Abstract
Subjects with schizophrenia (SZ) and bipolar disorder (BD) show decreased protein and transcript levels for mitochondrial complex I. In vitro results suggest antipsychotic and antidepressant drugs may be responsible. We measured complex I activity in BD, SZ, and controls and presence of antipsychotic and antidepressant medications, mitochondrial DNA (mtDNA) copy number, and the mtDNA “common deletion” in the brain. Complex I activity in the prefrontal cortex was decreased by 45% in SZ compared to controls (p = 0.02), while no significant difference was found in BD. Complex I activity was significantly decreased (p = 0.01) in pooled cases (SZ and BD) that had detectable psychotropic medications and drugs compared to pooled cases with no detectable levels. Subjects with age at onset in their teens and psychotropic medications showed decreased (p < 0.05) complex I activity compared to subjects with an adult age at onset. Both SZ and BD groups displayed significant increases (p < 0.05) in mtDNA copy number compared to controls; however, common deletion burden was not altered. Complex I deficiency is found in SZ brain tissue, and psychotropic medications may play a role in mitochondrial dysfunction. Studies of medication-free first-episode psychosis patients are needed to elucidate whether mitochondrial pathophysiology occurs independent of medication effects.
Keywords: Schizophrenia, Mitochondria, Antipsychotic drug, Antidepressant drug, Prefrontal cortex, Mitochondrial DNA common deletion, Complex I activity, mtDNA copy number
Introduction
Mitochondria are integrally important organelles for cellular energy functioning. Mitochondria have been studied in schizophrenia (SZ) and bipolar disorder (BD). These studies of mitochondrial dysfunction in neuropsychiatric disorders have shown alterations in calcium signaling and buffering, brain bioenergetic deficits in the production of adenosine triphosphate (ATP), and alterations in mitochondrial size and density, all of which can have an effect on synaptic transmission [1]. Mitochondria generate the majority of energy, for which there is a very high demand in the brain; it is estimated that a mouse neuron consumes approximately 106 ATP molecules each second due to the metabolic demands needed to fire action potentials and continuously restore sodium levels after firing [2]. The generation of ATP in mitochondria begins with complex I (NADH: ubiquinone oxidoreductase) and continues through complexes II-V, of the electron transport chain. Complex I is a large multimeric enzyme complex located in the inner mitochondrial membrane consisting of at least 45 protein subunits that have an aggregated molecular weight of 980 kDa. Deficiencies in complex I such as reduced enzyme activity and/or reduced subunit concentrations have been one classical sign of the cellular dysfunction associated with aging and neurodegeneration [3]. Mutations have been reported in 26 nuclear-encoded and mitochondrial DNA (mtDNA)-encoded genes of complex I that can lead to complex I deficiency disorders [4]. Prior studies have also found deficits in complex I from postmortem brain samples of SZ [5] and BD subjects [6]. Alterations in complex I subunit proteins and genes have also been reported in lymphoblastoid cell lines and fibroblasts from subjects with SZ [5] and BD [7].
Deletions in mtDNA can also cause complex I deficiency disorders and have been reported in brain and cells in both patient tissues and in aging tissues from healthy controls, suggesting they can serve as an indicator of mitochondrial dysfunction in some cases. Large deletions of the mtDNA genome can potentially cause mitochondria dysfunction due to decreased energy production because of decreased complexes I-V, or generation of more reactive oxygen species leading to more oxidative stress. Associations between the mtDNA 4,977-bp common deletion load in the brain and neuropsychiatric disorders have also been reported [8,9,10,11]. The mtDNA common deletion levels have been shown to increase in the brain with advanced aging [12,13]. However, we have previously observed that the age-related increase in the common deletion in brain was significant in control subjects but was not observed in SZ [13,14,15,16,17]. This SZ phenotype might be due to a lower activity of mitochondria in the brain of subjects with SZ that leads to a lower production of the mtDNA common deletion. Another speculation is that these results could be due to a lower number of mitochondria organelles per cell. In order to determine whether the common deletion is altered in the gross homogenate of brain tissue, we measured both the common deletion and the nondeleted wild-type (WT) mtDNA using a qPCR-based copy number assay.
We have previously reported alterations in mitochondria puncta co-localized with the postsynaptic density compartment of postmortem brain, suggesting mitochondrial density, or size may be affected in SZ [18]. In peripheral samples such as blood, mtDNA copy number has been observed to be decreased in first-episode neuroleptic-naïve subjects with SZ [19]. In addition, differences in copy number were observed between risperidone responders and nonresponders, i.e. after treatment with risperidone, the responders continued to show a decrease in mitochondria DNA copy number compared with nonresponders. In a large study of peripheral blood samples [18] we observed a significant reduction (p < 10−40) in mtDNA copy number in SZ subjects compared to controls, as well as a significant reduction in BD (p < 10−6). However, demographic data for these findings from the GAIN dataset have not been available for analysis. We also analyzed a Bulgarian family trios dataset [20] using the same approach, but did not find a significant difference in mtDNA copy number in SZ compared to unaffected controls [18]. These studies using peripheral tissues such as blood, however, do not necessarily correlate with what mtDNA changes may be occurring in SZ brains. In this current paper, we evaluated mtDNA copy number, relative levels of the common deletion, and complex I activity in postmortem brain tissue.
Prior studies of antipsychotic drugs (APDs) and antidepressant drugs (ADDs) have shown that both classes of drugs can cause functional mitochondrial impairments, such as decreased complex I activity of the electron transport respiration chain [21,22,23,24,25,26,27,28]. The current study aims to further address the role of APDs and ADDs on complex I activity in the human brain so that we may further understand if the mitochondrial dysfunction observed is exclusive to disease or medication effects, or may be a compounded effect of both.
Method
Subjects for this study were obtained from the UCI-Pritzker Brain Bank, and the UC Irvine Institutional Review Board (IRB) approved this study. Postmortem brains were screened for inclusion criteria [29,30] using quality indicators: agonal factor score = 0, average pH values of 6.6 (range 6.0–7.2) and RIN >6. Three groups of subjects were formed (SZ, BD, and controls) for a total of 98 subjects that had not been used in prior studies of mitochondria. About one-third of the subjects with SZ and BD committed suicide at 25 and 38%, respectively and had high rates of substance abuse or dependence of 57 and 85%, respectively (online suppl. Table 1; see www.karger.com/doi/10.1159/000484348 for all online suppl. material).
Prefrontal cortex (PFC) gray matter was sampled predominantly from medial BA9 and to ensure adequate amounts for additional planned studies such as immunohistochemistry, some dissected BA9 regions included a margin of posterior BA8. The genu of the corpus callosum was used as a guide to ensure anterior localization to the extent possible in BA9 and BA8. White matter was removed in microdissection, so that extraction of DNA and protein was subsequently from the gray matter.
PFC gray matter (100 mg per subject) was pulverized in liquid nitrogen, and stored for later analysis. Cerebellar tissue dissected from the lateral part of the posterior lobe of the cerebellum (∼8- 10 g) was sent for tissue toxicological evaluation at NMS (Willow Grove, PA, USA); 337 compounds were included in the Expanded 8052TI panel. Sufficient tissue was not available for toxicological testing of 4 subjects. NMS testing service for postmortem tissue requires a minimum of 8 g of brain tissue, 4 subjects did not have sufficient cerebellum tissue available, as being used in other studies, and we did not substitute another brain region which is more valuable and would not yield consistent comparisons for the remaining subjects.
PFC samples were extracted for genomic DNA assays for mtDNA copy number, albumin copy number, and mtDNA common deletion. Approximately 30 mg of the pulverized tissue was used with the Qiagen DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer's suggested protocol. Lysis time was limited to 3 h and elution volumes were 200 µL in Qiagen AE buffer; average recovery was 2.7 µg DNA according to Qubit measurements.
mtDNA Copy Number
Human Albumin Standard
A human control genomic DNA sample (50 ng) was used to amplify a 200-bp PCR product representing the human albumin gene (ALB) on chromosome 4. The primers used were: Alb forward (5′-CCGTGGTCCTGAACCAGTTA-3′) and Alb reverse (5′CTCCTTCTCAGAAAGTGTGCAT-3′). The PCR product was run on a 1% agarose gel, and the band was purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). The purified product was quantified on the Qubit 2.0 Fluorometer (Invitrogen, Life Technologies, CA, USA), then used to make a standard curve with dilutions ranging from 100,000 copies/µL to 97 copies/µL. Quantitative real-time PCR was run using the Applied Biosystems 7900HT real-time instrument (Waltham, MA, USA) in 12-µL reaction volumes. The following cycling parameters were used: 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min. Within the plate, 2 µL of each dilution of the 11 point standard curve and 2 µL of each PFC DNA at 1 ng/µL were run in triplicate. Each 12 µL PCR reaction included 0.25 µL of each 10 µM primer, Alb nested forward (5′-TGCATGAGAAAACGCCAGTA-3′), Alb nested reverse (5′-TCTGCATGGAAGGTGAATGT-3′), and 6.25 µL of Power SYBR Green PCR Master Mix (Life Technologies, Foster City, CA, USA).
Mitochondria Copy Number Standard
A human genomic DNA sample (50 ng) was PCR amplified with mitochondrial-specific primers Fuke forward [31] (5′-AGGCGCTATCACCACTCTGTTCG-3′) and Fuke reverse [31] (5′-AACCTGTGAGGAAAGGTATTCCTGC-3′) resulting in a 326-bp PCR product. This product was run on a 1% agarose gel, purified using the QIAquick Gel Extraction Kit and cloned into the pCR 2.1-TOPO vector (Invitrogen, Life Technologies, CA, USA) resulting in a 4,257-bp “WT” plasmid. The concentration of the plasmid was measured with the Qubit Fluorometer, and copies of WT mitochondria per microliter were determined. Subsequent dilutions of the 11-point standard curve ranged from 10,000,000 to 9,766 copies/µL. Quantitative real-time PCR conditions and volumes matched those used for the ALB SYBRGreen amplification above except that the following primers were used: WT nested F (5′-TTACAATCGGCATCAACCAA-3′) and WT nested R (5′- GCTAATGCTAGGCTGCCAAT-3′). Mitochondrial DNA copy number for the PFC samples was determined using the ratio of WT copy number/ALB copy number.
MtDNA Deletion
A 301-bp gBlock Gene Fragment (Integrated DNA Technologies, Coralville, IA, USA) was synthesized encompassing the mitochondria common deletion breakpoints (mt8224-8469 and mt13447-13501). An 11-point standard curve was created from this gBlock with copies ranging from 50,000 to 49 copies/µL. A TaqMan MGB Probe (Applied Biosystems, Foster City, CA, USA) was designed to MT: 8284–8313 to accurately measure the common deletion molecules for each sample. All standards and samples were run in triplicate using 2 µL DNA. For samples, this represents an amount of 2 ng of genomic DNA. The 12 µL total reaction volume consisted of: 0.9 µM each primer (deletion forward 5′-AGGGCCCGTATTTACCCTAT-3′ and deletion reverse 5′-GCTAATGCTAGGCTGCCAAT-3′), 0.25 µM TaqMan Probe 5′-6-FAM-CTAAGTTAGCTTTACAGTGGGCTCTAGAGG-MGBNFQ-3′, and 6 µL TaqMan Gene Expression Master Mix (Applied Biosystems). Cycling parameters were 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min on the Applied Biosystems 7900HT real-time machine, selecting ROX as the passive reference. Common deletion percentage was calculated using the following equation: % common deletion = (deletion copies)/(WT copies + deletion copies) × 100.
Complex I Concentration and Activity Measurements
Sample Isolation
PFC samples (60–100 mg frozen brain tissue) were washed with cold phosphate-buffered saline (PBS) without calcium and magnesium, pH 7.4 (21–040-CM, Corning, NY, USA) and resuspended in 300 µL PBS according to the manufacturer's protocol (AB178011, Abcam, Cambridge, UK). The brain samples were extracted by using a 1-mL Dounce homogenizer on ice for 30–40 strokes until samples were solubilized, then the solution was centrifuged at 3,000 rpm for 5 min at 4°C. The first supernatant was collected and measured for protein concentration on a Qubit to ensure adequate protein for both ELISA and activity assays. The input range of PFC for beginning protein extraction was 60–100 mg to ensure a recovery of 500-1,000 µg protein for the subsequent ELISA and activity assays to be run using duplicate samples. There was no difference in the total protein recovered among the three groups (mean % recovery ± SD), BD (1.69 ± 0.4), SZ (1.51 ± 0.4), and controls (1.63 ± 0.4).
The supernatant protein was then extracted by adding 10× detergent (Part 8201086, Abcam, Cambridge, UK) with a 1:10 ratio; incubated on ice for 30 min and centrifuged at 16,000 g at 4°C for 20 min. The resulting second supernatants were collected, aliquoted, and stored at −80°C for complex I ELISA and activity assays.
Complex I ELISA Assay
An ELISA kit (AB178011, Abcam, Cambridge, UK) was used for the quantitative measurement of NADH dehydrogenase protein (complex I) concentrations in human tissue extracts. The ELISA monoclonal capture and detection antibodies were raised against intact complex I isolates. The exact epitopes have not been determined; however, immunoprecipitation experiments with the capture antibody result in expected large molecular weight protein with expected subunit composition of human complex I [32,33]. The brain extracts of complex I were diluted 1:400 in 1× cell extraction buffer that resulted in values near the midpoint of the standard curve described below. All standards, samples, and controls were run in duplicate in 96-well plates precoated with a capture antibody provided in the Abcam kit. The materials and reagents were equilibrated to room temperature, and 50 µL of each sample, standard, and control were added to the plate wells. Next, 50 µL of antibody cocktail (capture antibody and detector antibody with antibody diluent 5B) was added to each well, the plate was sealed and then incubated for 1 h at room temperature with shaking at 400 rpm. After the 1 h, wells were aspirated or decanted, then washed with 1× wash buffer 3 times. The color substrate TMB (3,3′,5,5′-tetramethylbenzidine) was added to each well (100 μL) and incubated for 10 min in the dark while shaking at 400 rpm; then, 100 µL stop solution was added to each well and shaken for 1 min. The optical density (OD) at 450 nm was recorded in a 96-well spectrophotometer (Spectramax plus, Molecular Devices). Each assay included a standard curve prepared from the HeLa lyophilized cell extract (included in the ELISA kit), and additional standards were prepared from a postmortem human PFC brain extract. HeLa cell stock solution was diluted for the standard curve by reconstituting HeLa lyophilized cell extract with 40 µL water and performing 1:2 serial dilutions using 1× cell extraction buffer to yield a dilution series ranging from 400 to 12.5 µg/mL. The standard complex I protein curve was calculated from regression of the average OD 450 nm reading of each HeLa standard against their concentrations (online suppl. Fig. 1). A calibration human PFC extract was diluted 1:200, 1:300, 1:400, 1:500, 1:600, and 1:700 to determine the linear range of the ELISA assay (online suppl. Fig. 2). The assay appears linear within these dilutions.
Complex I Activity Assay
Midpoints in the activity assay curves were determined for 2 PFC samples at various protein levels (online suppl. Fig. 3). Each brain sample was then diluted to the same complex I protein (amount determined above by ELISA) to determine the rate of complex I activity. All samples and controls were run in duplicate; the samples were diluted to 200 µL with incubation solution (AB109721, Abcam, Cambridge, UK). The diluted 200 µL sample and control were loaded into each precoated antibody capture well in a 96-well plate and incubated for 3 h at room temperature according to the manufacturer's protocol (AB109721, Abcam, Cambridge, UK). After careful decanting and washing of each well with 300 µL 1× buffer solution twice, 200 µL of an activity assay solution was added to each well and bubbles were carefully removed. The activity assay working solution was 188 µL 1× buffer, 10 µL of 20× NADH and 2 µL of 100× dye per well. The plate was placed in a spectrophotometer (SpectraMax plus, Molecular Devices), and the OD was recorded at 450 nm for 30 min in 30-s intervals. The rate of activity was expressed as the change in absorbance per minute using this equation: Complex I activity for each sample (milli-optical density units, mOD/min) = (1,000 × ΔOD450/30 min) where Δ represents the final minus initial OD450.
Data Analysis
All analyses were conducted using JMP (JMP® Genomics 8.0, 64-bit) regression modeling incorporating diagnosis and covariates of age, sex, and pH. Least squares differences were detected with alpha = 0.05.
Results
The three groups (SZ, BD, and controls) were significantly different for covariates of pH, age, and sex (Table 1), while postmortem interval (PMI) was not significantly different between groups. As such, pH, age, and sex were included as covariates in all subsequent analyses of the three dependent variables in this study (mtDNA copy number, mtDNA common deletion, and complex I activity). The inclusion of PMI did not alter the results.
Table 1.
Demographics of three groups analyzed for mitochondria complex I, mtDNA copy number, and mtDNA common deletion level
| Bipolar disorder | Control | Schizophrenia | |
|---|---|---|---|
| Sex | |||
| Female, n1 | 13 | 6 | 3 |
| Male, n | 13 | 35 | 24 |
| pH | |||
| Mean | 6.68* | 6.86 | 6.64* |
| SD | 0.15 | 0.22 | 0.20 |
| Age, years | |||
| Mean | 46.0* | 57.9 | 41.2* |
| SD | 12.5 | 12.7 | 10.1 |
| PMI | |||
| Mean | 20.5 | 18.8 | 21.0 |
| SD | 6.61 | 7.12 | 7.39 |
| Number of medications | |||
| Mean | 3.7* | 0.9 | 2.1* |
| SD | 3.2 | 1.7 | 1.7 |
| Min | 0 | 0 | 0 |
| Max | 10 | 10 | 5 |
p < 0.05, significantly different from control group mean.
Indicates imbalance in the proportion of male and female subjects in design.
mtDNA Copy Number
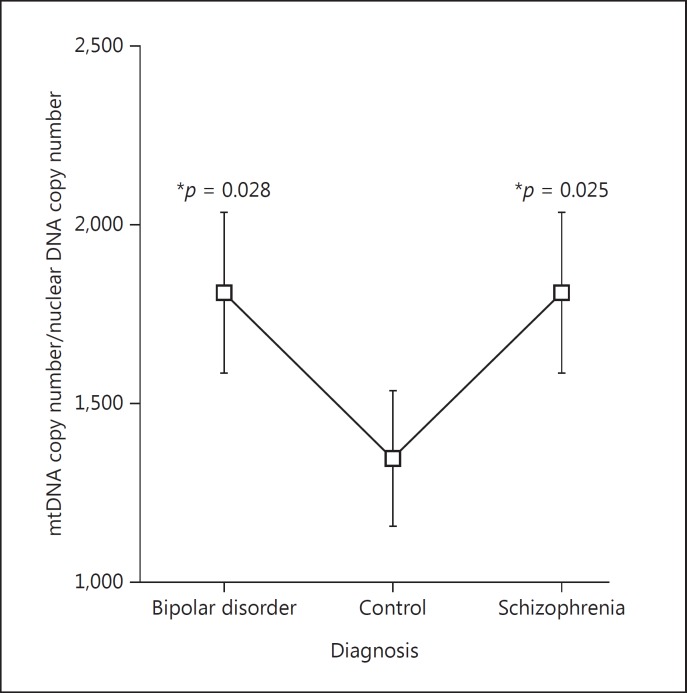
The mtDNA copy number was significant for the diagnosis main effect (p = 0.042), while pH (p = 0.81), sex (p = 0.21), and age (p = 0.81) were not significant. The mtDNA copy number in PFC showed a significant increase in both BD least square mean difference + standard error (SE) (369.3 ± 162.4, p = 0.025) and SZ least square mean difference + SE (369.6 ± 165.6, p = 0.028) compared to controls (Fig. 1; Table 2).
Fig. 1.
Mitochondrial DNA copy number was significantly increased in both bipolar disorder and schizophrenia compared to controls.
Table 2.
Effects of major factors on MtDNA copy number and group comparisons
| p value | ||||||
| A MtDNA copy number | ||||||
| Diagnosis | 0.042 | |||||
| Sex | 0.213 | |||||
| Age | 0.813 | |||||
| pH | 0.817 | |||||
| Group | Difference | SE diff. | Lower CL | Upper CL | p value | |
| B Diagnosis compared to | ||||||
| Schizophrenia | Control | 369.57 | 165.67 | 40.1 | 698.9 | 0.028* |
| Bipolar disorder | Control | 369.38 | 162.41 | 46.4 | 692.3 | 0.025* |
| Schizophrenia | Bipolar disorder | 0.19 | 156.39 | −310.7 | 311.1 | 0.999 |
The mtDNA copy number was increased in subjects with schizophrenia and bipolar disorder (B). The mtDNA copy number was normalized to the number of nuclear copies of DNA. SE, standard error; CL, confidence limit.
As in our previous work using SYBR Green qPCR [13], this redesigned SYBR Green mtDNA copy number assay shows similar ranges of mtDNA copy number estimates in DLPFC samples (range 632-3,513 mtDNA copies per single copy number of the nuclear gene ALB). The mean and standard deviation for the entire current sample was 1,608 ± 541 mtDNA copies per nuclear genome in PFC.
mtDNA Deletion
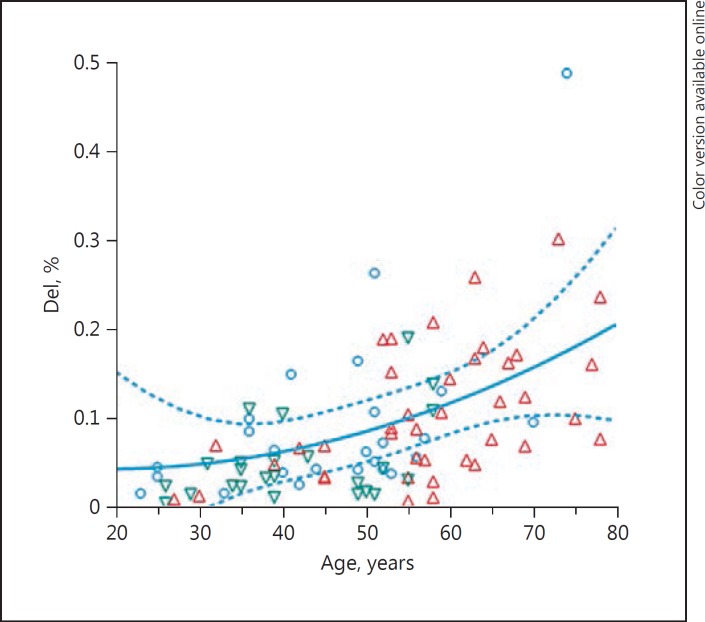
The percentage of mtDNA common deletion load was calculated as the ratio of number of deleted mtDNA molecules divided by the total number of mtDNA copies multiplied by 100. The total mtDNA copy number is the sum of mtDNA deletion copy number + WT mtDNA copy number. Individuals with SZ showed a significant decrease in mtDNA common deletion by 50% before correction for the other effects in the model. The percentage of mtDNA common deletion was not significantly altered in SZ or BD (Table 3A) using the model corrected for age, sex, and pH. However, as expected, the levels of the mtDNA common deletion significantly increased with age in the PFC samples (Fig. 2, p = 0.002). Across the entire cohort, females showed a larger percentage of common deletion compared to male brains (p = 0.03), shown in Table 3B. This amounted to a 60% increase in common deletion levels in female PFC compared to male PFC samples.
Table 3.
The percentage of mtDNA common deletion in the model including diagnosis, sex, age, and pH
| Least square means, % mthNA common deletion | SE | Unadjusted means, % mthNA common deletion | |
|---|---|---|---|
| A Diagnosis | |||
| Bipolar disorder | 0.126 | 0.019 | 0.115 |
| Control | 0.103 | 0.019 | 0.105 |
| Schizophrenia | 0.092 | 0.022 | 0.049 |
| B Sex | |||
| Female | 0.127 | 0.020 | 0.144 |
| Male | 0.079 | 0.011 | 0.075 |
The age variable was significant in the model (p = 0.002) while the diagnosis was not significant (A). Differences between sex (B) were significant (p = 0.03), female subjects having more mtDNA common deletion in PFC following adjustments for other effects. SE, standard error.
Fig. 2.
Percent levels of mitochondria common deletion increased with age in prefrontal cortex samples. Red triangle, control; blue circle, bipolar disorder; green triangle, schizophrenia.
Complex I Activity
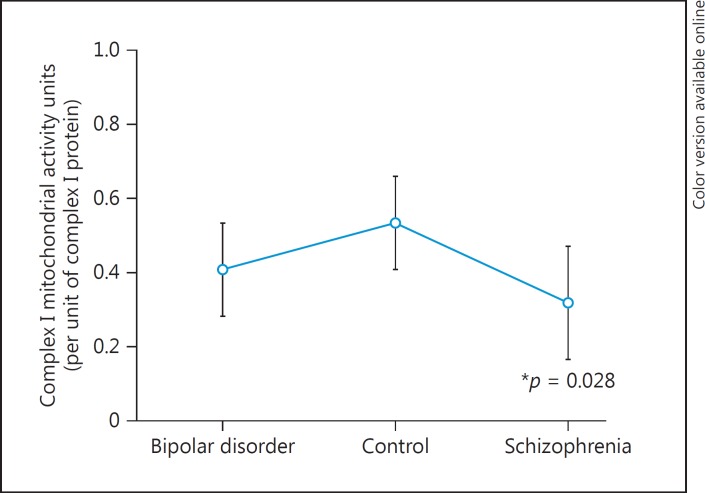
The concentrations of complex I protein were determined by ELISA. An equal amount of complex I protein (equivalent to the 0.1 mg/mL HeLa standard) was then assayed for complex I activity. In the activity assay, we report the units as mOD/min) of enzymatic NADH activity calculated based upon the slope of the activity curve for 30 min using positive HeLa cell extracts as controls (online suppl. Fig. 1). The complex I mOD/min activities were analyzed by the model that included diagnosis, sex, age, and pH. These variables were not significant for complex I activity using three groups in the diagnosis (Table 4A). The a priori planned least square mean comparison of SZ compared to controls was significant (p = 0.02) and showed a 53% reduction of the complex I activity in subjects with SZ (Table 4B, Fig. 3).
Table 4.
Effects of major variables on mitochondrial complex I activity and group comparisons
| p value | ||||||
| A Factors | ||||||
| pH | 0.081 | |||||
| Diagnosis | 0.085 | |||||
| Sex | 0.392 | |||||
| Age | 0.529 | |||||
| Level | Difference | SE diff. | Lower CL | Upper CL | p value | |
| B Level | ||||||
| Control | Schizophrenia | 0.213 | 0.095 | 0.02 | 0.40 | 0.028* |
| Control | Bipolar disorder | 0.124 | 0.094 | −0.06 | 0.31 | 0.196 |
| Bipolar disorder | Schizophrenia | 0.089 | 0.095 | −0.10 | 0.27 | 0.348 |
The effects of major variables on mitochondrial complex I activity were not significant (A) in the regression model. The mitochondrial complex I activity was decreased in subjects with schizophrenia by 53% in a planned group comparison (B). SE, standard error; CL, confidence limit.
Fig. 3.
Mitochondrial complex I activity for each diagnosis group. There was a significant reduction in schizophrenia compared to controls (p = 0.028).
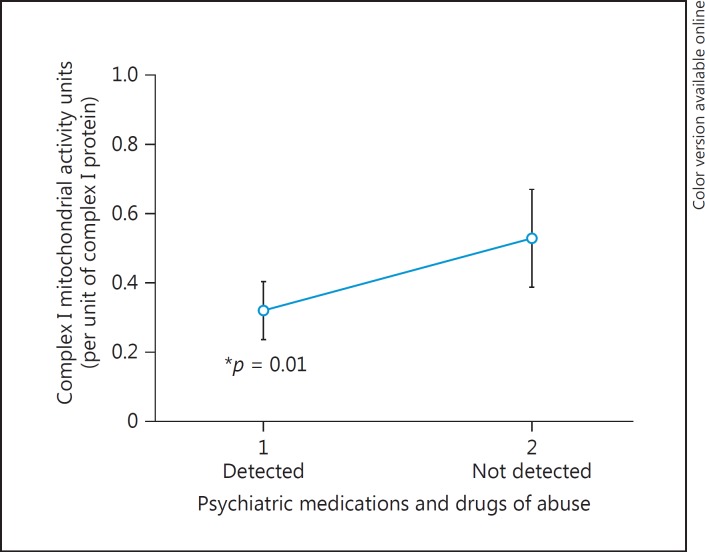
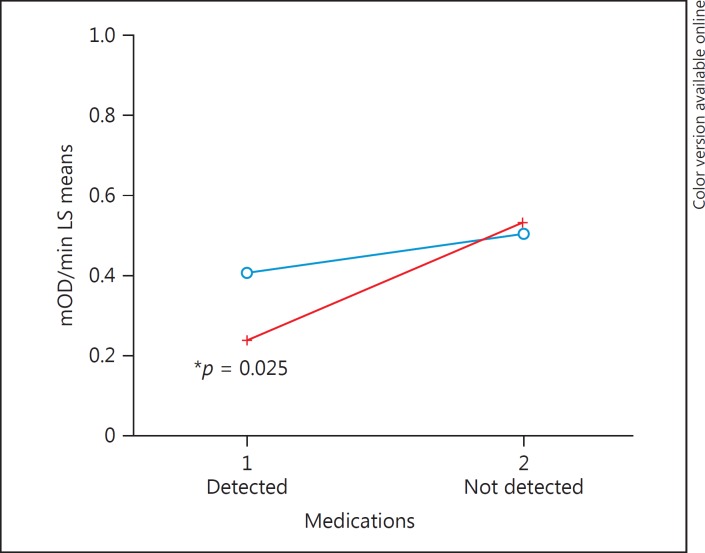
We next focused on a case analysis (Table 5) and compared subjects with and without psychotropic medications at the time of death and including the covariates of age, pH, and sex in the regression model of complex I activity. The list of all detected medications and compounds is shown in online supplementary Table 2. These compounds were detected by toxicological screening at NMS Labs of each subject's cerebellum tissue. There were no significant differences in cofactors such as age and pH between BD and SZ groups (Table 5A), whereas compared to controls these factors were significantly different (Table 1). Due to polypharmacy, an analysis of the different classes of individual medications was not conducted, as subjects could be taking antianxiety, antidepressant, and antipsychotic medications simultaneously, resulting in extremely small subgroups. There were subjects that had no psychotropic medications within the BD and SZ groups, and those were compared to subjects in the BD and SZ groups with detected psychotropic medications. Complex I activity significantly decreased in subjects that had psychotropic medications in their brains at the time of death compared to psychiatric subjects that did not have psychotropic medications detected in their brains (Fig. 4). This main effect of psychotropic medications appeared to be due to the subjects with SZ having psychotropic medications at the time of death showing a significantly decreased complex I activity compared to SZ not having psychotropic medications in the brain at the time of death (p = 0.025; Table 6; Fig. 5). This effect did not appear in subjects with BD (Table 6; Fig. 5). As mentioned, it was not possible to reliably determine which medication classes have the most pronounced impact on mitochondrial function as 53 compounds were detected in various combinations (online suppl. Table 2). Subjects with BD had significantly more medications (p = 0.028) detected at the time of death (mean ± SE, 3.76 ± 0.5) compared to SZ subjects (2.18 ± 0.49). When excluding subjects with zero compounds detected, BD subjects had an average of 5 compounds compared to 2.8 compounds in the brain at the time of death in SZ (p = 0.01) (Table 5B).
Table 5.
Case-case demographic analysis and pooled case analysis of study groups
| A Case-case demographic analysis | |||||||
| Psychotropic medications detected by postmortem toxicology | |||||||
| positive detection | no compounds detected | total | |||||
| female (n = 14) | male (n = 25) | sum (n = 39) | female (n = 2) | male (n = 12) | sum (n = 14) | (n = 53) | |
| Age, years | |||||||
| Mean | 48.43 | 42.28 | 44.49 | 47.00 | 40.17 | 41.14 | 43.60 |
| SD | 12.90 | 9.02 | 10.83 | 15.56 | 13.58 | 13.45 | 11.54 |
| pH | |||||||
| Mean | 6.63 | 6.68 | 6.66 | 6.81 | 6.66 | 6.68 | 6.66 |
| SD | 0.09 | 0.13 | 0.12 | 0.28 | 0.29 | 0.28 | 0.18 |
| Number of compounds | |||||||
| Sum | 66 | 88 | 154 | 0 | 31 | 3 | 157 |
| B Pooled case analysis | |||||||
| Cases | Number of subjects with detectable psychotropic compounds | Number of psychotropic compounds | |||||
| Mean | SD | SEM | lower 95% | upper 95% | |||
| Bipolar disorder | 19 | 5.0 | 2.9 | 0.66 | 3.5 | 6.4 | |
| Schizophrenia | 20 | 2.8 | 1.3 | 0.31 | 2.2 | 3.4 | |
Case-case demographic analysis combined groups of bipolar disorder and schizophrenia by psychotropic medications present at the time of death in the brain and excludes controls. There were no significant demographic differences between the positive and negative postmortem toxicology groups.
In the pooled case analysis, the BD group showed significantly more detected psychotropic compounds in the brain at the time of death compared to the SZ group. The numbers of BD (n =4) and SZ (n = 5) subjects that showed 0 psychotropic compounds were nearly the same. SD, standard deviation; SEM, standard error of the mean.
Caffeine only.
Fig. 4.
Complex I activity was lower in brains with detected psychotropic medications than those without. Case-case analysis matched for pH, age, and gender. Covarying with number of mitochondria did not change this result.
Table 6.
Interaction of diagnosis and psychotropics detected in the brain on complex I activity
| Level | Control level | Difference | SE difference | Lower CL | Upper CL | p value |
|---|---|---|---|---|---|---|
| Bipolar disorder, 1 | Schizophrenia, 2 | −0.124 | 0.112 | −0.396 | 0.148 | 0.532 |
| Bipolar disorder, 2 | Schizophrenia, 2 | −0.025 | 0.158 | −0.407 | 0.356 | 0.996 |
| Schizophrenia, 1 | Schizophrenia, 2 | −0.291 | 0.107 | −0.551 | −0.031 | 0.025* |
Psychotropic (1, detected one or more drugs of abuse or psychotropic medications; 2, no psychotropics except caffeine or nicotine only). SE, standard error; CL, confidence limit.
Fig. 5.
Complex I rate of activity was decreased in schizophrenia subjects that had psychotropic medications detected at time of death compared to the schizophrenia subjects that did not. These results were not seen in the bipolar subjects. Blue indicates bipolar disorder; red indicates schizophrenia.
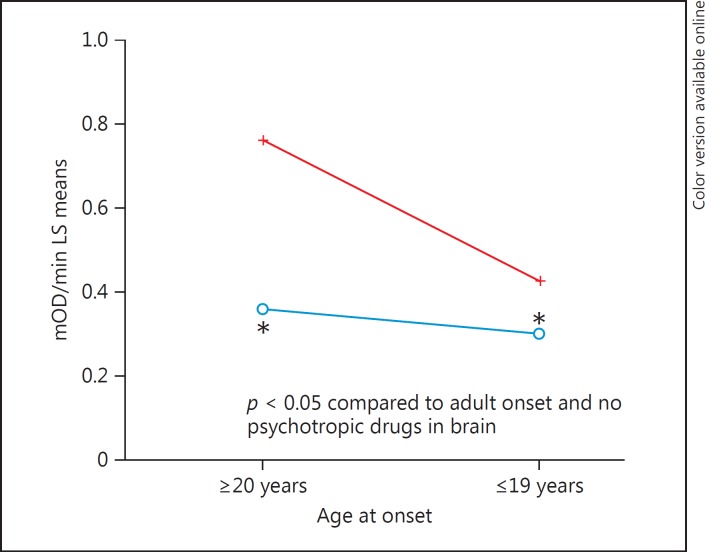
In the final analysis of complex I activity in BD and SZ cases, age at onset was utilized as a surrogate for illness severity. In combination with psychotropic medications, age at onset was included in a regression model with covariates (age, sex, and pH) to determine if both variables and interaction effects were significant. The result (Table 7) showed that psychotropic medications main effect was significant (p = 0.001), age at onset was significant (p = 0.016), while the interaction of psychotropic drugs and age at onset was at a trend level, but not significant (p = 0.08). These effects (Fig. 6) show that the highest complex I activity is found in adult-onset psychiatric cases that did not have psychotropic medications detected in the brain at the time of death compared to cases with age at onset during teenage years with psychotropic medications.
Table 7.
Interaction analysis of age at onset and psychotropic medication in a case analysis of complex I rate of activity
| Effect | p value |
|---|---|
| Psychotropic drugs | 0.001* |
| Age at onset | 0.016* |
| Psychotropic drugs × age at onset | 0.084 |
Fig. 6.
Complex I rate of activity and age at onset of disease (red, no psychotropic drugs in toxicology report; blue, yes psychotropic drugs in toxicology report). The highest complex I activity was found in adult-onset cases with no psychotropic medications in the brain.
The correlation of medications with complex I was significant in SZ (p = 0.042), but not in BD (p = 0.32). The lack of a correlation in BD (or perhaps stronger correlation in SZ) might be due to restricted range of number of medications. However, the slope of regression lines is parallel for SZ and BD (online suppl. Fig. 4), so it could be argued that the effects of medications are similar between groups. On the other hand, what is partly driving these correlations are subjects without medications having adult onset illness that show the highest complex I activity (Fig. 6), compared with those subjects with teen onset that have medications.
Discussion
We have studied different measures of mitochondrial function in postmortem brain of subjects with psychiatric disorders and compared to controls to extend prior results. There are no prior reports of complex I, mtDNA copy number and common deletion alterations, in psychiatric disorders evaluating these measures in the same studies.
As anticipated, age was a significant factor in the accumulation of the common deletion from prior work. The mitochondria common deletion levels increase with age in the brain, and the present results confirm those prior reports [9,10,13,31]. We did not find a significant decrease in mtDNA common deletion in SZ after correction for age in PFC. In our prior report, we also did not find a significant decrease in common deletion in DLPFC in SZ compared to controls [13]. This prior analysis combined 10 brain regions and showed there was a decrease in the common deletion in SZ compared to controls [13]; however, this analysis was not conducted herein since we analyzed a single brain region. We also show that female subjects have a 60% increase in the accumulation of the common deletion in the PFC consistent with previous reports in DLPFC showing increased common deletion levels in females compared to males [10,31,34]. The increase in the common deletion in female PFC could indicate that there might be higher levels of reactive oxygen produced due to more common deletion in mtDNA, while the reverse direction of cause could also be consistent. In general, we and others have reported higher levels of mitochondria (as measured by mtDNA) in brain and blood in female subjects. This could be a parsimonious explanation for increased mtDNA deletions in female brains. In aging, there can be an accumulation of the common deletion due to mitochondria dysfunction leading to a fusion of mitochondria into larger and less energetic organelles. There is also the possibility that shorter mtDNA molecules can replicate faster than larger full-length molecules, and that this will also cause the common deletion increases during aging. Thus, as aging occurs, both of these causes could lead to gradual accumulation of the common deletion, advancing more rapidly in brain regions with higher dopamine inputs and metabolic requirements.
The mtDNA copy number was significantly increased in both BD and SZ groups compared to controls in PFC; however, we normalized the amount of complex I protein as determined by ELISA prior to running the complex I activity assay. As such, any differences in copy number should not have a direct effect on the enzymatic activity analysis. The observation that mtDNA copy number is altered in the brains of subjects with SZ is in the opposite direction of those studies showing reduced density of mitochondria in SZ brain using different methods, i.e. electron microscopy and immunohistochemistry [18,35,36,37,38]. Our measure of mtDNA copy number is quantitative, but does not yield size and location, both of which may be critical aspects of the mitochondrial dysfunction in these cases. Based upon our findings with this cohort, we had an initial 60% power to detect a change of ∼360 mtDNA copies at alpha = 0.05. Our data suggest replication of this experiment with 90% power would require a larger cohort (n ≥180).
The current data could be explained by positing fewer functional mitochondria with larger amounts of mtDNA in the brains of both SZ and BD compared to controls. This is consistent with a report that subjects with mitochondrial deficiency show an increased copy number of mitochondrial DNA molecules indicating that replication of mtDNA can occur with concurrent reductions in functional activity [39]. Another potential mechanism to explain increased mtDNA copy number and decreased mitochondria activity is based upon observations of aged mitochondria that become hyperfused, thus increasing mitochondria elongation, and at the same time decreasing oxidative phosphorylation [40].
There were two significant effects observed on complex I activity in PFC. First, there was a significant decrease in complex I activity in subjects with psychotropic medications in brains at the time of death, and second, there was a significant decrease observed in subjects that had illness onset during teen age years compared to adult-onset illness. Taken together, psychotropic medications and early disease onset produced a significant decrease in complex I activity compared to adult onset illness with no detectable psychotropic medications. Therefore, we can conclude that pooled cases with BD or SZ show significant decline in complex I activity concomitant to medications and early onset of illness. Since both factors are highly significant, it requires further research to determine whether prior to onset of either BD or SZ and medications there is a mitochondrial dysfunction.
Prior studies of the frontal cortex of subjects with BD [41] and SZ [42] have reported reductions in complex I activity. These reductions might be the result of an overrepresentation of subjects with chronic antipsychotic medication that reduces complex I activity. To date, very few studies of deficits in complex I have been reported in medication-free subjects as reviewed [5,6,42]. The reports of a deficit in complex I are found in studies of medicated subjects, e.g. we have previously found a reduction of mtDNA-encoded gene expression in complex I. Of the 7 mtDNA-encoded transcripts in complex I, 6 were significantly decreased in the expression in SZ [9].
The pharmacological profile of the medications in the present cohort strongly suggests that beyond membrane-bound receptor targets, these medications have effects on mitochondria activity. The effects on mitochondria might not be direct, as the brain activity such as neurotransmission could be reduced due to medications and illness, and this could be an explanation of the measures of reduced mitochondria complex I activity.
The argument for medication effects on complex I mitochondria activity is supported by animal and in vitro studies that show antipsychotics impair complex I. Typical APDs are more potent inhibitors compared to atypical APDs [43]. Haloperidol and clozapine tend to inhibit anterograde transport of mitochondria stranding them in perinuclear locations [44,45]. This sequestration of mitochondria away from axon and spines could reduce the amount of energy released by mitochondria, which directly affects neurotransmission and adequate calcium buffering in synapses.
Another possibility is that different pharmacological treatments can interact with each other in certain patients further lowering mitochondria activity. A more severe form of illness might require more medications, further impairing complex I. In contrast, patients that had no psychotropic medications in the brain and an adult onset showed an increase in complex I activity of 0.76 ± 0.1, compared to healthy controls 0.52 ± 0.05 (mean mOD/min ± SEM). Thus, in the nonmedicated state, it is possible that patients may have increased complex I activity. Or it could be argued that these subjects had milder forms of illness or had been responsive to very low and intermittent medication dosage, and in stopping medication the mitochondria complex I activity rebounded to pretreatment levels. In a post hoc analysis of the number of psychiatric hospitalizations, there was no difference in hospitalizations for subjects with and without detectable psychotropic medications (mean number of hospitalizations ± SD; 2.4 ± 1.8; 2.3 ± 1.7, respectively). The illness severity was also evaluated by using age at onset, which showed that subjects with adult-onset illness had similar numbers of hospitalizations compared to subjects with teenage-onset illness (mean number of hospitalizations ± SD: 2.1 ± 1.6; 2.7 ± 1.9, respectively). Reductions in complex I activity for teen-age onset cases compared to adult-onset cases were not due differences in illness severity as measured by the proxy variables of the number of hospitalizations or the number of medications. Overall, the subjects with BD had significantly more medications than subjects with SZ, yet the SZ and BD groups did not show a significant difference in complex I activity when compared to one another.
Two putative sites of the inhibitory action of antipsychotics and dopamine itself are located in complex I at different locations [5,42]. Since both dopamine and APDs can inhibit complex I, we suggest that drug-treated subjects with hyperdopaminergic etiology may have a dual inhibition of complex I through separate sites in complex I. The net effect of inhibiting complex I could result in less O2 consumption in nerve terminals and less ATP production [5,42], which is supported by bioenergetic imaging studies of DLPFC in subjects with SZ. Impaired complex I function could also lead to increased oxidative stress by transferring more electrons to O2 and creating reactive oxygen species.
There are over 1,200 genes listed in MitoCarta (www.broadinstitute.org/pubs/MitoCarta) with potential mitochondrial function [46]. In a recent review of proteomic, transcriptomic, and genetic studies relevant to SZ, we reported that >250 MitoCarta genes had been shown to be significant in at least one study relevant to SZ [18]. Alterations in these genes could cause mitochondrial dysfunction, although predicting the exact direction and aspect of mitochondrial dysfunction is difficult. Conceivably, regulators of complex I might be altered and might account for the present findings independent of medication effects. In addition, other conditions present in patients might lead to complex I alterations such as metabolic syndrome. It is known that small molecules such as glucose, glutamate, and arachidonic acid can negatively regulate complex I activity [reviewed in [6]. These speculations will require a cohort of prospective subjects to undergo bioenergetics measurements using noninvasive methods and prior to medications to determine if lower or higher mitochondrial function precedes treatment. Alternatively, neurons from induced pluripotent stem cells could be generated and studied for the potential effects of antipsychotic medications on mitochondria as shown in the work on BD and lithium [47].
Potential limitations to the interpretation of this study are in two areas. First, our sampling technique for gross homogenate predominantly included gray matter prefrontal cortical samples from BA9. We plan to analyze complex I activity among other prefrontal, occipital, and temporal regions to address differences in cases and controls. Further, some differences in projections to adjacent prefrontal regions could influence the complex I activity levels. Second, the current findings for a single brain region indicate that the PFC has both medication effects on complex I activity, and perhaps effects due to psychopathology. However, the PFC subserves multiple anatomical connections that require different ranges of synaptic activity andconsequently varying levels of energy required to maintain the balance of inhibition and excitatory tone. There is, for example, more input from frontal eye fields in BA8 compared to BA9, while there are more interconnections with anterior cingulate to BA9 than to BA8. So, these adjacent regions have two circuits that presumably will have varying bioenergetic capacity and utilization. These baseline differences might be augmented further by distribution differences of medication among brain regions. Since medications can differentially be absorbed in different circuits, the use of a cerebellum as a proxy for brain levels would be better addressed by individual toxicological analysis of each brain region. This regional difference in distribution for an antidepressant medication (venlafaxine) and metabolite O-desmethylvenlafaxine between regions substantiates the need to measure individual regions [48]. This is costly, so presently we have chosen the cerebellum and presence of the compounds as a proxy measure. The semiquantitative gene expression levels for 3 transporters where medications are likely to have an impact do not differ appreciably along major cortical ribbon for SLC6A2, SLC6A3, SLC6A4, the norepinephrine, dopamine, and serotonin transporters, respectively. Thus, the metabolism and distribution of drugs that affect these transporters can have a substantial impact on the cellular function, where inhibition of complex I was shown in the present study. As the findings of Murrell et al. [48] indicate that different cortical and cerebellar regions have varying ratios of venlafaxine and O-desmethylvenlafaxine, this corroborates differences in distribution, metabolism, and uptake. These, in turn, would differentially affect mitochondrial activity, and thus medication effects might be more pronounced in different brain regions. Thus, we cannot accurately predict the regional heterogeneity of brain regions for medication effects, baseline mitochondrial activity, and regional differences due to brain activation and bioenergetic requirements. We have planned additional brain regions to experimentally determine these effects and correlations between regions involved in the psychopathology of these disorders.
Future analyses aimed to identify individuals at-risk for SZ and BD should include the analysis of mtDNA variants as well as nuclear gene variant analysis, and whether certain individuals with varying baseline mitochondrial function develop impaired mitochondrial function both in responders and nonresponders to drug treatments [47]. Drugs that enhance mitochondrial function and localization may be an efficacious strategy in individuals that have a lower mitochondrial function.
Conclusions
The present study suggests that in chronic patients with SZ and BD there is a significant reduction in mitochondrial activity, particularly in subjects that had taken recent APD/ADD medications. One indicator of illness severity, age at onset, showed that subjects with a younger age at onset showed more reduction in complex I activity than patients with adult-onset illness. Present data show an increase in copies of mtDNA in SZ and BD, a surrogate for the actual numbers of mitochondria in brain. These results support prior studies showing that APD/ADD treatments can lead to mitochondrial hypofunction and suggest that drugs which target improvement in mitochondrial function may be a beneficial therapeutic strategy for some SZ and BD patients with early age at onset.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors are members of the Pritzker Neuropsychiatric Disorders Research Foundation.
Author Contributions
Brandi Rollins and Ling Z. Morgan conducted all laboratory experiments; Ling Z. Morgan conducted all complex I assays; Brandi Rollins conducted all real-time qPCR measurements. Adolfo Sequeira, Brandi Rollins, and Ling Z. Morgan conducted microdissection of tissue for all analyses. Brooke E. Hjelm assisted in development and optimization of the qPCR assays described in addition to drafting and editing of the manuscript. William E. Bunney was involved in the collection of subjects; Alan F. Schatzberg, Jack D. Barchas, Francis Lee, Richard M. Myers, Stanley J. Watson, William E. Bunney, and Huda Akil were involved in the manuscript preparation, reading, and editing; Marquis P. Vawter designed the experimental approach and conducted data analysis.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We are grateful to the donors' families for the brain donation, and David Walsh, Preston Cartagena, Richard Stein, Joe Davis, and Kathleen Burke, for characterizing, acquiring, and storing the postmortem human brains. We also acknowledge the many hours of dissection from the University of Michigan team and UCI team. The Pritzker Neuropsychiatric Disorders Research Fund LLC (PNDRF), William L. Penzner Foundation, and NIH grant (MPV R01MH08580) funded this research.
References
- 1.Devaraju P, Zakharenko SS. Mitochondria in complex psychiatric disorders: lessons from mouse models of 22q11.2 deletion syndrome: hemizygous deletion of several mitochondrial genes in the 22q11.2 genomic region can lead to symptoms associated with neuropsychiatric disease. Bioessays. 2017;39:1600177. doi: 10.1002/bies.201600177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sengupta B, Stemmler M, Laughlin SB, Niven JE. Action potential energy efficiency varies among neuron types in vertebrates and invertebrates. PLoS Comput Biol. 2010;6:e1000840. doi: 10.1371/journal.pcbi.1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard AK, Craig EL, Chakrabarti L. Mitochondrial complex 1 activity measured by spectrophotometry is reduced across all brain regions in ageing and more specifically in neurodegeneration. PLoS One. 2016;11:e0157405. doi: 10.1371/journal.pone.0157405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodenburg RJ. Mitochondrial complex I-linked disease. Biochim Biophys Acta. 2016;1857:938–945. doi: 10.1016/j.bbabio.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Shachar D. Mitochondrial multifaceted dysfunction in schizophrenia; complex I as a possible pathological target. Schizophr Res. 2017;187:3–10. doi: 10.1016/j.schres.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Duong A, Che Y, Ceylan D, Pinguelo A, Andreazza AC, Trevor Young L, Berk M. Regulators of mitochondrial complex I activity: a review of literature and evaluation in postmortem prefrontal cortex from patients with bipolar disorder. Psychiatry Res. 2016;236:148–157. doi: 10.1016/j.psychres.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Viswanath B, Jose SP, Squassina A, Thirthalli J, Purushottam M, Mukherjee O, Vladimirov V, Patrinos GP, Del Zompo M, Jain S. Cellular models to study bipolar disorder: a systematic review. J Affect Disord. 2015;184:36–50. doi: 10.1016/j.jad.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 8.Szolnoki Z, Szekeres M, Szaniszlo I, Balda G, Bodor A, Kondacs A, Mandi Y, Somogyvari F. Decreased number of mitochondria in leukoaraiosis. Arch Med Res. 2015;46:604–608. doi: 10.1016/j.arcmed.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequeira A, Martin MV, Rollins B, Moon EA, Bunney WE, Macciardi F, Lupoli S, Smith EN, Kelsoe J, Magnan CN, van Oven M, Baldi P, Wallace DC, Vawter MP. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan KJ, Ratnaike TE, De Gruyter HL, Jaros E, Turnbull DM. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer's disease. Neurobiol Aging. 2012;33:2210–2214. doi: 10.1016/j.neurobiolaging.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Mao P, Gallagher P, Nedungadi S, Manczak M, Shirendeb UP, Kohama SG, Ferguson B, Park BS, Reddy PH. Mitochondrial DNA deletions and differential mitochondrial DNA content in Rhesus monkeys: implications for aging. Biochim Biophys Acta. 2012;1822:111–119. doi: 10.1016/j.bbadis.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamdani F, Rollins B, Morgan L, Sequeira PA, Vawter MP. The somatic common deletion in mitochondrial DNA is decreased in schizophrenia. Schizophr Res. 2014;159:370–375. doi: 10.1016/j.schres.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 15.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortopassi GA, Shibata D, Soong NW, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci USA. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43:645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G, Gall CM, Sequeira A, Vawter MP. Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia. Mol Neuropsychiatry. 2015;1:201–219. doi: 10.1159/000441252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Hu M, Zong X, He Y, Wang D, Dai L, Dong M, Zhou J, Cao H, Lv L, Chen X, Tang J. Association of telomere length and mitochondrial DNA copy number with risperidone treatment response in first-episode antipsychotic-naive schizophrenia. Sci Rep. 2015;5:18553. doi: 10.1038/srep18553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O'Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner-Lavie H, Klein E, Ben-Shachar D. Mitochondrial complex I as a novel target for intraneuronal DA: modulation of respiration in intact cells. Biochem Pharmacol. 2009;78:85–95. doi: 10.1016/j.bcp.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeld M, Brenner-Lavie H, Ari SG, Kavushansky A, Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry. 2011;69:980–988. doi: 10.1016/j.biopsych.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem. 2002;83:1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 24.Bachmann E, Zbinden G. Effect of antidepressant and neuroleptic drugs on respiratory function of rat heart mitochondria. Biochem Pharmacol. 1979;28:3519–3524. doi: 10.1016/0006-2952(79)90394-0. [DOI] [PubMed] [Google Scholar]
- 25.Sagara Y. Induction of reactive oxygen species in neurons by haloperidol. J Neurochem. 1998;71:1002–1012. doi: 10.1046/j.1471-4159.1998.71031002.x. [DOI] [PubMed] [Google Scholar]
- 26.Casademont J, Garrabou G, Miro O, Lopez S, Pons A, Bernardo M, Cardellach F. Neuroleptic treatment effect on mitochondrial electron transport chain: peripheral blood mononuclear cells analysis in psychotic patients. J Clin Psychopharmacol. 2007;27:284–288. doi: 10.1097/JCP.0b013e318054753e. [DOI] [PubMed] [Google Scholar]
- 27.Balijepalli S, Boyd MR, Ravindranath V. Inhibition of mitochondrial complex I by haloperidol: the role of thiol oxidation. Neuropharmacology. 1999;38:567–577. doi: 10.1016/s0028-3908(98)00215-9. [DOI] [PubMed] [Google Scholar]
- 28.Adzic M, Brkic Z, Bulajic S, Mitic M, Radojcic MB. Antidepressant action on mitochondrial dysfunction in psychiatric disorders. Drug Dev Res. 2016;77:400–406. doi: 10.1002/ddr.21332. [DOI] [PubMed] [Google Scholar]
- 29.Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, Overman K, Stein R, Tomita H, Potkin S, Myers R, Watson SJ, Jones EG, Akil H, Bunney WE, Jr, Vawter MP. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods. 2007;163:295–309. doi: 10.1016/j.jneumeth.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE., Jr Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuke S, Kametani M, Kato T. Quantitative analysis of the 4,977-bp common deletion of mitochondrial DNA in postmortem frontal cortex from patients with bipolar disorder and schizophrenia. Neurosci Lett. 2008;439:173–177. doi: 10.1016/j.neulet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Murray J, Zhang B, Taylor SW, Oglesbee D, Fahy E, Marusich MF, Ghosh SS, Capaldi RA. The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunopurification. J Biol Chem. 2003;278:13619–13622. doi: 10.1074/jbc.C300064200. [DOI] [PubMed] [Google Scholar]
- 33.Schilling B, Bharath MMS, Row RH, Murray J, Cusack MP, Capaldi RA, Freed CR, Prasad KN, Andersen JK, Gibson BW. Rapid purification and mass spectrometric characterization of mitochondrial NADH dehydrogenase (Complex I) from rodent brain and a dopaminergic neuronal cell line. Mold Cell Proteomics. 2005;4:84–96. doi: 10.1074/mcp.M400143-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Kakiuchi C, Ishiwata M, Kametani M, Nelson C, Iwamoto K, Kato T. Quantitative analysis of mitochondrial DNA deletions in the brains of patients with bipolar disorder and schizophrenia. Int J Neuropsychopharmacol. 2005;8:515–522. doi: 10.1017/S1461145705005213. [DOI] [PubMed] [Google Scholar]
- 35.Uranova N, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, Vostrikov V, Rachmanova V. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 36.Kung L, Roberts RC. Mitochondrial pathology in human schizophrenic striatum: a postmortem ultrastructural study. Synapse. 1999;31:67–75. doi: 10.1002/(SICI)1098-2396(199901)31:1<67::AID-SYN9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 37.Somerville SM, Conley RR, Roberts RC. Mitochondria in the striatum of subjects with schizophrenia. World J Biol Psychiatry. 2011;12:48–56. doi: 10.3109/15622975.2010.505662. [DOI] [PubMed] [Google Scholar]
- 38.Somerville SM, Lahti AC, Conley RR, Roberts RC. Mitochondria in the striatum of subjects with schizophrenia: relationship to treatment response. Synapse. 2011;65:215–224. doi: 10.1002/syn.20838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu-Wai-Man P, Lai-Cheong J, Borthwick GM, He L, Taylor GA, Greaves LC, Taylor RW, Griffiths PG, Turnbull DM. Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles. Invest Ophthalmol Vis Sci. 2010;51:3347–3353. doi: 10.1167/iovs.09-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rana A, Oliveira MP, Khamoui AV, Aparicio R, Rera M, Rossiter HB, Walker DW. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HK, Andreazza AC, Elmi N, Chen W, Young LT. Nod-like receptor pyrin containing 3 (NLRP3) in the post-mortem frontal cortex from patients with bipolar disorder: a potential mediator between mitochondria and immune-activation. J Psychiatr Res. 2016;72:43–50. doi: 10.1016/j.jpsychires.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Bergman O, Ben-Shachar D. Mitochondrial oxidative phosphorylation system (OXPHOS) deficits in schizophrenia: possible interactions with cellular processes. Can J Psychiatry. 2016;61:457–469. doi: 10.1177/0706743716648290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modica-Napolitano JS, Lagace CJ, Brennan WA, Aprille JR. Differential effects of typical and atypical neuroleptics on mitochondrial function in vitro. Arch Pharm Res. 2003;26:951–959. doi: 10.1007/BF02980205. [DOI] [PubMed] [Google Scholar]
- 44.Corena-McLeod M, Walss-Bass C, Oliveros A, Gordillo Villegas A, Ceballos C, Charlesworth CM, Madden B, Linser PJ, Van Ekeris L, Smith K, Richelson E. New model of action for mood stabilizers: phosphoproteome from rat pre-frontal cortex synaptoneurosomal preparations. PLoS One. 2013;8:e52147. doi: 10.1371/journal.pone.0052147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corena-McLeod M. Comparative pharmacology of risperidone and paliperidone. Drugs R D. 2015;15:163–174. doi: 10.1007/s40268-015-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucl Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, Zheng Y, Diffenderfer KE, Zhang J, Soltani S, Eames T, Schafer ST, Boyer L, Marchetto MC, Nurnberger JI, Calabrese JR, Odegaard KJ, McCarthy MJ, Zandi PP, Alda M, Nievergelt CM, Mi S, Brennand KJ, Kelsoe JR, Gage FH, Yao J. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murrell MD, Cruz DA, Javors MA, Thompson PM. Distribution of venlafaxine, O-desmethylvenlafaxine, and O-desmethylvenlafaxine to venlafaxine ratio in postmortem human brain tissue. J Forensic Sci. 2014;59:683–689. doi: 10.1111/1556-4029.12393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data