Abstract
Direct isolation of human neural and glioma stem cells from fresh tissues permits their biological study without prior culture and may capture novel aspects of their molecular phenotype in their native state. Recently, we demonstrated the ability to prospectively isolate stem cell populations from fresh human germinal matrix and glioblastoma samples, exploiting the ability of cells to bind the Epidermal Growth Factor (EGF) ligand in fluorescence-activated cell sorting (FACS). We demonstrated that FACS-isolated EGF-bound neural and glioblastoma populations encompass the sphere-forming colonies in vitro, and are capable of both self-renewal and multilineage differentiation. Here we describe in detail the purification methodology of EGF-bound (i.e., EGFR+) human neural and glioma cells with stem cell properties from fresh postmortem and surgical tissues. The ability to prospectively isolate stem cell populations using native ligand-binding ability opens new doors for understanding both normal and tumor cell biology in uncultured conditions, and is applicable for various downstream molecular sequencing studies at both population and single-cell resolution.
Keywords: EGF, EGFR, FACS, Stem cells, Neural, Glioblastoma, Germinal matrix, Human
Background
Understanding the intrinsic biology of human neural and glioma stem cells has been challenging, due to the lack of universal neural and glioma stem cell markers ( Lathia et al., 2015 ) and the frequent reliance on cultured cells rather than those isolated directly from the tissue. The transmembrane glycoprotein Prominin, or CD133, is one of the best described and frequently used stem cell markers for the isolation of neural ( Uchida et al., 2000 ) and glioma stem cells (GSC) ( Singh et al., 2003 ; Singh et al., 2004 ; Lathia et al., 2015 ), with its utility being demonstrated in both acutely sorted human tissues and neurospheres. However, some recent studies have pointed out that CD133-negative cells isolated from human glioblastoma (GBM) also harbor stem cell properties ( Beier et al., 2007 ; Wang et al., 2008 ; Tome- Garcia et al., 2017 ). Other cell surface markers used to isolate GSC include CD44 ( Anido et al., 2010 ), CD15 ( Son et al., 2009 ), A2B5 ( Ogden et al., 2008 ), integrin alpha ( Lathia et al., 2010 ) or EGFR ( Mazzoleni et al., 2010 ), but these antibody-based methodologies have also lacked the ability to capture all sphere-forming populations or their use has been limited to cultured cells.
A direct comparison of the molecular phenotypes between non-neoplastic and tumoral stem cell niches can provide novel insight into the developmental pathways co-opted during tumor formation and may uncover more comprehensive stem cell markers. In the context of gliomagenesis, several developmental pathways important for the growth and proliferation of normal neural progenitors have been demonstrated to be aberrantly reactivated during gliomagenesis ( Sanai et al., 2005 ; Canoll and Goldman, 2008; Chen et al., 2012 ; Tsankova and Canoll, 2014; Lathia et al., 2015 ). Among these is the Epidermal Growth Factor Receptor (EGFR) tyrosine kinase pathway. Expression of EGFR is high during human neural development, especially within the germinal matrix stem cell niche, diminishing significantly in adulthood ( Weickert et al., 2000 ; Sanai et al., 2011 ; Erfani et al., 2015 ), and it is aberrantly re-activated in GBM ( Verhaak et al., 2010 ; Brennan et al., 2013 ). Recently, we adapted a mouse fluorescence-activated cell sorting (FACS) strategy, which selects for EGFR+ cells based on their native binding to EGF ligand ( Ciccolini et al., 2005 ; Pastrana et al., 2009 ; Codega et al., 2014 ), and isolated human EGFR+ populations from fresh germinal matrix (GM) dissections and GBM tissues. This allowed us to directly compare the functional properties and whole-transcriptome signatures in developing and neoplastic neural populations (Tome- Garcia et al., 2017 ). EGFR+ populations from both GM and GBM tissues captured all sphere-forming cells in vitro, displayed similar proliferative stem cell properties, and shared transcriptome signatures related to cell growth and cell-cycle regulation. EGFR+ GBM populations also displayed tumor initiation in vivo (Tome- Garcia et al., 2017 ). Below, we describe this prospective purification strategy for neural and glioblastoma populations with stem cell properties from primary human samples, detailing the steps of tissue dissociation, ligand/antibody incubation, FACS, and in vitro functional stem cell property analysis.
Materials and Reagents
Pipette tips (Fisher Scientific, FisherbrandTM, catalog numbers: 02-707-404; 02-707-430; 02-707-432)
American Safety Razors PersonnaTM PalTM Single Edge Blade (AccuTec Blades, catalog number: 620177)
Falcon tissue culture dish (Corning, Falcon®, catalog number: 353002)
Falcon® 15 ml conical centrifuge tubes (Corning, Falcon®, catalog number: 352099)
40 μm cell strainer (Corning, Falcon®, catalog number: 352340)
1.7 ml Microcentrifuge snap-cap tubes (Corning, Costar®, catalog number: 3621)
Ultra Low attachment 96-well plates (Corning, catalog number: 3474)
0.2 μm filter (Corning, catalog number: 431219)
10 ml serological pipets (Corning, catalog number: 357530)
60 ml syringes (BD, catalog number: 309653)
Micro cover glass (VWR, catalog number: 48393-081)
Papain (Worthington Biochemical, catalog number: LS003119)
10x Red Blood Cells lysis buffer (Thermo Fisher Scientific, eBioscienceTM, catalog number: 00-4300-54)
Trypan blue solution, 0.4% (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061)
4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D1306)
-
Antibodies
Anti-CD24 PE-conjugated (BD, BD Biosciences, catalog number: 560991, working dilution 1:10)
Anti-CD34 PE-conjugated (BD, BD Biosciences, catalog number: 550619, working dilution 1:10)
Anti-CD45 PE-conjugated (BD, BD Biosciences, catalog number: 555483, working dilution 1:10)
Donkey anti-Rabbit Cy3 conjugated (Jackson ImmunoResearch, catalog number: 711-165-152, working dilution 1:250)
Donkey anti-Rat AF647 conjugated (Jackson ImmunoResearch, catalog number: 712-605-153, working dilution 1:500)
EGF ligand-AF647 conjugated (Thermo Fisher Scientific, InvitrogenTM, catalog number: E35351)
Goat anti-Mouse-AF488 IgM, µ Chain Specific (Jackson ImmunoResearch, catalog number: 115-545-075, working dilution 1:500)
Mouse anti-O4 (Millipore Sigma, catalog number: MAB345, working dilution 1:200)
Rabbit anti-TuJ1 (Covance, catalog number: PRB-435P, working dilution 1:1,000)
Rat anti-GFAP (Thermo Fisher Scientific, InvitrogenTM, catalog number: 13-0300, working dilution 1:1,000)
Laminin 1 mg/ml (Thermo Fisher Scientific, GibcoTM, catalog number: 23017015)
Aqua-Poly/Mount (Polysciences, catalog number: 18606-20)
Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: S392212)
-
JustPure PIPES (Fisher Scientific, catalog number: BP292450)
Note: This product has been discontinued.
D-Glucose (Fisher Scientific, catalog number: D16-1)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333-500G)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-1)
Ultrapure distilled water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977023)
Phenol red solution, 0.02% (Fisher Scientific, catalog number: S25464)
Antibiotic-Antimycotic 100x (Thermo Fisher Scientific, GibcoTM, catalog number: 15240062)
L-Cysteine hydrochloride (Sigma-Aldrich, catalog number: C1276-10G)
Ethylenediaminetetraacetic acid (EDTA) (Fisher Scientific, catalog number: BP118-500)
Deoxyribonuclease I (DNase I) (Worthington Biochemical, catalog number: LS002139)
Trypsin inhibitor, Ovomucoid (Sigma-Aldrich, catalog number: T9253-250MG)
Phosphate buffered saline, (PBS) pH 7.4 (Fisher Scientific, catalog number: BP665-1)
Percoll (MP Biomedicals, catalog number: 0219536925)
Bovine serum albumin (BSA) (Fisher Scientific, catalog number: BP9704100)
1x Hank’s balanced salt solution (HBSS), no calcium, no magnesium (Thermo Fisher Scientific, GibcoTM, catalog number: 14175103)
B-27 Supplement (50x), serum-free (Thermo Fisher Scientific, GibcoTM, catalog number: 17504044)
N-2 Supplement (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 17502048)
L-Glutamine (200 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 25030149)
100x Insulin-Transferrin-Selenium (ITS-X) (Thermo Fisher Scientific, GibcoTM, catalog number: 51500056)
Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM/F-12) (Thermo Fisher Scientific, GibcoTM, catalog number: 11320082)
Human recombinant basic Fibroblast Growth Factor (bFGF) (STEMCELL Technologies, catalog number: 78003)
Human recombinant Epidermal Growth Factor (EGF) (STEMCELL Technologies, catalog number: 78006.1)
16% paraformaldehyde (PFA) (Alfa Aesar, catalog number: 43368)
Normal donkey serum (NDS) (Jackson ImmunoResearch, catalog number: 017-000-121)
1 M HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (Thermo Fisher Scientific, GibcoTM, catalog number: 15630080)
Triton X-100 (Fisher Scientific, catalog number: BP151-500)
10 N sodium hydroxide (NaOH) (see Recipes)
1 M PIPES (see Recipes)
10x sodium chloride (NaCl)/potassium chloride (KCl) (NaCl/KCl) salt solution (see Recipes)
30% D-glucose solution (see Recipes)
1 N NaOH (see Recipes)
PIPES solution (see Recipes)
Activating solution for papain (see Recipes)
10 mg/ml DNase I solution (see Recipes)
7 mg/ml ovomucoid (see Recipes)
10x phosphate buffered saline (PBS) (see Recipes)
22% Percoll solution (see Recipes)
10% bovine serum albumin (BSA) (see Recipes)
1% BSA/0.1% glucose/1x HBSS (see Recipes)
1x PBS (see Recipes)
Neurosphere media (see Recipes)
Epidermal Growth Factor (EGF) (20μg/ml) (see Recipes)
Basic Fibroblast Growth Factor (bFGF) (20μg/ml) (see Recipes)
10% normal donkey serum (NDS)/0.5% Triton X-100 (see Recipes)
1% normal donkey serum (NDS)/0.25% Triton X-100 (see Recipes)
4% paraformaldehyde (PFA) (see Recipes)
Equipment
Biological safety cabinet (Labconco, model: Class II, Type A2)
P2 pipetman (Gilson, catalog number: F144801)
P20 pipetman (Gilson, catalog number: F123600)
P200 pipetman (Gilson, catalog number: F123601)
P1000 pipetman (Gilson, catalog number: F123602)
Motorized pipette controller (Accupet, catalog number: PH01)
Milligram balance (Sartorius, model: Entris 323)
Minidizer hybridization oven (UVP, model: HB-500)
37 °C, 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i)
Refrigerated centrifuge (Eppendorf, model: 5804 R)
Hemocytometer chamber (Hausser Scientific, catalog number: 3110)
Water bath (Fisher Scientific, model: Isotemp 2340)
Thermo ScientificTM NuncTM Lab-Tek® Chamber Slide System (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 178599)
Fluorescence-activated cell sorter (BD, model: FACSARIATM III)
Inverted microscope (Motic, model: AE20)
Confocal microscope (ZEISS, model: LSM 710)
Beaker (Fisher Scientific, catalog numbers: FB10050, FB100100, FB100150)
Autoclave (Getinge, model: PACS 2000)
Procedure
-
Fresh brain tissue dissociation into single cell suspension
Notes:
Tissue processing and FACS are performed in a Biological Safety Level (BSL-2) laminar flow hood, using universal precautions for bloodborne human pathogens. All tissues are collected de-identified, under approved IRB protocols.
It is recommended that tissues/cells are kept on ice as much as possible, unless otherwise noted for specific incubation steps.
Collect fresh brain tissue (GM postmortem dissection or GBM surgical resection) in cold, freshly prepared PIPES solution (see Recipes).
-
Dissolve papain in activating solution (1 ml of activating solution per 0.003 g papain for each 50-200 mg tissue) (see also Note 7).
Note: The papain must be activated for at least 20 min but no more than 30 min at room temperature, prior to tissue incubation.
-
Mechanical digestion
While the papain is activating, mince the tissue with a razor blade in a small Petri dish, containing a small volume of cold PIPES solution (1-2 ml), until the tissue is finely minced (Figure 1A). The process of mincing should not last more than 5 min. Once the tissue is minced, the Petri dish should be placed on ice.
-
Transfer the minced tissue into a 15 ml Falcon tube and add additional cold PIPES solution up to 9 ml. Add 1 ml of activated papain for a final volume of 10 ml (final concentration of papain 300 μg/ml).
Note: Papain digestion solution (10 ml total) = 0.003 g papain + 1 ml activating solution + 50-200 mg tissue in 9 ml cold PIPES.
-
Enzymatic digestion
Incubate the tissue with activated papain, 13 min at 37 °C, in a rotating incubator (rocking speed: 12 rpm). After the first 5 min, add 50 μl of DNase I (10 mg/ml, see Recipes), mix the digesting tissue by shaking, and then re-incubate at 37 °C for the remaining 8 min.
Centrifuge the digested tissue in a refrigerated centrifuge, 10 min at 310 × g at 4 °C.
-
Stop the enzymatic digestion
After centrifugation, there will be a pellet of cells (Figure 1B). Gently decant the supernatant, taking care not to disturb the pellet, which may be loose. Resuspend the cells in 1 ml of ice cold DMEM/F-12 with additional 50 μl of DNAse I (10 mg/ml) and 100 μl of papain/trypsin inhibitor (ovomucoid) (7 mg/ml, see Recipes).
-
Single cell suspension
Triturate the cell pellet continuously first with a P1000 pipette until no tissue chunks are visible (Video 1). Then, triturate with a P200 pipette, approximately 50 times, or until a murky, homogenous cell suspension is obtained without visible debris (Figure 1C) (Video 2). Avoid foaming.
-
Eliminate extracellular debris
Add 4 ml of cold 22% Percoll solution to new, sterile 15 ml tubes (two tubes per sample). Take approximately half of the cell suspension (~650 μl) and layer it on top of the Percoll in a continuous drop-by-drop manner in order to prevent as much as possible mixing between the cell suspension and the Percoll solution (see Recipes and Note) (Video 3) (Figure 1D). Layer the remaining half of the cell suspension into another tube with 4 ml of cold 22% Percoll (split each cell suspension sample into two Percoll tubes).
Note: Tilting the Percoll tube at 45° while adding the cell suspension may help to prevent mixing.
Centrifuge the sample, 10 min at 594 × g in a refrigerated centrifuge at 4 °C, with ‘no brake’ or ‘free deceleration’ setup.
After centrifugation, myelin and other extracellular and fibrillar debris will be contained within the Percoll supernatant, while intact cells will be pelleted at the bottom of the tube (Figure 1E). Carefully decant the entire supernatant and resuspend each pellet in 450 μl of cold 1% BSA/0.1% glucose/1x HBSS (see Recipes). Re-combine every two resuspended pellets (that were split during Step A9) into a new 15 ml Falcon tube.
-
Eliminate red blood cells:
Add 100 μl (10x) of Red blood cell lysis (RBL) buffer and incubate at room temperature, 10 min.
-
Wash the RBL
After erythrocyte lysis, add 9 ml of cold 1% BSA/0.1% glucose/1x HBSS and centrifuge, 5 min at 310 × g at 4 °C.
-
Count live cells
Decant the supernatant and resuspend the cell pellet in 550 μl of cold 1% BSA/0.1% glucose/1x HBSS. Count the number of live cells using trypan blue exclusion in a hemocytometer. Mix 5 μl of trypan blue with 5 μl of sample, and load in a hemocytometer chamber to count live cells, which exclude trypan blue, under the microscope. Calculate the number of live cells in the stock solution.
-
Ligand and antibody incubation for FACS:
Aliquot out 5% of the total cell suspension for each single color/no color controls (approximately 30 μl for each) and use the remaining cell suspension for the experimental sample.
For the experimental sample, add EGF-Alexa Fluor 647 ligand (5 μg/106 live cells) and exclusion antibody markers (anti CD24-PE, 1:10; anti CD34-PE, 1:10; anti CD45-PE, 1:10) to the remaining cell suspension in the same 15-ml tube (approximately 450 μl). Mix gently by flicking the tube.
For single color controls, add EGF-Alexa Fluor 647 ligand only or CD24-PE/CD34-PE/CD45-PE only to each 30 μl (5%) cell suspension aliquot in a new 15-ml tube, using the same ligand and antibody dilution used for the experimental sample. For the no color (DAPI only) control, incubate the 30 μl aliquot on ice without adding anything and add DAPI only during Step A18.
Incubate on ice, 30 min, in darkness. Re-mix gently by flicking the tube after the first 15 min.
-
Wash unbound antibodies/ligand
Wash all tubes with 10 ml of cold 1% BSA/0.1% glucose/1x HBSS and centrifuge, 5 min at 310 g at 4 °C.
Decant the supernatant and resuspend the cell pellet in 1 ml of cold 1% BSA/0.1% glucose/1x HBSS with DAPI (1:1,000).
-
Achieve single cell suspension and minimize clumping:
Pass the solution through a 40 μm filter.
-
Fluorescence-Activated Cell Sorting (FACS)
Note: Technical support during sorting is highly recommended for untrained personnel.
Gate and sort cells following gating procedure described in Tome-Garcia et al. (2017) and in Figure 2. No color (DAPI only), single color, and fluorescence minus one color (if using more than two colors) controls should be performed with each experiment to establish negative/positive cut-off values for the specific cell populations tested, and to ensure consistency.
-
After proper gating, collect EGF-bound (i.e., EGFR+) live (DAPI- and DAPIlow) cells for molecular or functional downstream analysis. For cell culture, collect cells in a 1.5 ml microcentrifuge tube containing sterile and freshly prepared neurosphere media (see Recipes) supplemented with EGF (20 ng/ml) and bFGF (20 ng/ml).
Note: Neurosphere growth is also observed in the absence of ligand supplementation.
-
Functional validation of in vitro stem cell behavior
Note: Culture validation is recommended for all GBM tumors, since in rare cases, tumors without a well-defined EGFR+ population may show sphere growth in both positive and negative fractions (Tome-Garcia et al., 2017) (see also Note 4).
Seed the cells in 96-well low-attachment plates at a clonal density of 10 cells/μl (200 μl/well), ( Pastrana et al., 2011 ), in triplicate wells at minimum. Let the cells form neurospheres for one week and then change media every three days (replace only ~1/3-1/2 of the media volume, removing it carefully from the edge of the well without disturbing the neurospheres). Neurosphere formation can be checked under an inverted microscope as soon as 1 week after seeding (Figures 3A-3B). Consider letting cells grow for 12-21 days or until neurospheres reach at least 40 μm diameter before passaging them into secondary neurospheres or testing their multipotency for differentiation (Figure 3).
-
To differentiate neurospheres, pick up single neurospheres with a 200 μl pipette and seed them on laminin-coated chambers. Let the neurospheres attach to the laminin for 4-5 h in a 37 °C cell incubator. Once attached, remove the old neurosphere media, and add new neurosphere media without B27, EGF, bFGF growth factors supplement. Let the neurospheres differentiate in a 37 °C cell incubator for 1 week with no change of media.
Note: Prior to neurosphere seeding, Lab-Tek® chambers should be pre-coated with laminin (10 μg/ml), incubated overnight at 37 °C, and then washed three times with 1x PBS.
After 1 week of differentiation, fix the cells using 4% PFA solution (see Recipes), 10 min at room temperature. Wash the cells with 1x PBS (see Recipes), three times.
To perform immunofluorescence studies, incubate the differentiated cells with blocking solution (10% normal donkey serum (NDS)/0.5% Triton X-100) (see Recipes), 1 h at room temperature, and then incubate with primary antibodies in 1% NDS/0.25% Triton X-100 (see Recipes), overnight at 4 °C. Perform three washes with 1x PBS, 5 min each, and incubate with species-appropriate fluorochrome-conjugated secondary antibody in 1% NDS/0.25% Triton X-100 (see Recipes), 4 h at room temperature. Wash the secondary antibody with 1x PBS, 5 min each, counterstain with DAPI (1:1,000), and mount with Aqua-Poly/Mount aqueous mounting medium.
Specifically, for O4, immunofluorescence is performed on live, unfixed cells. Incubate cells with primary anti-O4 antibody, 40 min at 4 °C, perform three washes with DMEM/F-12 media, 5 min each at room temperature, and incubate with anti-mouse IgM secondary antibody, 40 min at 4 °C. Wash three times with DMEM/F-12 media at room temperature, 5 min each time, and fix cells for 10 min with 4% PFA solution.
Check immunoreactivity for GFAP (astrocytes), TuJ1 (neurons) and O4 (oligodendrocytes) under a confocal or epifluorescence microscope (Figure 3C).
Figure 1. Tissue dissociation from fresh human brain tissue.
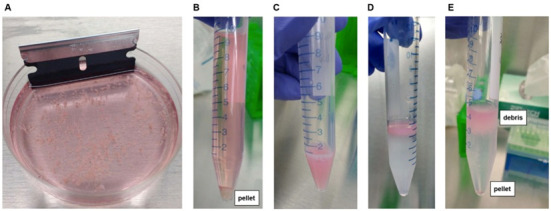
A. Mechanical tissue dissociation using a razor blade, prior to adding papain. B. Enzymatic tissue dissociation using papain, with resultant cell pellet after centrifugation. C. Cell suspension formed after trituration of dissociated tissue. D. Layering of cell suspension onto 22% Percoll solution. E. Separation of cells from extracellular debris after gentle centrifugation in 22% Percoll. Myelin and other extracellular debris are contained within the supernatant (top) while live cells are pelleted (bottom).
Video 1. Primary trituration of dissociated tissue suspension using P1000.
After enzymatic digestion, the dissociated cell pellet is triturated continuously first with a P1000 pipette until no large tissue chunks are visible.
Video 2. Secondary trituration of dissociated tissue suspension using P200.
Subsequent trituration with a P200 pipette is also performed in order to obtain a homogenous cell suspension without any visible debris.
Video 3. Elimination of extracellular debris using Percoll solution.

The cell suspension is carefully layered onto a 22% cold Percoll solution in a continuous drop-by-drop manner, with the tube being tilted at 45°, minimizing contact between the Percoll solution and the cell suspension. This step allows elimination of myelin and other extracellular debris.
Figure 2. Isolation of EGFR+ cells from freshly dissociated GM and GBM tissues by FACS.
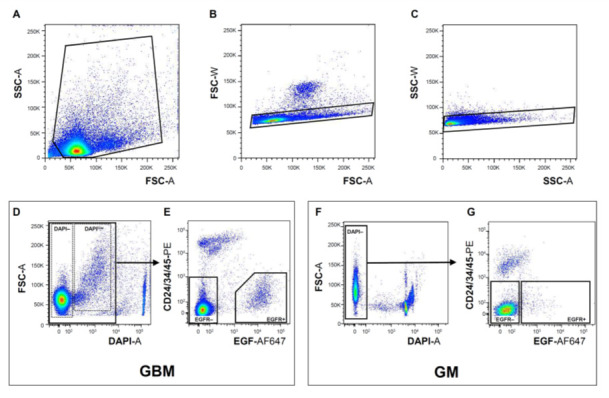
Representative examples of sequential gating during FACS in order to eliminate tissue debris (A) (GBM sample), exclude cell doublets (B and C) (GBM sample), remove dead/dying cells using DAPI (D and F) (GBM and GM samples, respectively), and exclude ependymal/neuroblasts/endothelial/inflammatory cells using CD24/34/45 antibody cocktail (E and G) (GBM and GM samples, respectively, see also Note 6). In GBM, but not in GM sorts, a subset of live cells displays a shift in DAPI fluorescence (DAPIlow) (D). The EGFR+ DAPIlow GBM population contains the majority of cells with stem cell properties (Tome- Garcia et al., 2017 ) (see also Notes). Violet-A, PE-A, and APC-A light filters were used to visualize DAPI, bound CD24/34/45-PE antibodies, and bound EGF-AF647 ligand, respectively.
Figure 3. In vitro functional analysis of stem cell properties of acutely isolated EGFR+ and EGFR- GM and GBM cells.
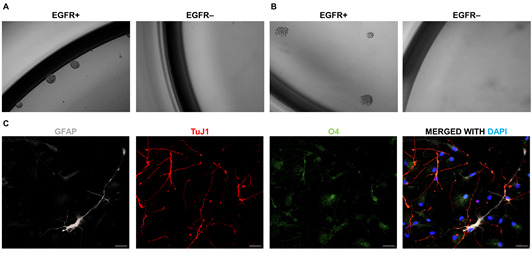
A-B. FACS-isolated EGFR+ and EGFR- primary cells are seeded at clonal density (10 cells/μl). Neurosphere formation, assessed at day 6 of seeding, in GM EGFR+/EGFR- populations (A), and at day 12 in GBM EGFR+/EGFR- populations (B). C. Illustration of trilineage differentiation of primary GM neurospheres using immunofluorescence, with EGFR+ cells showing differentiation into astrocytic (GFAP+, white); neuronal (TuJ1+, red) and oligodendroglial (O4+, green) lineages. DAPI (blue) is used to counterstain cell nuclei. Magnification of neurosphere pictures, 10x. Pictures have been adapted from Tome- Garcia et al., 2017 . Scale bars = 20 μm.
Data analysis
To ensure statistical significance and reproducibility in the neurosphere and differentiation assays, all experiments were performed in three technical replicates and were repeated at least three independent times. To determine statistical significance for the number of neurospheres between EGFR+ and EGFR- cell populations, unpaired two-tailed Student’s t-test was applied.
Notes
Due to potentially low cell viability in tissues with longer surgical ischemia or postmortem times, samples should be immersed in freshly made (same day) cold PIPES solution immediately after being removed from the brain, and the PIPES-immersed sample should be kept cold (on ice) until it can be processed in the laboratory.
Human germinal matrix specimens display a wide range of viable cell yield (8.9%-96% DAPI- live cells out of total cells) and EGFR+ cells (1%-25% of total viable cells), depending on gestational age and postmortem time. Human GBM samples also display a wide range of viable cell yield (36%-99%) and EGFR+ cells (1.3%-77% of total viable cells), which we attribute to intra- and intertumoral molecular heterogeneity of glioblastoma samples, in addition to post-surgical ischemia time. For detailed information on percent EGFR+ cell yield under the different abovementioned variables, refer to Supplemental Figure 1 in the original report (Tome- Garcia et al., 2017 ).
Gating of EGFR+ and EGFR- populations can affect the selectivity of stem cell isolation. While we recommend following the gates illustrated in Figure 2, some modifications may be necessary for individual samples. Particularly, for GBM sorts, the proportion of DAPI- and DAPIlow cells can be quite variable, depending on cell aneuploidy, post-surgical ischemia time, and the percentage of glioma cells with stem cell properties in the tissue sample. In our experience, the majority of tumor cells with stem cell properties are captured in the DAPIlow (EGFR+) population, although occasional DAPI- (EGFR+) cells also showed sphere formation in some tumor samples. Thus, depending on whether specificity or sensitivity is a priority for the downstream analysis, the user may choose to sort DAPI- separately from DAPIlow, or to combine both as illustrated in Figure 2.
The use of EGF ligand for FACS isolation of sphere-forming human glioma populations has been validated for most de novo glioblastoma resections, both IDH-wildtype and IDH-mutant, more than 90% of which show expression of EGFR, even in cases when EGFR amplification is not detected ( Erfani et al., 2015 ; Tome- Garcia et al., 2017 ). This methodology may not be applicable for GBMs with strong amplification of other receptor tyrosine kinases, such as the platelet-derived growth factor receptor alpha (PDGFRalpha) seen in ~10% of GBMs ( Brennan et al., 2013 ), especially if the tumor lacks significant EGFR expression altogether (2/19 GBMs tested in our cohort) (Tome- Garcia et al., 2017 [Figure S2]). This methodology has not been validated for recurrent GBMs and lower-grade gliomas.
In germinal matrix tissues of early gestation age (< 18 weeks), rare small sphere formations have been observed in the EGFR- population, which were unable to undergo trilineage differentiation (Tome- Garcia et al., 2017 [Figure S2]).
In the outlined experiments above and previously published (Tome- Garcia et al., 2017 ), CD24 was used as an exclusion marker of neuroblasts and ependymal cells in GM sorts, and was also included in GBM sorts in order to mimic FACS conditions between GM and GBM. Inclusion of CD24 in GBM sorts is optional, but omitting it has not been fully tested.
The weight of tissue processed can vary, depending on GM gestational age and the extent of GBM resection. In our experiments, we calibrated the papain concentration based on a range of 50-200 mg of tissue. If more than 200 mg of tissue was used, we digested the additional tissue in a separate papain solution (0.003 g papain per 50-200 mg of additional tissue). Most of our processed samples had a starting weight of ~300 mg. For processing of tissue less than 50 mg, papain concentration may need to be optimized.
Recipes
-
10 N sodium hydroxide (NaOH)
20 g of NaOH pellets
Bring the volume to 50 ml with ultrapure distilled water
Note: As a precaution use a beaker and keep it on ice during the process.
-
1 M piperazine-N,N’-bis(2-ethanesulfonic acid) (PIPES)
Dissolve 30.22 g of PIPES in 90 ml of ultrapure distilled water
Add NaOH pellets until reaching pH 6.7
Add 10 N NaOH dropwise to reach pH 6.9
Bring the volume to 100 ml with ultrapure distilled water
Filter with a 0.2 μm filter
-
10x sodium chloride (NaCl)/potassium chloride (KCl) (NaCl/KCl) salt solution
3.5 g sodium chloride (NaCl)
0.186 g potassium chloride (KCl)
Bring the volume to 50 ml with ultrapure distilled water
Filter with a 0.2 μm filter
-
30% D-glucose solution
15 g D-glucose
Bring the volume to 50 ml with ultrapure distilled water
Filter with a 0.2 μm filter and store at 4 °C
-
1 N NaOH
2 g of NaOH pellets
Bring the volume to 50 ml with ultrapure distilled water
Note: As a precaution use a beaker and keep it on ice during the process.
-
PIPES solution
1 ml 1 M PIPES
5 ml 10x NaCl/KCl salt solution
750 μl 30% glucose
1,250 μl 0.02% phenol red
500 μl 100x antibiotic/antimycotic
Bring the volume to 50 ml with ultrapure distilled water
Add 75 μl 1 N NaOH to turn the solution from salmon to pink (pH 7.4, if it turns magenta-to-purple, start over)
Filter with a 0.2 μm filter
-
Activating solution for papain
0.00878 g Cysteine HCl
0.0093 g EDTA
Bring the volume to 50 ml with ultrapure distilled water
Filter with a 0.2 μm filter and store at 4 °C
-
10 mg/ml DNase I solution
10 mg DNase I
Bring the volume to 1,000 μl with ultrapure distilled water
Aliquot and keep frozen at -20 °C
-
7 mg/ml ovomucoid
7 mg ovomucoid
Bring the volume to 1,000 μl with ultrapure distilled water
Aliquot and keep frozen at -20 °C
-
10x phosphate buffered saline (PBS)
98.9 g 10x concentrated powder PBS
Bring the volume to 1 L with ultrapure distilled water
Dissolve and autoclave
-
22% Percoll solution
11 ml Percoll
5 ml 10x PBS
Bring the volume to 34 ml with ultrapure distilled water
Filter with 0.2 μm filter and store at 4 °C
-
10% bovine serum albumin (BSA)
5 g BSA
Bring the volume to 50 ml with 1x HBSS
Store in the fridge until it dissolves completely
Filter with 0.2 μm filter and store at 4 °C
-
1% BSA/0.1% glucose/1x HBSS
50 ml 10% BSA
1.67 ml 30% D-glucose
448.3 ml 1x HBSS
Filter with a 0.2 μm filter and store at 4 °C
-
1x PBS
100 ml 10x PBS
900 ml ultrapure distilled water
-
Neurosphere media
500 μl N-2 Supplement (100x)
1,000 μl B-27 Supplement (50x)
500 μl 200 mM L-glutamine
500 μl 100x antibiotic-antimycotic
500 μl 100x Insulin-Transferrin-Selenium (ITS-X)
μl 1 M HEPES
1 ml 30% D-glucose
Bring the volume to 50 ml with DMEM/F12
-
Epidermal Growth Factor (EGF) (20 μg/ml)
Resuspend the lyophilized product in enough volume of ultrapure distilled water to have a final stock concentration of 20 μg/ml
Dilute 1:1,000 in Neurosphere media to reach a working concentration of 20 ng/ml
-
Basic Fibroblast Growth Factor (bFGF) (20 μg/ml)
Resuspend the lyophilized product in enough volume of ultrapure distilled water to have a final stock concentration of 20 μg/ml
Dilute 1:1,000 in Neurosphere media to reach a working concentration of 20 ng/ml
-
10% normal donkey serum (NDS)/0.5% Triton X-100
100 μl NDS
5 μl Triton X-100
μl 1x PBS
-
1% normal donkey serum (NDS)/0.25% Triton X-100
10 μl NDS
2.5 μl Triton X-100
987.5 μl 1x PBS
-
4% paraformaldehyde (PFA)
Dilute 10 ml of 16% PFA with 30 ml of 1x PBS
Acknowledgments
These experimental procedures have been previously published and are herein adapted and modified from Tome- Garcia et al., 2017 . The work was partially supported by R03NS104669 (N.T.). We thank members of the Pathology department and Biorepository Core at the Icahn School of Medicine at Mount Sinai (ISMMS) for facilitating tissue collection and procurement and the ISMMS Flow Cytometry CORE for expert advice and accommodation of fresh human tissue sorts 24 h/day. The research was supported by Mount Sinai seed grant funds. The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Anido J., Saez-Borderias A., Gonzalez-Junca A., Rodon L., Folch G., Carmona M. A., Prieto-Sanchez R. M., Barba I., Martinez-Saez E., Prudkin L., Cuartas I., Raventos C., Martinez-Ricarte F., Poca M. A., Garcia-Dorado D., Lahn M. M., Yingling J. M., Rodon J., Sahuquillo J., Baselga J. and Seoane J.(2010). TGF-β receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma . Cancer Cell 18(6): 655-668. [DOI] [PubMed] [Google Scholar]
- 2.Beier D., Hau P., Proescholdt M., Lohmeier A., Wischhusen J., Oefner P. J., Aigner L., Brawanski A., Bogdahn U. and Beier C. P.(2007). CD133+ and CD133- glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles . Cancer Res 67(9): 4010-4015. [DOI] [PubMed] [Google Scholar]
- 3.Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., Van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O'Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., Chin L. and Network T. R.(2013). The somatic genomic landscape of glioblastoma. Cell 155(2): 462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canoll P. and Goldman J. E.(2008). The interface between glial progenitors and gliomas. Acta Neuropathol 116(5): 465-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., McKay R. M. and Parada L. F.(2012). Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 149(1): 36-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccolini F., Mandl C., Holzl-Wenig G., Kehlenbach A. and Hellwig A.(2005). Prospective isolation of late development multipotent precursors whose migration is promoted by EGFR. Dev Biol 284(1): 112-125. [DOI] [PubMed] [Google Scholar]
- 7.Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A. R., Deleo A. M., Pastrana E. and Doetsch F.(2014). Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche . Neuron 82(3): 545-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erfani P., Tome-Garcia J., Canoll P., Doetsch F. and Tsankova N. M.(2015). EGFR promoter exhibits dynamic histone modifications and binding of ASH2L and P300 in human germinal matrix and gliomas. Epigenetics 10(6): 496-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lathia J. D., Gallagher J., Heddleston J. M., Wang J., Eyler C. E., Macswords J., Wu Q., Vasanji A., McLendon R. E., Hjelmeland A. B. and Rich J. N.(2010). Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell 6(5): 421-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lathia J. D., Mack S. C., Mulkearns-Hubert E. E., Valentim C. L. and Rich J. N.(2015). Cancer stem cells in glioblastoma. Genes Dev 29(12): 1203-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzoleni S., Politi L. S., Pala M., Cominelli M., Franzin A., Sergi Sergi L., Falini A., De Palma M., Bulfone A., Poliani P. L. and Galli R.(2010). Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res 70(19): 7500-7513. [DOI] [PubMed] [Google Scholar]
- 12.Ogden A. T., Waziri A. E., Lochhead R. A., Fusco D., Lopez K., Ellis J. A., Kang J., Assanah M., McKhann G. M., Sisti M. B., McCormick P. C., Canoll P. and Bruce J. N.(2008). Identification of A2B5+CD133- tumor-initiating cells in adult human gliomas . Neurosurgery 62(2): 505–514.; discussion 514-505. [DOI] [PubMed] [Google Scholar]
- 13.Pastrana E., Cheng L. C. and Doetsch F.(2009). Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A 106(15): 6387-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastrana E., Silva-Vargas V. and Doetsch F.(2011). Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8(5): 486-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanai N., Alvarez-Buylla A. and Berger M. S.(2005). Neural stem cells and the origin of gliomas. N Engl J Med 353(8): 811-822. [DOI] [PubMed] [Google Scholar]
- 16.Sanai N., Nguyen T., Ihrie R. A., Mirzadeh Z., Tsai H. H., Wong M., Gupta N., Berger M. S., Huang E., Garcia-Verdugo J. M., Rowitch D. H. and Alvarez-Buylla A.(2011). Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478(7369): 382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh S. K., Clarke I. D., Terasaki M., Bonn V. E., Hawkins C., Squire J. and Dirks P. B.(2003). Identification of a cancer stem cell in human brain tumors. Cancer Res 63(18): 5821-5828. [PubMed] [Google Scholar]
- 18.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D. and Dirks P. B.(2004). Identification of human brain tumour initiating cells. Nature 432(7015): 396-401. [DOI] [PubMed] [Google Scholar]
- 19.Son M. J., Woolard K., Nam D. H., Lee J. and Fine H. A.(2009). SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 4(5): 440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tome-Garcia J., Tejero R., Nudelman G., Yong R. L., Sebra R., Wang H., Fowkes M., Magid M., Walsh M., Silva-Vargas V., Zaslavsky E., Friedel R. H., Doetsch F. and Tsankova N. M.(2017). Prospective isolation and comparison of human germinal matrix and glioblastoma EGFR+ populations with stem cell properties . Stem Cell Reports 8(5): 1421-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsankova N. M. and Canoll P.(2014). Advances in genetic and epigenetic analyses of gliomas: a neuropathological perspective. J Neurooncol 119(3): 481-490. [DOI] [PubMed] [Google Scholar]
- 22.Uchida N., Buck D. W., He D., Reitsma M. J., Masek M., Phan T. V., Tsukamoto A. S., Gage F. H. and Weissman I. L.(2000). Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A 97(26): 14720-14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhaak R. G., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., Hayes D. N. and Cancer Genome Atlas Research N.(2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1): 98-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Sakariassen P. O., Tsinkalovsky O., Immervoll H., Boe S. O., Svendsen A., Prestegarden L., Rosland G., Thorsen F., Stuhr L., Molven A., Bjerkvig R. and Enger P. O.(2008). CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer 122(4): 761-768. [DOI] [PubMed] [Google Scholar]
- 25.Weickert C. S., Webster M. J., Colvin S. M., Herman M. M., Hyde T. M., Weinberger D. R. and Kleinman J. E.(2000). Localization of epidermal growth factor receptors and putative neuroblasts in human subependymal zone. J Comp Neurol 423(3): 359-372. [DOI] [PubMed] [Google Scholar]


