Abstract
Background
For more than 25 years, the golden mussel, Limnoperna fortunei, has aggressively invaded South American freshwaters, having travelled more than 5000 km upstream across 5 countries. Along the way, the golden mussel has outcompeted native species and economically harmed aquaculture, hydroelectric powers, and ship transit. We have sequenced the complete genome of the golden mussel to understand the molecular basis of its invasiveness and search for ways to control it.
Findings
We assembled the 1.6-Gb genome into 20 548 scaffolds with an N50 length of 312 Kb using a hybrid and hierarchical assembly strategy from short and long DNA reads and transcriptomes. A total of 60 717 coding genes were inferred from a customized transcriptome-trained AUGUSTUS run. We also compared predicted protein sets with those of complete molluscan genomes, revealing an exacerbation of protein-binding domains in L. fortunei.
Conclusions
We built one of the best bivalve genome assemblies available using a cost-effective approach using Illumina paired-end, mate-paired, and PacBio long reads. We expect that the continuous and careful annotation of L. fortunei’s genome will contribute to the investigation of bivalve genetics, evolution, and invasiveness, as well as to the development of biotechnological tools for aquatic pest control.
Keywords: Amazon, binding domain, bivalves, genomics, TLR, transposon
Data Description
The golden mussel Limnoperna fortunei is an Asian bivalve that arrived in the southern part of South America about 25 years ago [1]. Research suggests that L. fortunei was introduced in South America through ballast water of ships coming from Hong Kong or Korea [2]. It was found for the first time in the estuary of the La Plata River in 1991 [1]. Since then, it has moved ∼5000 km, invading upstream continental waters and reaching northern parts of the continent [3], leaving behind a track of great economic impact and environmental degradation [4]. The latest infestation was reported in 2016 in the São Francisco River, one of the main rivers in the northeast of Brazil, with a 2700-km riverbed that provides water to more than 14 million people. At Paulo Afonso, one of the main hydroelectric power plants in the São Francisco River, maintenance due to clogging of pipelines and corrosion caused by the golden mussel is estimated to cost U$700 000 per year (personal communication, Mizael Gusmã, Chief Maintenance Engineer for Centrais Hidrelétricas do São Francisco [CHESF]).
A recent review has shown that, before arriving in South America, L. fortunei was already an invader in China. Originally from the Pearl River Basin, the golden mussel has traveled 1500 km into the Yang Tse and Yellow River basins, being limited further north only by the extreme natural barriers of Northern China [5]. Today, L. fortunei is found in the Paraguaizinho River, located only 150 km from the Teles-Pires River that belongs to the Alto Tapajós River Basin and is the first to directly connect with the Amazon River Basin [6]. Due to its fast dispersion rates, it is very likely that L. fortunei will reach the Amazon River Basin in the near future.
The reason why some freshwater bivalves, such as L. fortunei, Dreissena polymorpha, and Corbicula fluminea, are aggressive invaders is not fully understood. These bivalves present characteristics such as (i) tolerance to a wide range of environmental variables, (ii) short life span, (iii) early sexual maturation, and (iv) high reproductive rates that allow them to reach densities as high as 150 000 ind.m−2 over a year [7, 8] that may explain the aggressive behavior. On the other hand, these traits are not exclusive to invasive freshwater bivalves and do not explain how they outcompete native species and disperse so widely.
To the best of our knowledge, there are no reports of strategies successful at controlling the expansion of mussel invasion in industrial facilities. Bivalves can sense chemicals in the water and close their valves as a defensive response [9], making them tolerant to a wide range of chemical substances, including strong oxidants like chlorine [10]. Microencapsulated chemicals have shown better results in controlling mussel populations in closed environments [10, 11], but it is unlikely they would work in the wild. Currently, there is no effective and efficient approach to control the invasion by L. fortunei.
The genome sequence is one of the most relevant and informative descriptions of species biology. The genetic substrate of invasive populations, upon which natural selection operates, can be of primary importance to understanding and controlling a biological invader [12, 13].
We have partially funded the golden mussel genome sequencing through a pioneer crowdfunding initiative in Brazil [14]. In this campaign, we were able to raise around USD$20 000.00 at the same time as we promoted scientific education and awareness in Brazil.
Here we present the first complete genome dataset for the invasive bivalve Limnoperna fortunei, assembled from short and long DNA reads and using a hybrid and hierarchical assembly strategy. This high-quality reference genome represents a substantial resource for further studies of genetics and evolution of mussels, as well as for the development of new tools for plague control.
Genome sequencing in short Illumina and long PacBio reads
Limnoperna fortunei mussels were collected from the Jacui River, Porto Alegre, Rio Grande do Sul, Brazil (29°59΄29.3″S 51°16΄24.0″W). Voucher specimens were housed at the zoological collection (specimen number: 19 643) of the Biology Institute at the Universidade Federal do Rio de Janeiro, Brazil. For the genome assembly, a total of 3 individuals were sampled for DNA extraction from gills and to produce the 3 types of DNA libraries used in this study. DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) to prepare libraries for Illumina Nextera paired-end reads, with ∼180-bp and ∼500-bp insert sizes, (ii) Illumina Nextera mate-paired reads with insert sizes ranging from 3 to 15 Kb, and (iii) Pacific Biosciences long reads (Table 1). Illumina libraries were sequenced, respectively, in a HiScanSQ or HiSeq 1500 machine, and Pacific Biosciences reads were produced with the P4C6 chemistry and sequenced in 10-SMRT Cells. All Illumina reads were submitted to quality analysis with FastQC (FastQC, RRID:SCR_014583) followed by trimming with Trimmomatic (Trimmomatic, RRID:SCR_011848) [15]. Pacific Biosciences adaptor-free subread sequences were used as input data for the genome assembly.
Table 1:
DNA reads produced for L. fortunei genome assembly
| Library technology | Raw data | Trimmed data* | ||||
|---|---|---|---|---|---|---|
| Reads insert size | Pairs | Number of reads | Number of bases | Number of reads | Number of bases | |
| Illumina Nextera | Paired-end – 180 bp | R1 | 209 542 721 | 21 060 365 702 | 209 036 571 | 21 001 101 404 |
| R2 | 209 542 721 | 21 049 308 698 | 209 036 571 | 20 991 650 008 | ||
| Paired-end – 500 bp | R1 | 153 948 902 | 15 472 966 961 | 153 482 290 | 15 423 123 500 | |
| R2 | 153 948 902 | 15 462 883 157 | 153 482 290 | 15 414 813 589 | ||
| Mate-paired 3 – 12 Kb | R1 | 178 392 944 | 18 017 687 344 | 58 157 933 | 5 822 572 152 | |
| R2 | 178 392 944 | 18 017 687 344 | 58 157 933 | 5 811 310 412 | ||
| Pacific Biosciences | P4C – 10/SMTRC | Subreads | 1 663 730 | 11 171 487 485 |
*Trimmomatic parameters for Illumina reads—ILLUMINACLIP: NexteraPE-PE.fa:2:30:10 SLIDINGWINDOW:4:2 LEADING:10 TRAILING:10 CROP:101 HEADCROP:0 MINLEN:80.
For transcriptome sequencing, RNA was sampled from 4 tissues (gills, adductor muscle, digestive gland, and foot) of 3 different golden mussel specimens. RNA was extracted using the NEXTflex Rapid Directional RNA-Seq Kit (Bio Scientifics, TX, USA) and 12 barcodes from NEXTflex Barcodes compatible with Illumina NexSeq Machine. Resulting reads (Supplementary Table S1) were submitted to FastQC quality analysis and trimmed with Trimmomatic for all NEXTflex adaptors and barcodes. A total of 3 sets of de novo assembled transcriptomes were generated using Trinity (Trinity, RRID:SCR_013048) (Table 2); 1 set for each specimen was a pool of the 4 tissue samples to avoid assembly bias due to intraspecific polymorphism [16].
Table 2:
Trinity assembled transcripts used in the assembly and annotation of L. fortunei genome
| Number of reads | Number of | Number of | Average | GC | ||
|---|---|---|---|---|---|---|
| Sample | Pooled tissues | prior assembly | trinity transcripts | trinity genes | contig length | % |
| Mussel 1 | Gills, mantle, digestive gland, foot | 406 589 144 | 433 197 | 303 172 | 854 | 34 |
| Mussel 2 | Gills, mantle, digestive gland, foot | 376 577 660 | 435 054 | 298 117 | 824 | 34 |
| Mussel 3 | Gills, mantle, digestive gland, foot | 334 316 116 | 499 392 | 351 649 | 844 | 34 |
Genome assembly using a hybrid and hierarchical strategy
Jellyfish software (Jellyfish, RRID:SCR_005491) [17] was used to count and determine the distribution frequency of lengths 25 and 31 kmers (Fig. 1) for the Illumina DNA paired-end and mate-paired reads (Table 1). The genome size was estimated to be 1.6 Gb by using the 25-kmer distribution plot as total kmer number and then subtracting erroneous reads (starting kmer counts from ×12 coverage) to further divide by the homozygous coverage-peak depth (×45 coverage), as performed by Li et al. (2010) [18]. A double-peak kmer distribution was used as evidence of genome diploidy (Fig. 1) and high heterozygosity. The rate of heterozygosity was estimated to be 2.3%, and it was calculated as described by Vij et al. (2016) [19], using as input data the 17-kmer distribution plot for reads from 1 unique specimen.
Figure 1:

Kmer distribution of Limnoperna fortunei Illumina DNA reads (Table 1).
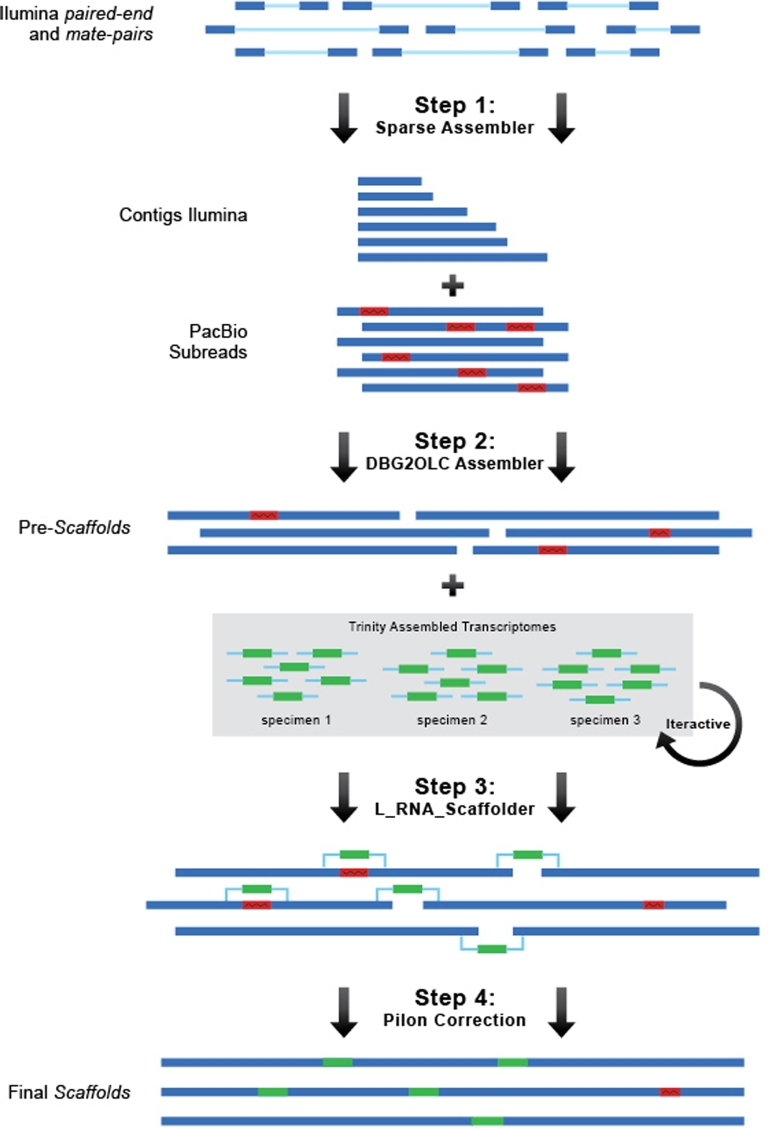
Initially, we attempted to assemble the golden mussel genome using only short Illumina reads of different insert sizes (paired-end and mate-paired) (Table 1) using traditional de novo assembly software such as ALLPATHS (ALLPATHS-LG, RRID:SCR_010742) [20], SOAPdenovo (SOAPdenovo, RRID:SCR_010752) [21], and MaSuRCA (MaSuRCA, RRID:SCR_010691) [22]. All these attempts resulted in very fragmented genome drafts, with an N50 no higher than 5 Kb and a total of 4 million scaffolds. To reduce fragmentation, we further sequenced additional long reads (10 PacBio SMTR Cells) (Table 1) and performed a hybrid and hierarchical de novo assembly, described below and depicted in Fig. 2.
Figure 2:
Hierarchical assembly strategy employed for the golden mussel genome assembly. Trimmed Illumina reads were assembled to the level of contigs with the Sparse Assembler algorithm (Step 1). Then, Illumina contigs and PacBio reads were used to build scaffolds with the DBG2OLC assembler, which anchors Illumina contigs to erroneous PacBio subreads, correcting them and building longer scaffolds (Step 2), followed by transcriptome joining scaffolds using L_RNA_scaffolder (Step 3). Final scaffolds were corrected by re-aligning all Illumina DNA and RNA-seq reads back to them and calling consensus with Pilon software (Step 4). In bold is the bioinformatics software used in each step. Red blocks indicate PacBio errors, which are represented by insertions and/or deletions, found in approximately 12% of PacBio subreads.
First, (i) trimmed paired-end and mate-paired DNA Illumina reads (Table 1) were assembled into contigs using the software Sparse Assembler [23] with parameters LD 0 NodeCovTh 1 EdgeCovTh 0 k 31 g 15 PathCovTh 100 GS 1 800 000 000. Next, (ii) the resulting contigs were assembled into scaffolds using Pacific Biosciences long subreads data and the PacBio-correction-free assembly algorithm DBG2OLC [24] with parameters LD1 0 k 17 KmerCovTh 10 MinOverlap 20 AdaptiveTh 0.01. Finally, (iii) resulting scaffolds were submitted to 6 iterative runs of the program L_RNA_Scaffolder [25], which uses exon distance information from de novo assembled transcripts (Table 2) to fill gaps and connect scaffolds whenever appropriate. At the end, (iv) the final genome scaffolds were corrected for Illumina and Pacific Biosciences sequencing errors with the software PILON [26]: All DNA and RNA short Illumina reads were re-aligned back to the genome with BWA aligner (BWA, RRID:SCR_010910) [27], and resulting SAM files were BAM-converted, sorted, and indexed with the SAMTOOLs package (SAMTOOLS, RRID:SCR_002105) [28]. Pilon [26] identifies INDELS and mismatches by coverage of reads and yields a final corrected genome draft. Pilon was run with parameters –diploid –duplicates.
The final genome was assembled in 20 548 scaffolds, with an N50 of 312 Kb and a total assembly length of 1.6 Gb (Table 3).
Table 3:
Assembly statistics for Limnoperna fortunei’s genome
| Parameter | Value |
|---|---|
| Estimated genome size by kmer analysis, Gb | 1.6 |
| Total size of assembled genome, Gb | 1.673 |
| Number of scaffolds | 20 548 |
| Number of contigs | 61 093 |
| Scaffold N50, Kb | 312 |
| Maximum scaffold length, Mb | 2.72 |
| Percentage of genome in scaffolds >50 Kb | 82.55 |
| Masked percentage of total genome | 33 |
| Mapping percentage of Illumina reads back to scaffolds | 91 |
The golden mussel genome presents 81% of all Benchmarking Universal Single Copy Orthologs (BUSCO version 3.3 analysis with Metazoa database; BUSCO, RRID:SCR_015008) (Table 4) and, compared with the mollusk genomes currently available [29–36], it represents one of the best assemblies of molluscan genomes so far also in terms of scaffold N50 and contiguity (Table 5).
Table 4:
Summary statistics of BUSCO analysis for L. fortunei genome run for Metazoans
| Categories | Number of genes | Percentage |
|---|---|---|
| Total BUSCO groups searched | 978 | – |
| Complete BUSCOs | 801 | 81.9 |
| Complete and single-copy BUSCOs | 769 | 78.62 |
| Complete and duplicated BUSCOs | 32 | 3.27 |
| Fragmented BUSCOs | 72 | 7.36 |
| Missing BUSCOs | 105 | 10.73 |
Table 5:
Comparison of genome assembly statistics for molluscan genomes
| Haliotis | Lottia | Aplysia | Ruditapes | Patinopecten | Crassostrea | Mytillus | Bathymodiolus | Modiolus | Limnoperna | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| discus hannai | gigantea | californica | philippinarum | yessoensis | gigas | Pinctadafucata | galloprovincialis | platifrons | philippinarum | fortunei | |
| Estimated genome size | 1.65 Gb | 359.5 Mb | 1.8 Gb | 1.37 Gb | 1.43 Gb | 545 Mb | 1.15 Gb | 1.6 Gb | 1.64 Gb | 2.38 Gb | 1.6 Gb |
| Number of scaffolds | 80 032 | 4475 | 8766 | 223 851 | 82 731 | 11 969 | 7997 | 1746 447 | 65 664 | 74 575 | 20 548 |
| Total size of scaffolds | 1 865 475 499 | 359 512 207 | 715 791 924 | 2 561 070 351 | 987 685 017 | 558 601 156 | 915 721 316 | 1 599 211 957 | 1 659 280 971 | 2 629 649 654 | 1 673 125 894 |
| Longest scaffold | 2 207 537 | 9 386 848 | 1 784 514 | 572 939 | 7 498 238 | 1 964 558 | 5 897 787 | 67 529 | 2 790 175 | 715 382 | 2 720 304 |
| Shortest scaffold | 854 | 1000 | 5001 | 500 | 200 | 100 | 1807 | 100 | 292 | 205 | 558 |
| Number of scaffolds >1 K nt (%) | 79 923 (99.9) | 4471 (99.9) | 8766 (100) | 138 771 (61.9) | 16 004 (19.3) | 5788 (48.4) | 7997 (100) | 393 685 (22.5) | 38 704 (58.9) | 44 921 (60.2) | 20 547 (100) |
| Number of scaffolds >1 M nt (%) | 67 (0.1) | 98 (2.2) | 27 (0.3) | 0 (0.0) | 248 (0.3) | 60 (0.5) | 27 (0.3) | 0 (0.0) | 164 (0.2) | 0 (0) | 95 (0.5) |
| Mean scaffold size | 23 309 | 80 338 | 81 655 | 11 441 | 11 939 | 46 671 | 114 508 | 916 | 25 269 | 35 262 | 81 425 |
| Median scaffold size | 1697 | 3622 | 13 763 | 1327 | 362 | 824 | 14 683 | 258 | 1284 | 13 722 | 22 134 |
| N50 scaffold length | 200 099 | 1 870 055 | 264 327 | 48 447 | 803 631 | 401 319 | 345 846 | 2651 | 343 373 | 100 161 | 312 020 |
| Sequencing coverage | ×322 | ×8.87 | ×11 | ×39.7 | ×297 | ×155 | ×234 | ×32 | ×319 | ×209.5 | ×60 |
| Sequencing Technology | Illumina + PacBio | Sanger | Sanger | Illumina | Illumina | Illumina | Illumina + BACs | Illumina | Illumina | Illumina | Illumina + PacBio |
One main challenges of assembling bivalve genomes lies in the high heterozygosity and amount of repetitive elements these organisms present: (i) the mussels L. fortunei and Modiolus philippinarum and the oyster Crassostrea gigas genomes were estimated to have heterozygosity rates of 2.3%, 2.02%, and 1.95%, respectively, which are substantially higher than other animal genomes [30], and (ii) repetitive elements correspond to at least 30% of the genomes of all studied bivalves so far (Table 3) [29–32, 34–36]. Also, retroelements might be active in some species such as L. fortunei (refer to the “Retroelements” section of this paper) and C. gigas [30], allowing genome rearrangements that may hinder genome assembly. One exception seems to be the deep-sea mussel B. platifrons, which has lower heterozygosity rates compared with other bivalves [32]. Sun et al. [32] suggested that it might be due to recurrent population bottlenecks that happened after events of population extinction and recolonization in the extreme environment [32]. Nevertheless, most of the bivalve genome projects relying only on short Illumina reads are likely to present fragmented initial drafts [29, 31]. PacBio long reads allowed us to increase the N50 to 32 Kb and to reduce the number of scaffolds from millions to 61 102, using the DBG2OLC [24] assembler. Finally, interactive runs of L_RNA_scaffolder [25] using the transcriptomes (Table 2) rendered the final result of N50 312 Kb in 20 548 scaffolds. It is important to note that assembly statistics can perform better for genomes assembled with reads generated with DNA extracted from 1 unique individual. This, however, was not possible for L. fortunei’s genome due to the high amount of high-quality DNA necessary to produce Illumina mate-pairs and PacBio long reads. In this study, the challenge of assembling the high polymorphic regions between haplotypes was enhanced by the difficulties of assembling reads that originated from highly polymorphic regions across individuals. However, the golden mussel assembly presented here shows that the use of Illumina contigs, low coverage of PacBio long reads, and transcriptome and Illumina re-mapping for final correction (Fig. 2) represent an option for cost-efficient assembly of highly heterozygous genomes of nonmodel species such as bivalves.
Around 10% of repetitive elements are transposons
Initial masking of L. fortunei genome was done using the RepeatMasker program (RepeatMasker, RRID:SCR_012954) [37] with the parameter -species bivalves and masked 3.4% of the total genome. This content was much lower than the masked portion of other molluscan genomes, 34% in C. gigas [30] and 36% in M. galloprovincialis [29], suggesting that the fast evolution of interspersed elements limits the use of repeat libraries from divergent taxa [38]. Thus, we generated a de novo repeat library for L. fortunei using the program RepeatModeler (RepeatModeler, RRID:SCR_015027) [39] and its integrated tools RECON [40], TRF [41], and RepeatScout [42]. This de novo repeat library was the input to RepeatMasker, together with the first masked genome draft of L. fortunei, and resulted in a final masking of 33.4% of the genome. Even though more than 90% of the repeats were not classified by RepeatMasker (Supplementary Table S2), 8.85% of the repeats were classified as LINEs, Class I transposable elements. In addition, large numbers of reverse-transcriptases (824 counts, Pfam RVT_1 PF00078), transposases (177 counts, Pfam HTH_Tnp_Tc3_2 PF01498), integrases (501 counts, Pfam Retroviral integrase core domain PF00665), and other related elements were detected; more than 98% of these had detectable transcripts.
More than 30 000 sequences were identified by gene prediction and automated annotation
To annotate the golden mussel genome, we sequenced a number of transcriptomes (Table S1), de novo assembled (Table 2) and aligned these transcriptomes to the genome scaffolds, and created gene models with the PASA pipeline [37]. These models were used to train and run the ab initio gene predictor AUGUSTUS (Augustus: Gene Prediction, RRID:SCR_008417) (Supplementary Fig. S1) [38]. The complete gene models yielded by PASA [43] were BLASTed (e-value 1e-20) against the Uniprot database (UniProt, RRID:SCR_002380), and those with 90% or more of their sequences showing in the BLAST hit alignment were considered for further analysis. Next, all the necessary filters to run an AUGUSTUS [44] personalized training were performed: (i) only gene models with more than 3 exons were maintained, (ii) sequences with 90% or more overlap were withdrawn and only the longest sequences were retained, and (iii) only gene models free of repeat regions, as indicated by BLASTN similarity searches with de novo library of repeats, were maintained. These curated data yielded a final set of 1721 gene models on which AUGUSTUS [36] was trained in order to predict genes in the genome using the default AUGUSTUS [44] parameters. Once the gene models were predicted, a final step was performed by using the PASA pipeline [43] once again in the update mode (parameters -c -A -g -t). This final step compared the 55 638 gene models predicted by AUGUSTUS [44] with the 40 780 initial transcript-based gene-structure models from PASA [43] to generate the final set of 60 717 gene models for L. fortunei. Of those, 58% had transcriptional evidence based on RNA Illumina reads (Table S2) re-mapping, a rate that was expected as our RNA-Seq libraries were constructed for only 4 tissues of adult golden mussel specimens without any environmental stress induction (Table 2). Therefore, these libraries lack transcripts for developmental stages for some other cell types (i.e., hemocytes) and stress-inducible genes. Finally, 67% of the gene models were annotated by homology searches against Uniprot or NCBI NR (Table 6).
Table 6:
Summary of gene annotation against various databases for L. fortunei whole-genome-predicted genes
| Total number of genes | 60 717 |
| Total number of exons | 220 058 |
| Total number of proteins | 60 717 |
| Average protein size, aa | 304 |
| Number of protein BLAST hits* with Uniprot | 26 198 |
| Number of protein BLAST hits* with NR NCBI (no hits with Uniprot) | 14 810 |
| Number of protein HMMER hits* with Pfam.A | 24 513 |
| Number with proteins with KO assigned by KEGG | 8387 |
| Number of proteins with BLAST hits* with EggNOG | 36 868 |
*All considered hits had a minimum e-value of 1e-05.
Protein clustering indicates evolutionary proximity among mollusk species
Gene family relationships were assigned using reciprocal best BLAST and OrthoMCL software (version 1.4) [45] between L. fortunei proteins and the total protein set predicted for 9 other mollusks: the mussels M. galloprovincialis, M. philippinarum, and B. platifrons, the clam Ruditapes philippinarum, the scallop Patinopecten yessoensis, the pacific oyster C. gigas, the pearl oyster Pinctada fucata (genome version from Du et al. [36]), and the gastropods Lottia gigantea and Haliotis discus hannai (see Supplementary Table S3 for detailed information on the comparative data). Figure 3A presents orthologs relationships for 5 of the bivalves analyzed. A total of 6337 ortholog groups are shared among the 5 bivalve species.
Figure 3:
(A) Gene family assigned with OrthoMCL for the total set of proteins predicted from 5 mussel genome projects. Outside the Venn diagram, the species name is represented, and below it is the number of proteins/number of clustered proteins/number of clusters. (B) Phylogeny of the concatenated dataset using 44 single-copy orthologs extracted from 10 molluscan genomes. The VT model was estimated to be the best-fitting substitution model with ProtTest 3.4.2. We reconstructed the phylogeny using PhyML and 100 bootstrap repetition.
Of all the orthologs found for the total 10 species, 44 groups are composed of single-copy orthologs containing 1 representative protein sequence of each species. These sequences were used to reconstruct a phylogeny: the single-copy ortholog sequences were concatenated and aligned with CLUSTALW [46], with a resulting alignment 30 755 sites in length (Fig. 3B). ProtTest 3.4.2 [47] was used to estimate the best-fitting substitution model, which was VT+G+I+F [48]. With this alignment and model, we reconstructed the phylogeny using PhyML [49] and 100 bootstrap repetition; the resulting tree is shown in Fig. 3B.
Protein domain analysis shows expansion of binding domain in L. fortunei
We performed a quantitative comparison of protein domains predicted from whole-genome projects of 10 molluscan species. The complete protein sets of M. galloprovincialis, M. philippinarum, B. platifrons, Ruditapes philippinarum, Patinopecten yessoensis, C. gigas, Pinctada fucata, Lottia gigantean, and Haliotis discus hannai (Supplementary Table S3) were submitted to domain annotation using HMMER against the Pfam-A database (e-value 1e-05). Protein expansions in L. fortunei were rendered using the normalized Pfam count value (average) obtained from the other 9 mollusks, according to a model based on the Poisson cumulative distribution. Bonferroni correction (P ≤ 0.05) was applied for false discovery, and absolute frequencies of Pfam-assigned domains were initially normalized by the total count number of Pfam-assigned domains found in L. fortunei to compensate for discrepancies in genome size and annotation bias.
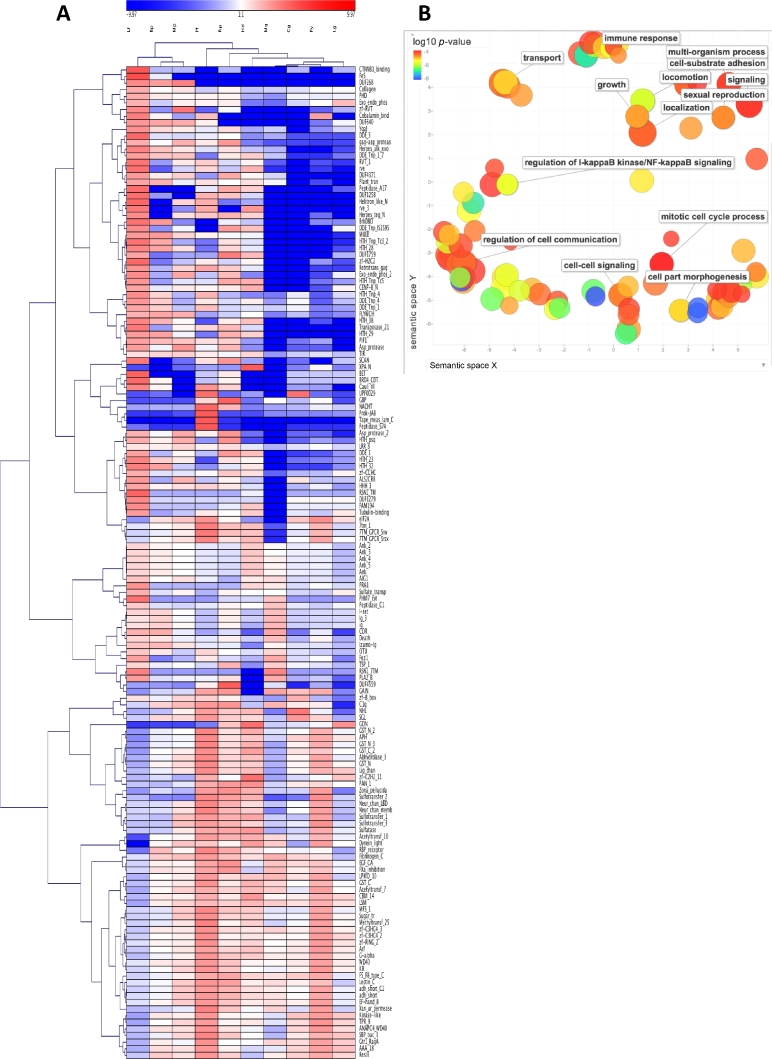
For L. fortunei, the annotation against Pfam-A classified 40 127 domains in 24 513 gene models, of which 83 and 67 were expanded or contracted, respectively, in comparison with the other mollusks (Fig. 4A; Supplementary Table S4 and S5). The 83 overrepresented domains were further analyzed for functional enrichment using domain-centric Gene Ontology (Fig. 4B). The analysis shows a prominent expansion of binding domains in L. fortunei, such as Thrombospondin (TSP_1), Collagen, Immunoglobulins (Ig, I-set, Izumo-Ig Ig_3), and Ankyrins (Ank_2, Ank_3, and Ank_4). These repeats have a variety of binding properties and are involved in cell-cell, protein-protein, and receptor-ligand interactions driving the evolutionary improvement of complex tissues and the immune defense system in metazoans [50–54]. An evolutionary pressure toward the development of a diversified innate immune system is also suggested by the high amount of leucine rich repeats (LRR) and Toll/interleukin-1 receptor homology domains (TIR). Death, another over-represented Pfam, is also part of TLR signaling, being present in several docking proteins such as Myd88, Irak4, and Pelle [55]. Interestingly, BLAST analysis of L. fortunei gene models against Uniprot identified 2 types of TLRs whose prototypical architecture of N-terminal extracellular LRR motifs including either a single or multiple cysteine cluster domain, a C-terminal TIR domain spaced by a single transmembrane-spanning domain [56], could be correctly identified using the Simple Modular Architecture Research Tool (SMART) [57]. Indeed, we confirmed 141 sequences with similarity to single cysteine clusters TLRs (scc) typical of vertebrates and 29 sequence hits with the multiple cysteine cluster TLRs (mcc) typical of Drosophila [56]. Phylogenetic analysis of all sequences (using PhyML [49], model JTT) (Supplementary Fig. S2) shows evidence for TLRs clade separation in L. fortunei; the scc TLRs exhibit a higher degree of amino acid changes, higher molecular evolution, and diversification than the mcc TLRs. Overall, the expansion of these gene families might suggest an improved resistance to infections. It is, however, equally curious that other immune-related gene families such as Fribinogen_C and C1q seem to be contracted (Supplementary Table S5). This feature may depend on the evolution-driven, yet random fate of the L. fortunei genome, a consequence of different specific duplicate genes in other species. Also, other protein families involved in toxin metabolism, especially glutathione-based processes and sulfotransferases, are clearly contracted (Table S5).
Figure 4:
Gene family representation analysis in the L. fortunei genome. (A) Pfam hierarchical clustering, heatmap. Features were selected according to a model based on the Poisson cumulative distribution of each Pfam count in the golden mussel genome vs the normalized average values found in the other 9 molluscan genomes (Bonferroni correction, P ≤ 0.05). Transposable elements were included in the analysis. Colors depict the log2 ratio between Pfam counts found in each single genome and the corresponding mean values. The hierarchical clustering used the average dot product for the data matrix and complete linkage for branching. Abbreviations: Bp: Bathymodioulus platifrons; Cg: Crassostrea gigas; Hd: Haliotus discus hannai; Lf: L. fortunei; Lg: Lottia gigantean; Mg: Mytilus galloprovincialis; Mp: Modioulus philippinarum; Pf: Pinctada fucata; Py: Patinopecten yessoensis; Rp: Ruditapes philippinarum. (B) Gene Ontology analysis of expanded gene families, semantic scatter plot. Shown are cluster representatives after redundancy reduction in a 2-dimensional space applying multidimensional scaling to a matrix of semantic similarities of GO terms. Color indicates the GO enrichment level (legend in upper left-hand corner); size indicates the relative frequency of each term in the UNIPROT database (larger bubbles represent less specific processes).
Final considerations
Here we have described the first version of the golden mussel complete genome and its automated gene prediction, which were funded through a crowdfunding initiative in Brazil. This genome contains valuable information for further evolutionary studies of bivalves and metazoa in general. Additionally, our team will further search for the presence of proteins of biotechnology interest such as the adhesive proteins produced by the foot gland that we have described elsewhere [58] or genes related to the reproductive system that have been shown to be very effective for invertebrate plague control [59]. The golden mussel genome and the predicted proteins are available for download in the GigaScience repository, and the scientific community is welcome to further curate the gene predictions.
As the golden mussel advances towards the Amazon River Basin, the information provided in this study may be used to help develop biotechnological strategies that may control the expansion of this organism in both industrial facilities and open environment.
Availability of supporting data
Limnoperna fortunei's genome and transcriptome data are available in the Sequence Read Archive (SRA) as BioProject PRJNA330677 and under the accession numbers SRR5188384, SRR5195098, SRR518800, SRR5195097, SRR5188315, SRR5181514. This Whole Genome Shotgun Project has been deposited in the DDBJ/ENA/GenBank under accession number NFUK00000000. The version described in this paper is version NFUK01000000. Supporting data, also including annotations and BUSCO results, are available via the GigaScience repository, GigaDB [60].
Additional files
Supplementary Table S1. RNA raw reads sequenced for 3 L. fortunei specimens, 4 tissues each.
Supplementary Table S2. RepeatMasker classification of repeats predicted in the L. fortunei genome.
Supplementary Table S3. Details of the online availability of the data used for ortholog assignment and protein domain expansion analysis.
Supplementary Table S4. Expanded protein families in the L. fortunei genome.
Supplementary Table S5. Contracted protein families in the L. fortunei genome.
Supplementary Table S6. Fantasy names given to L. fortunei genes and proteins from the backers that supported us through crowdfunding (www.catarse.me/genoma).
Supplementary Figure 1. Steps performed for the prediction and annotation of the L. fortunei genome.
Supplementary Figure 2. Phylogenetic tree of Toll-like (TLRs) receptors found in the L. fortunei genome.
Abbreviations
BUSCO: Benchmarking Universal Single-Copy Orthologs; KEGG: Kyoto Encyclopedia of Genes and Genomes; SRA: Sequence Read Archive.
Ethics approval
Limnoperna fortunei specimens used for DNA extraction and sequencing were collected in the Jacuí River (29°59΄29.3″S 51°16΄24.0″W), southern Brazil. This bivalve is an exotic species in Brazil and is not characterized as an endangered or protected species.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by the Brazilian Government agencies CAPES (PVE 71/2013), FAPERJ APQ1 (2014), and FAPERJ/DFG (39/2014). Also, this work was funded through crowdfunding with the support of 346 people (www.catarse.me/genoma).
Author contributions
Conceived and designed the experiments: M.R., M.U., T.O., C.M., F.D. Performed the experiments: M.U., J.A. Analyzed the data: M.U., T.O., C.M., F.D., F.P., N.C., I.C., M.R. Contributed reagents/materials/analysis tools: M.R., F.P., C.M. Wrote the paper: M.U., F.D., M.R. All authors read and approved the final manuscript.
Supplementary Material
31 Jul 2017 Reviewed
08 Nov 2017 Reviewed
23 Aug 2017 Reviewed
Acknowledgements
We thank Susan Mbedi and Kirsten Richter from BeGenDiv for RNA-Seq library preparation and sequencing. We thank Dr. Loris Bennett for IT support while performing bioinformatics analysis.
We especially want to thank the 346 backers that supported the sequencing of the golden mussel through crowdfunding in a 2013 campaign that raised U$20 000.00 (www.catarse.me/genoma). We decided to give fantasy names to the genes and proteins that we found in the genome to thank the backers for their support. The name list is available in Supplementary Table S6.
References
- 1. Pastorino G, Darrigran G, Maris MS et al. Limnoperna fortunei (Dunker, 1857) (Mytilidae), nuevo bivalvo invasor em águas Del Rio de la plata. Neotropica 1993;39:101–2. [Google Scholar]
- 2. Darrigran G. Potential impact of filter-feeding invaders on temperate inland freshwater environments. Biol Invasions 2002;4(1/2):145–56. [Google Scholar]
- 3. Uliano-Silva M, Fernandes F da C, Holanda IBB et al. Invasive species as a threat to biodiversity: the golden mussel Limnoperna fortunei approaching the Amazon River basin. In: Alodi S, ed. Exploring Themes on Aquatic Toxicology. Research Signpost, India; 2013. [Google Scholar]
- 4. Boltovskoy D, Correa N. Ecosystem impacts of the invasive bivalve Limnoperna fortunei (golden mussel) in South America. Hydrobiologia 2015;746(1):81–95. [Google Scholar]
- 5. Xu M. Distribution and spread of Limnoperna fortunei in China. In: Boltovskoy D, ed. Limnoperna fortunei. Cham, Switzerland: Springer International Publishing; 2015:313–20. [Google Scholar]
- 6. Oliveira M, Hamilton S, Jacobi C. Forecasting the expansion of the invasive golden mussel Limnoperna fortunei in Brazilian and North American rivers based on its occurrence in the Paraguay River and Pantanal wetland of Brazil. Aquat Invasions 2010;5(1):59–73. [Google Scholar]
- 7. Karatayev AY, Boltovskoy D, Padilla DK, Burlakova LE. The invasive bivalves Dreissena polymorpha and Limnoperna fortunei: parallels, contrasts, potential spread and invasion impacts. J Shellfish Res 2007;26(1):205–13. [Google Scholar]
- 8. Orensanz JM (Lobo), Schwindt E, Pastorino G et al. No longer the pristine confines of the world ocean: a survey of exotic marine species in the Southwestern Atlantic. Biol Invasions 2002;4(1/2):115–43. [Google Scholar]
- 9. Claudi R, Mackie GL. Practical manual for zebra mussel monitoring and control. Boca Raton, FL: Lewis Publishers; 1994:227 [Google Scholar]
- 10. Calazans SHC, Americo JA, Fernandes F da C et al. Assessment of toxicity of dissolved and microencapsulated biocides for control of the Golden Mussel Limnoperna fortunei. Mar Environ Res 201391:104–8. [DOI] [PubMed] [Google Scholar]
- 11. Aldridge DC, Elliott P, Moggridge GD. Microencapsulated biobullets for the control of biofouling zebra mussels. Environ Sci Technol 2006;40(3):975–9. [DOI] [PubMed] [Google Scholar]
- 12. Cox GW. Alien species and evolution: the evolutionary ecology of exotic plants, animals, microbes, and interacting native species. Washington DC: Island Press; 2004:377. [Google Scholar]
- 13. Hall MR, Kocot KM, Baughman KW et al. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature 2017;544(7649):231–4. [DOI] [PubMed] [Google Scholar]
- 14. www.catarse.me/genoma. [Google Scholar]
- 15. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;1;170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trapnell C, Roberts A, Goff L et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of Kmers. Bioinformatics 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li R, Fan W, Tian G et al. The sequence and de novo assembly of the giant panda genome. Nature 2010;463(7279):311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vij S, Kuhl H, Kuznetsova IS et al. Chromosomal-level assembly of the asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet 2016;12(4):e1005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gnerre S, Maccallum I, Przybylski D et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A 2011;108(4):1513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo R, Liu B, Xie Y et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zimin AV, Marçais G, Puiu D et al. The MaSuRCA genome assembler. Bioinformatics 2013;29(21):2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye C, Ma Z, Cannon CH et al. Exploiting sparseness in de novo genome assembly. BMC Bioinformatics 2012;13(Suppl 6):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye C, Hill CM, Wu S et al. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep 2016;6:31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue W, Li J-T, Zhu Y-P et al. L_RNA_scaffolder: scaffolding genomes with transcripts. BMC Genomics 2013;14(1):604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker BJ, Abeel T, Shea T et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014;9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Handsaker B, Wysoker A et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murgarella M, Puiu D, Novoa B et al. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS One 2016;11(3):e0151561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang G, Fang X, Guo X et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012;490(7418):49–54. [DOI] [PubMed] [Google Scholar]
- 31. Takeuchi T, Kawashima T, Koyanagi R et al. Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res 2012;19(2):117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun J, Zhang Y, Xu T et al. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol 2017;1(5):0121. [DOI] [PubMed] [Google Scholar]
- 33. Nam B-H, Kwak W, Kim Y-O et al. Genome sequence of pacific abalone (Haliotis discus hannai): the first draft genome in family Haliotidae. Gigascience 2017;6(5):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang S, Zhang J, Jiao W et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol 2017;1(5):0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mun S, Kim Y-J, Markkandan K et al. The whole-genome and transcriptome of the manila clam (Ruditapes philippinarum). Genome Biol Evol 2017;9(6):1487–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Du X, Fan G, Jiao Y et al. The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 2017;6(8):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smit AF, Hubley R, Green PJ. RepeatMasker Open-3.0. 1996. 2010. [Google Scholar]
- 38. Fu H, Dooner HK. Intraspecific violation of genetic colinearity and its implications in maize. Proc Natl Acad Sci U S A 2002;99(14):9573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith AFA, Hubley R. RepeatModeler Open-1.0. 2014. http://www.repeatmasker.org. Accessed June 2016. [Google Scholar]
- 40. Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 2002;12(8):1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999;27(2):573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics 2005;21(Suppl 1):i351–8. [DOI] [PubMed] [Google Scholar]
- 43. Haas BJ, Delcher AL, Mount SM et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 2003;31(19):5654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stanke M, Diekhans M, Baertsch R et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinforma Oxf Engl 2008;24(5):637–44. [DOI] [PubMed] [Google Scholar]
- 45. Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 1994;22(22):4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Darriba D, Taboada GL, Doallo R et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 2011;27(8):1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Müller T, Vingron M. Modeling amino acid replacement. J Comput Biol 2000;7(6):761–76. [DOI] [PubMed] [Google Scholar]
- 49. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52(5):696–704. [DOI] [PubMed] [Google Scholar]
- 50. Björklund ÅK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comp Biol 2006;2(8):e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rennemeier C, Hammerschmidt S, Niemann S et al. Thrombospondin-1 promotes cellular adherence of gram-positive pathogens via recognition of peptidoglycan. FASEB J 2007;21(12):3118–32. [DOI] [PubMed] [Google Scholar]
- 52. Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Devel 2009;23(2):147–56. [DOI] [PubMed] [Google Scholar]
- 53. Pancer Z, Amemiya CT, Ehrhardt GRA et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004;430(6996):174–80. [DOI] [PubMed] [Google Scholar]
- 54. Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol 2004;36(6):969–74. [DOI] [PubMed] [Google Scholar]
- 55. Park HH, Lo YC, Lin SC et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol 2007;25(1):561–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leulier F, Lemaitre B. Toll-like receptors — taking an evolutionary approach. Nat Rev Genet 2008;3(3):165–78. [DOI] [PubMed] [Google Scholar]
- 57. Schultz J, Milpetz F, Bork P et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 1998;95(11):5857–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uliano-Silva M, Americo JA, Brindeiro R et al. Gene discovery through transcriptome sequencing for the invasive mussel Limnoperna fortunei. PLoS One 2014;9(7):e10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hammond A, Galizi R, Kyrou K et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol 2015;34(1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uliano-Silva M, Dondero F, Otto TD et al. A hybrid-hierarchical genome assembly strategy to sequence the invasive golden mussel, Limnoperna fortunei. GigaScience Database 2017. http://dx.doi.org/10.5524/100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
31 Jul 2017 Reviewed
08 Nov 2017 Reviewed
23 Aug 2017 Reviewed