's linguistic phenotype may result from changes in a functional network important for language development and evolution.
Keywords: Cognitive delay, Language impairment, MED13L, Missense mutation
Rare conditions involving language deficits which result from point mutations in the coding region of genes provide crucial evidence of how the linguistic brain is wired in response to genetic guidance. The MED13L gene encodes a subunit of the mediator complex (a scaffolding protein device that promotes the assembly of functional preinitiation complexes with RNA polymerase II and the general transcription factors) and seemingly helps regulate the transcription of targets of the Wnt, Notch, and Shh signaling pathways [Asadollahi et al., 2013; Davis et al., 2013; Utami et al., 2014]. Among other organs, MED13L is highly expressed in the fetal and adult brain [Muncke et al., 2003; Musante et al., 2004]. MED13L alterations were initially reported to cause complex heart defects only, but it is now clear that mutations in MED13L underlie a phenotypic gradient, also giving rise to facial dysmorphisms, hypotonia, abnormal MRI and/or EEG findings, intellectual disability, delayed speech and language, and autistic features [Adegbola et al., 2015; Cafiero et al., 2015; Asadollahi et al., 2017]. It has been hypothesized that these diverse effects may result from the involvement of this gene in neural crest induction [for discussion, see Asadollahi et al., 2013].
As noted by Cafiero et al. [2015], MED13L haploinsufficiency, resulting from copy number variations (CNVs) or loss-of-function intragenic variants, causes a recognizable intellectual disability syndrome with distinctive facial features and with or without heart defects. Nevertheless, the clinical relevance of missense mutations in MED13L is poorly understood, and only some cases have been described. This is particularly true regarding their effect on language and cognition. As stressed by Asadollahi et al. [2017], detailed patient descriptions and potential genotype-phenotype correlations are needed in order to identify mutational hotspots along the gene. In this study, we report on a boy with a missense mutation affecting the N-terminal region of the MED13L protein and provide a detailed characterization of his clinical profile with a focus on his linguistic and cognitive features. We discuss our findings in the context of current studies of the genetic basis of language.
Materials and Methods
Linguistic, Cognitive, and Behavioral Assessment
Global development of the child was assessed with the Spanish version of the Battelle Developmental Inventories [De la Cruz and González, 2011]. This test comprises 341 items and evaluates gross and fine motor skills, adaptive abilities, personal/social development, cognitive development, and receptive and expressive communication skills.
The perceptual and comprehension abilities of the child were also assessed with the Spanish version of the ComFor Test [Verpoorten et al., 2014]. This test measures perception and sense-making at the levels of presentation and representation, with the aim of designing accurate augmentative communication interventions for children with profound language impairment. It encompasses 36 items, arranged in 5 series and 2 levels. Level I comprises series 1–3 and evaluates perception: objects or pictures have to be sorted according to perceptible features (shape, color, matter, and size). Level II comprises series 4 and 5, and evaluates representation: objects or pictures have to be sorted on the basis of nonperceptible features.
Molecular and Cytogenetic Analyses
DNA from the patient and his parents was extracted from 100 μL of EDTA-anticoagulated whole blood with MagNA Pure (Roche Diagnostics, West Sussex, UK) and used for subsequent analyses.
Fragile X Syndrome Analysis
CGG expansions in the FMR1 gene (the main determinant for fragile X syndrome) were analyzed according to standard protocols. Polymerase chain reaction (PCR) of the fragile site was performed with specific primers and the trinucleotide repeat size of the resulting fragments was evaluated.
Microrrays for CNV Search and Chromosome Aberrations Analysis
The patient's DNA was hybridized on a CGH platform (Cytochip Oligo ISCA 60K). The derivative log ratio spread value was >0.10. The platform included 60,000 probes. Data were analyzed with Agilent Genomic Workbench 7.0 and the ADM-2 algorithm (threshold = 6.0; the aberrant regions had more than 5 consecutive probes).
Whole-Exome Sequencing
The panel used for preparing the library was designed by SureSelectXT Human All Exon V5 (Agilent Technologies, Santa Clara, CA, USA). It captures >35,000 exons from >19,000 genes as well as the splicing flanking (5 bp) regions. The size was approximately 50 Mb. Sequencing was performed with the HiSeq 2500 SystemTM (Illumina, San Diego, CA, USA) sequencer. The obtained reads were then filtered, based on quality parameters, and aligned to the reference genome (build 37 of genome hg19), using the BWA alignment program (version 0.7.12). Sequence variations with respect to the reference genome were detected using the GATK algorithm (version 2.3–9). Variants were annotated with the latest available version of the ION Reporter (Life Technologies, Carlsbad, CA, USA) and with an in-house developed pipeline, aimed to register the patient's variants according to a genetic type classification. The analysis focused on variants found in coding or splicing regions. Registered changes included missense, nonsense, and stop-loss mutations as well as nucleotide insertions or deletions, only if they were found in >40% of the sequences.
Sanger Sequencing
In order to know whether the patient's parents bear the variants of unknown significance identified by the whole-exome sequencing study in the proband, a PCR was performed using specific primers for exon 2 of MED13L and exon 37 of ANK3, according to the reference sequences NM_15335 and NM020987, respectively. Double-sense sequencing of the PCR products was performed in an automatic sequencer ABI 3130 DNA Analyzer (Applied Biosystems, Foster City, CA, USA), and sequence variants were subsequently studied using the seqScape v2.5 software (Life Technologies).
In silico Analysis of Language Dysfunction Associated with MED13L Alterations
In order to delve into the molecular causes of the speech and language problems exhibited by the proband, we surveyed the relevant literature via PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) to learn more about the biological functions of MED13L in the brain and the language profile of people bearing pathological changes in the gene. We also examined the DECIPHER database (https://decipher.sanger.ac.uk/) to discern more about the clinical presentation of missense mutations in MED13L. Finally, we used String 10.5 (www.string-db.org) for examining potential functional links between MED13L and the products of known candidates for language disorders (dyslexia, specific language disorder), cognitive diseases entailing language deficits (autism-spectrum disorder (ASD) and schizophrenia), and language evolution. String 10.5 predicts physical and functional associations between proteins relying on different sources (genomic context, high-throughput experiments, conserved coexpression, and text mining) [Szklarczyk et al., 2015]. For dyslexia, we relied on Paracchini et al. [2016]. For specific language impairment, we chose of Pettigrew et al. [2016] and Chen et al. [2017]. For language deficits in ASD and schizophrenia, we relied on Benítez-Burraco and Murphy [2016] and Murphy and Benítez-Burraco [2016], [2017]. Finally, for language evolution, we considered the genes discussed by Boeckx and Benítez-Burraco [2014] as well as Benítez-Burraco and Boeckx [2015].
Results
Clinical History
The proband is a boy born after 38 weeks of gestation by vaginal delivery to a healthy 37-year-old female. At birth, his weight was 2,755 g (10th percentile), height was 49 cm (31st percentile), and his OFC was 34 cm (35th percentile). The Apgar scores were 9 (at 1 min) and 10 (at 5 min). The child exhibited 2 achromatic spots on the body side and suffered from neonatal sepsis (with bilirubin levels of 3.1 mg/dL at the 4th day). Also, bilateral cryptorchidism was observed.
No signs of cardiac disease were detected in the proband. Auricular and ventricular volumes were concordant, and no abnormal signs of dilatation were observed. Auriculoventricular and sigmoid valves showed normal morphology and kinetics, and no cardiac insufficiency was detected. Global and segmental ventricular contraction was normal. The child exhibited a type I septum IV and a mild, nonpathological hypertrabeculation in the apical zone of the left ventricle. M-Mode measurements were as follows: ejection fraction: 61%, interventricular septum end-diastolic thickness: 6 mm, left ventricular end-diastolic diameter: 33 mm, left ventricular end-systolic diameter: 22.5 mm, and left-ventricular posterior wall thickness: 5.6 mm.
Cranial MRI, performed at the age of 3 years and 5 months, yielded normal results. An EEG study with sleep deprivation, performed at the age of 4 years and 9 months, revealed frequent epileptiform discharges during sleep in the left parietotemporal region and in the right centrotemporal region in absence of continuous spikes and waves during slow-wave sleep.
Developmental anomalies were soon reported by the parents, including lack of crying, absence of feeding demands, sucking weakness, reduced eye contact, hypotonia, and neck rigidity. Still, the child seemed to enjoy physical contact and smiled regularly. Motor development was also delayed. Generalized hypotonia was confirmed at 6 months. When the proband was 2 years old, mild axial hypotonia was still present, whereas at the age of 4 years and 10 months some hypertonic symptoms were detected by his neuropediatrician. Self-hurting behavior was reported at the age of 4 years and 10 months. The child started to walk without aid when he was 3 years old. Stereotypic movements were reported from that age too. Dysmorphic features included reduced palpebral fissures, nasal base enlargement, enlarged plane philtrum, a thin upper lip, low-set ears, and a prominent columella (Fig. 1). Hypermetropia was diagnosed when the boy was 4 years and 10 months old.
Fig. 1.

Photography of the proband.
When the child was 16 months old, he started attending a normal nursery school, but a month later, he was transferred to an early care center for disabled children, where he was reported to interact normally with his peers. At present (5 years and 4 months), the child is attending a normal school because of the lack of a clear diagnosis. Self-hurting behavior is still observed. The child has not yet mastered sphincter control. Feeding problems are still common, to the extent that he is only able to eat pureed food.
Language and Cognitive Development
Early language milestones were achieved quite normally by the child, including cooing (observed at 6 months), babbling (observed at 12 months), and the utterance of a first word (“agua,” water, reported shortly after). Nonetheless, no further progress was observed, and he soon lost this first word. At the age of 2 years, the child did not interact adequately and was unable to understand simple commands.
When the child was 1 year and 5 months old, speech therapy was initiated 4 times a week. Comprehension abilities improved, and at present, he is able to understand simple commands. However, he only utters nonsense disyllabic sequences with no clear communicative function, which resemble those generated during the canonical babbling stage, reinforcing the view that the child may have experienced some kind of language regression. Still, some communicative intentionality is apparent in his behavior. For instance, for requesting some object of interest, he usually holds the interlocutor's hand and leads him/her to the place where the object is located. However, deictic pointing has not been reported. Also, no signs of symbolic behavior have been observed, including differed imitation, drawing, or playing with objects representing animals or persons. Echolalias are absent, but hypersensibility to loud, unfamiliar sounds has been frequently reported by the parents. When the child was 4 years and 10 months old, the neuropediatrician diagnosed him with syndromic ASD with no language and with intellectual disability.
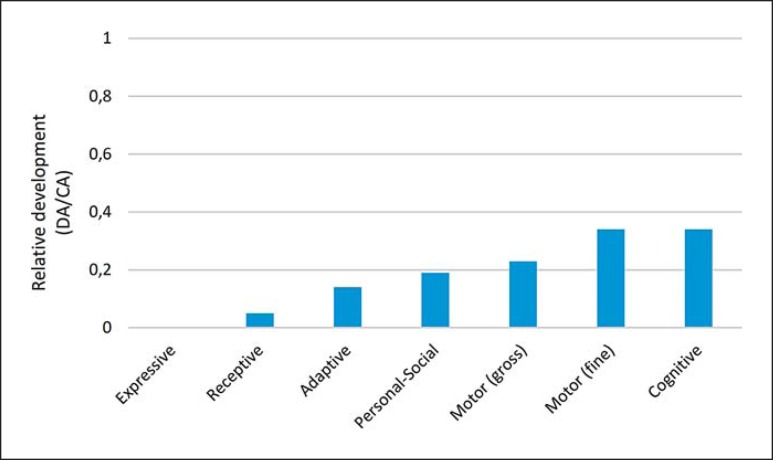
At the age of 5 years and 4 months, we assessed the global developmental profile of the proband with the Spanish version of the Battelle Developmental Inventories. The resulting scores were suggestive of a broad developmental delay, mostly impacting on his language abilities. Accordingly, communication skills were severely impaired, adaptive and personal-social abilities were impaired, whereas motor and cognitive skills were the less-affected domains (Fig. 2).
Fig. 2.
The proband's developmental profile at the age of 5 years and 4 months according to the Battelle Developmental Inventories. In order to make reliable comparisons, the resulting scores are shown as relative values referred to the expected scores according to the chronological age of the child. CA, chronological age; DA, developmental age.
In the ComFor test, which evaluates precursors of communication, the child performed correctly in series 1–3 at Level I, but failed at Level II tasks. This outcome suggests that his representational abilities are severely affected.
Molecular and Cytogenetic Analyses
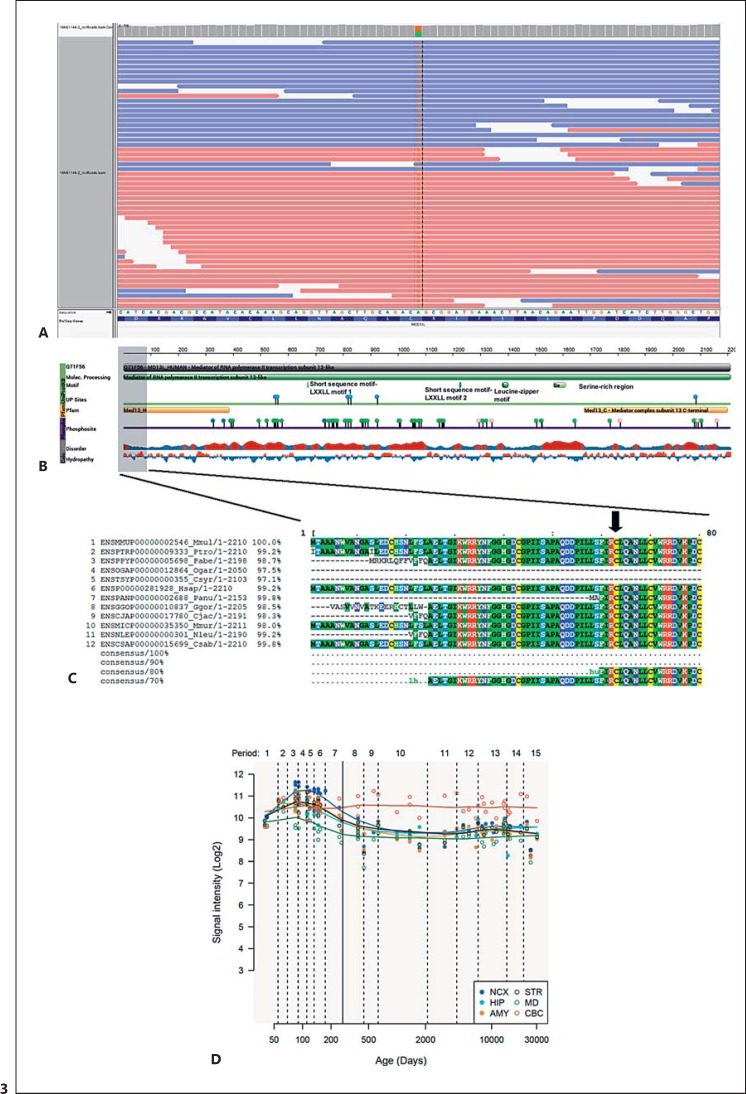
Routine molecular and cytogenetic analyses of the child were performed at the age of 2 years and 7/8 months. PCR analysis of the FMR1 site was normal, as far as the patient exhibited a normal allele of 30 CGG repeats. No chromosomal rearrangements or CNVs were observed in the array CGH. Whole-exome sequencing analysis revealed several variants in genes previously associated with clinical conditions sharing some or most of the pathological features exhibited by the child (Table 1). Considering the clinical profile of the proband, the most promising variant was the c.187T>C transition (NM_015335.4) in heterozygous state in exon 2 of the MED13L gene (Fig. 3A), resulting in a p.Cys63Arg change in the MED13L N-terminal domain of the protein (http://pfam.xfam.org/protein/Q71F56) (Fig. 3B). This cysteine residue is conserved across all primates, except in bushbabies and tarsiers, and is found in >70% of bony vertebrates (Fig. 3C). The change is predicted as probably damaging, according to PolyPhen, with a score of 0.998 (sensitivity: 0.27; specificity: 0.99), and according to SIFT, with a SIFT score of 0 (median conservation = 2.77). The NPS@ tool for protein sequence analysis [Combet et al., 2000] predicts that the change affects an alpha helix stretch within the N-terminal portion of the protein, between amino acids 57 and 73. This is a novel variant not previously reported in the literature or in public databases (dbSNP, ExAC, HGMD, LOVD, and NHLBI ESP). The segregation study performed in the patient's parents revealed that this is a de novo substitution. Finally, our in silico analysis reveals that MED13L is highly expressed in the cerebellum after birth (Fig. 3D).
Table 1.
Missense mutations of clinical interest in the proband as revealed by WES analysis
| Coordinates (GRCh37)/genotype | Gene | Nucleotide change | Missense change | OMIM | Frequency in ExAC (%) | De novo or inherited |
|---|---|---|---|---|---|---|
| chr12:116675396A>G | MED13L | c.187T>C (NM_015335.4) | p.Cys63Arg | 608771 | 0 | de novo |
| chr10:61830642T>A | ANK3 | c.9997A>T (NM_020987.3) | p.Thr3333Ser | 600465 | 0.002 | inherited |
| chr10:61829951T>C | ANK3 | c.10688A>G (NM_020987.3) | p.Gln3563Gly | 600465 | 0.002 | inherited |
| chr7:117351728T>G | CTTNBP2 | c.4855A>C (NM_033427.2) | p.Lys1619Gln | 609772 | 0.002 | NA |
| chr12:49427108G>A | KMT2D | c.11380C>T (NM_033427.2) | p.Pro3794Ser | 602113 | 0.001 | NA |
| chr13:51519581G>A | RNASEH2B | c.529G>A (NM_024570.3) | p.Ala177Thr | 610326 | 0.001 | NA |
| chr18:31319101C>T | ASXL3 | c.1733C>T (NM_030632.1) | p.Ser578Phe | 615115 | 0.00001 | NA |
| chr20:31024797T>C | ASXL1 | c.4282T>C (NM_015338.5) | p.Ser1428Pro | 612990 | 0.0001 | NA |
NA, not available; WES, whole-exome sequencing.
Fig. 3.
Molecular aspects of the MED13L alteration in our proband. A Outcome of the whole-exome sequencing analysis of the patient showing the missense change in position chr12:116237591. B Main structural features of the MED13L protein. The scheme was obtained from the Protein Feature View tool of the Protein Data Bank (http://www.rcsb.org/pdb/protein/Q71F56?evtc=Suggest&evta=ProteinFeatureView&evtl=autosearch_SearchBar_querySuggest). The vertical color bar on the left side indicates data provenance. Data in green originate from UniProtKB. Data in yellow originate from Pfam, by interacting with the HMMER3 website. Data in purple originate from Phosphosite. Data in gray have been calculated using BioJava. Protein disorder predictions are based on JRONN (Troshin and Barton, unpublished data), a Java implementation of RONN (red: potentially disordered region; blue: probably ordered region). Hydropathy was calculated using a sliding window of 15 residues and summing up scores from standard hydrophobicity tables (red: hydrophobic; blue: hydrophilic). C Alignment of primate MED13L proteins using the MView tool of EMBL-EBI (http://www.ebi.ac.uk/Tools/msa/mview/). Conserved amino acids are displayed at the bottom. The black arrow at the top of the figure points to Cys changed to Arg in the proband. D Expression pattern of the MED13L gene in the brain according to the Human Brain Transcriptome (http://hbatlas.org/hbtd/images/wholeBrain/MED13L.pdf). Six different brain regions are considered: the amygdala (AMY), the cerebellar cortex (CBC), the hippocampus (HIP), the mediodorsal nucleus of the thalamus (MD), the striatum (STR), and 11 areas of the neocortex (NCX).
In addition to this probably pathogenic mutation, in MED13L 2 heterozygous missense changes in exon 37 of the ANK3 gene, reported with low frequency in the ExAC database, were also identified: c.9997A>T (p.Thr3333Ser) and c.10688A>G (p.Glu3563Gly) (NM_020987.3). These variants in the ANK3 gene have been inherited from his asymptomatic mother and, therefore, were reclassified as probably benign variants. Finally, the whole-exome sequencing analysis also revealed 5 additional low-frequency variants (<1% in the general population) of potential interest: c.4282T>C (p.Ser1428Pro) (NM_015338.5) in ASXL1, c.1733C>T (p.Ser578Phe) (NM_030632.1) in ASXL3, c.4855A>C (p.Lys1619Gln) (NM_033427.2) in CTTNBP2, c.11380C>T (p.Pro3794Ser) (NM_033427.2) in KMT2D, and c.529G>A (p.Ala177Thr) (NM_024570.3) in RNASEH2B. These genes are involved in brain development and function, and their mutation gives rise to some of the symptoms or distinctive features observed in our proband. For example, both ASXL1 and ASXL3 are candidates for Bohring-Opitz syndrome (OMIM 605039) and Bainbridge-Ropers syndrome (OMIM 615485), 2 conditions showing an autosomal dominant pattern of inheritance and entailing intellectual disability and dysmorphic features [Shashi et al., 2016]. However, the clinical profile of the patient and the nature of these variants led us to reclassify them as not relevant for genotype-phenotype correlations.
In silico Analysis of Language Dysfunction Associated with the Mutation of MED13L
We surveyed the relevant literature, as well as curated databases of candidate genes for cognitive diseases entailing language deficits and of clinical conditions resulting from genomic variants, to learn more about the potential contribution of the mutation found in our proband to his language disabilities. We found that MED13L is a candidate for ASD [Iossifov et al., 2012, 2014, 2015], a condition characterized by a distinctive abnormal language profile [for review, see Benítez-Burraco and Murphy, 2016]. Likewise, we found that several other missense mutations in the gene have been associated with speech and language problems (Table 2). Finally, several proteins important for language development and language evolution are predicted to be direct interactors of MED13L, in particular, EP300, CEBPB, and CREBBP.
Table 2.
Summary table with the most relevant clinical features of the proband compared to other cases involving missense mutations of MED13L, as well as the common presentation of the MED13L haploinsufficiency syndrome
| p.Cys63Arga | p.Asp860Glyb | p.Asp860Glyc | p.Pro866Leud | p.Pro866Alae | p.Cys1160Argf | p.Arg1416Hisg | p.Gly1899Argh | p.Gly1899Argi | p.Ser2002Leuj | p.Thr2162Metk | Key phenotype features of MED13L-associated diseasel | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cardiac anomalies | − | ? | − | ? | + | + | ? | ? | ? | ? | ? | + |
| Visual problems | + | ? | − | ? | + | ? | ? | ? | ? | + | ? | |
| Facial dysmorphisms | + | + | + | + | + | ? | ? | + | + | + | + | + |
| Motor problems | ||||||||||||
| Motor delay | + | ? | + | ? | ? | ? | ? | ? | + | ? | ||
| Hypotonia | + | ? | + | ? | ? | + | ? | ? | + | + | ? | + |
| Skull anomalies | − | ? | ? | + | + | ? | ? | + | ? | ? | ? | + |
| Structural brain anomalies | − | ? | + | ? | + | ? | ? | ? | ? | + | ? | |
| EEG anomalies | + | + | + | ? | + | + | ? | ? | ? | ? | ? | |
| Cognitive impairment | ||||||||||||
| Behavioral problems | + | ? | + | ? | ? | ? | ? | ? | + | ? | + | + |
| Developmental delay | + | ? | + | ? | + | + | ? | ? | ? | ? | ? | |
| Intellectual disability | + | + | + | + | ? | ? | + | ? | + | + | + | + |
| Speech/language | ||||||||||||
| impairment | + | ? | + | ? | ? | ? | ? | ? | ? | ? | ? |
The 4 missense mutations found by Muncke et al. [2003] (p.Lys686Gln, p.Glu251Gly, Arg1872His, and Asp2023Gly) are reported to entail cardiac problems only; no behavioral or cognitive deficits are mentioned in their paper. +, present; –, absent; ?, unknown.
Our proband.
Trio 22 from Gilissen et al. [2014].
Patient 2 from Asadollahi et al. [2017].
DECIPHER patient 258131.
DECIPHER patient 268019.
DECIPHER patient 265953.
Family M142 from Najmabadi et al. [2011].
Patient 2 from Caro-Llopis et al. [2016] (DECIPHER patient 262545).
DECIPHER patient 323183.
DECIPHER patient 260542.
DECIPHER patient 272205.
Summarized from Adegbola et al. [2015] (Table 3) and Asadollahi et al. [2017] (Table 2).
Discussion
The development of next-generation sequencing facilities has exponentially increased the number of new genes and variants of known genes associated with clinical conditions entailing problems with language. In this study, we have characterized the linguistic and cognitive profile of a boy with a novel heterozygous missense mutation in the MED13L gene, which is predicted to alter the N-terminal domain of the protein. Our proband exhibits most of the cognitive and behavioral features of the MED13L haploinsufficiency syndrome, without congenital heart defect, and particularly, all the abnormal features linked to missense mutations in the gene (Table 2).
At present, language impairment related to MED13L dysfunction has been mostly reported in patients bearing point nonsense mutations (DECIPHER patients 258675 and 261065), frameshift mutations (DECIPHER patients 260083, 273003, and 323645), or partial deletions of the gene (DECIPHER patients 283932 and 322949) [see also 1 in Asadollahi et al., 2017]. Recently, a missense mutation resulting in a p.Asp860Gly change has been found to entail absent speech with some preserved receptive and reading abilities [patient 2 from Asadollahi et al., 2017]. The occurrence of language problems in an unrelated patient with the same missense mutation is unknown [trio 22 from Gilissen et al., 2014]. It is thus of interest to know more about the effects of point changes in the functional domains of MED13L on language abilities of the affected individuals. We have performed a detailed analysis of the language profile of our proband and found that his speech is severely affected, and his verbal comprehension is limited to simple, direct commands. We also predict problems for acquiring an augmentative and alternative communication, in view of the results achieved by the child in the ComFor test.
The fact that MED13L is a candidate for ASD could explain the ASD-like phenotype of our proband, which includes language regression, no deictic pointing, and absence of symbolic behavior. Additionally, it is of interest that MED13L is highly expressed in the cerebellum after birth, because this brain region plays a pivotal role in many aspects of speech and language processing, including syntax, phonology, lexical semantics and speech perception and planning [Mariën et al., 2014]. Interestingly, the cerebellum has also been related to many of the processes that are impaired in people with ASD, from language and communication to social interaction and behavior [Hampson and Blatt, 2015]. Moreover, the circumstance that the MED13L protein is functionally related to the products of EP300 and CREBBP [Krebs et al., 2010; Galbraith et al., 2013] may help understand the problems with language experienced by our proband. Accordingly, both EP300 and CREBBP serve as a link between the FOXP2 and ROBO1 networks, which play a central role in the development and function of the brain areas involved in speech and language processing [see Boeckx and Benítez-Burraco, 2014]. Also, both EP300 and CREBBP are candidates for different subtypes of Rubinstein-Taybi syndrome (OMIM 613684 and OMIM 180849, respectively), a clinical condition entailing intellectual disability and growth retardation, skeletal abnormalities, and brain abnormalities mostly impacting on the rolandic area, which is crucial for language processing [Sener, 1995; Viosca et al., 2010]. Interestingly, Crebbp+/- mice exhibit altered vocal behavior [Wang et al., 2010], whereas the knockout of the gene in postmitotic neurons of the forebrain results in problems for object recognition [Valor et al., 2011]. Finally, we wish to highlight the role played by MED13L in neural crest induction and the fact that neurocristopathies, i.e., clinical conditions resulting from neural crest defects, usually entail skull, cognitive, and language abnormalities [for discussion, see Benítez-Burraco et al., 2016] as well as occasional autistic traits, as observed in CHARGE syndrome (OMIM 214800) [Fernell et al., 1999].
Conclusion
Although the exact molecular causes of the language and cognitive impairment exhibited by our proband remain to be fully elucidated, we expect that they result from the change we have uncovered in a conserved residue of the N-terminal domain of the MED13L protein. To the best of our knowledge, this missense mutation is the second missense mutation in MED13L which has been explicitly linked to language problems. In this study we provide a detailed characterization of the language deficits linked to this novel mutation. We also hypothesize that the resulting amino acid change may have altered the functionality of the protein and/or its movement to the nucleus, ultimately reducing the available amount of functional MED13L and impacting on the functions performed by the protein as part of the mediator complex, both during embryonic development and after birth. Finally, we suggest that the negative effect of this mutation on language abilities may result from the functional connections that MED13L has with key genes for language development. Overall, we expect that this clinical case helps to improve our current knowledge of missense variants in this important regulatory gene and, ultimately, of the genetic background of human language.
Statement of Ethics
Ethics approval for this research was granted by the Comité Ético del Hospital “Juan Ramón Jiménez”. Written informed consent was obtained from the proband's parents for publication of this case report and of any accompanying tables and images.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgment
We would like to thank the proband and his family for their participation in this research and for their enduring aid during the writing of the manuscript. Preparation of this work was supported by funds from the Spanish Ministry of Economy and Competitiveness, grant FFI2016-78034-C2-2-P P (AEI/FEDER, UE), to A.B.B.
References
- Adegbola A, Musante L, Callewaert B, Maciel P, Hu H, et al. Redefining the MED13L syndrome. Eur J Hum Genet. 2015;23:1308–1317. doi: 10.1038/ejhg.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi R, Oneda B, Sheth F, Azzarello-Burri S, Baldinger R, et al. Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet. 2013;21:1100–1104. doi: 10.1038/ejhg.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi R, Zweier M, Gogoll L, Schiffmann R, Sticht H, et al. Genotype-phenotype evaluation of MED13L defects in the light of a novel truncating and a recurrent missense mutation. Eur J Med Genet. 2017;60:451–464. doi: 10.1016/j.ejmg.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Benítez-Burraco A, Boeckx C. Possible functional links among brain- and skull-related genes selected in modern humans. Front Psychol. 2015;6:794. doi: 10.3389/fpsyg.2015.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Burraco A, Murphy E. The oscillopathic nature of language deficits in autism: from genes to language evolution. Front Hum Neurosci. 2016;10:120. doi: 10.3389/fnhum.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Burraco A, Theofanopoulou C, Boeckx C.Globularization and domestication. Topoi DOI: 10.1007/s11245-016-9399-7 (2016).
- Boeckx C, Benítez-Burraco A. Globularity and language-readiness: generating new predictions by expanding the set of genes of interest. Front Psychol. 2014;5:1324. doi: 10.3389/fpsyg.2014.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafiero C, Marangi G, Orteschi D, Ali M, Asaro A, et al. Novel de novo heterozygous loss-of-function variants in MED13L and further delineation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet. 2015;23:1499–1504. doi: 10.1038/ejhg.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Llopis A, Roselló M, Orellana C, Oltra S, Monfort S, et al. (2016) De novo mutations in genes of mediator complex causing syndromic intellectual disability: mediatorpathy or transcriptomopathy? Pediatr Res. 2016;80:809–815. doi: 10.1038/pr.2016.162. [DOI] [PubMed] [Google Scholar]
- Chen XS, Reader RH, Hoischen A, Veltman JA, Simpson NH, et al. Next-generation sequencing identifies novel gene variants and pathways involved in specific language impairment. Sci Rep. 2017;7:46105. doi: 10.1038/srep46105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with mediator. Genes Dev. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz MV, González M. Madrid: TEA Ediciones; 2011. Adaptación española del inventario de Desarrollo Battelle. [Google Scholar]
- Fernell E, Olsson VA, Karlgren-Leitner C, Norlin B, Hagberg B, Gillberg C. Autistic disorders in children with CHARGE association. Dev Med Child Neurol. 1999;41:270–272. doi: 10.1017/s0012162299000572. [DOI] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- Hampson DR, Blatt GJ. Autism spectrum disorders and neuropathology of the cerebellum. Front Neurosci. 2015;9:420. doi: 10.3389/fnins.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Levy D, Allen J, Ye K, Ronemus M, et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci USA. 2015;112:E5600–5607. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Demmers J, Karmodiya K, Chang NC, Chang AC, Tora L. ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep. 2010;11:541–547. doi: 10.1038/embor.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariën P, Ackermann H, Adamaszek M, Barwood CH, Beaton A, et al. Consensus paper: language and the cerebellum: an ongoing enigma. Cerebellum. 2014;13:386–410. doi: 10.1007/s12311-013-0540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muncke N, Jung C, Rüdiger H, Ulmer H, Roeth R, et al. Missense variants and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- Murphy E, Benítez-Burraco A. Bridging the gap between genes and language deficits in schizophrenia: an oscillopathic approach. Front Hum Neurosci. 2016;10:422. doi: 10.3389/fnhum.2016.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Benítez-Burraco A. Language deficits in schizophrenia and autism as related oscillatory connectomopathies: an evolutionary account. Neurosci Biobehav Rev. 2017;83:742–764. doi: 10.1016/j.neubiorev.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Musante L, Bartsch O, Ropers HH, Kalscheuer VM. cDNA cloning and characterization of the human THRAP2 gene which maps to chromosome 12q24, and its mouse ortholog Thrap2. Gene. 2004;332:119–127. doi: 10.1016/j.gene.2004.02.044. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Diaz R, Stein J. 2016. Advances in dyslexia genetics – new insights into the role of brain asymmetries. In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics. Vol. 96. London: Academic Press; 2016. pp. 53–97. [DOI] [PubMed] [Google Scholar]
- Pettigrew KA, Frinton E, Nudel R, Chan MT, Thompson P, et al. Further evidence for a parent-of-origin effect at the NOP9 locus on language-related phenotypes. J Neurodev Disord. 2016;8:24. doi: 10.1186/s11689-016-9157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener RN. Rubinstein-Taybi syndrome: cranial MR imaging findings. Comput Med Imaging Graph. 1995;19:417–418. doi: 10.1016/0895-6111(95)00023-2. [DOI] [PubMed] [Google Scholar]
- Shashi V, Pena LD, Kim K, Burton B, Hempel M, et al. De novo truncating variants in ASXL2 are associated with a unique and recognizable clinical phenotype. Am J Hum Genet. 2016;99:991–999. doi: 10.1016/j.ajhg.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utami KH, Winata CL, Hillmer AM, Aksoy I, Long HT, et al. Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum Mutat. 2014;35:1311–1320. doi: 10.1002/humu.22636. [DOI] [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jiménez-Minchán M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci. 2011;31:1652–1663. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpoorten R, Noens I, van Berckelaer-Onnes I. Manual ComFor: precursores de la comunicación. Ávila: Autismo Ávila. 2014 [Google Scholar]
- Viosca J, López-Atalaya JP, Olivares R, Eckner R, Barco A. Syndromic features and mild cognitive impairment in mice with genetic reduction on p300 activity: differential contribution of p300 and CBP to Rubinstein-Taybi syndrome etiology. Neurobiol Dis. 2010;37:186–194. doi: 10.1016/j.nbd.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Wang J, Weaver IC, Gauthier-Fisher A, Wang H, He L, et al. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Dev Cell. 2010;18:114–125. doi: 10.1016/j.devcel.2009.10.023. [DOI] [PubMed] [Google Scholar]