Abstract
Long-chain fatty acyl-CoA synthetases (ACSLs) are homologs of firefly luciferase, but are incapable of emitting light with firefly luciferin. Recently, we found that an ACSL from the fruit fly Drosophila melanogaster is a latent luciferase that will emit light with the synthetic luciferin CycLuc2. Here we have profiled a panel of three insect ACSLs with a palette of >20 luciferin analogues. An ACSL from the nonluminescent beetle Agrypnus binodulus (AbLL) was found to be a second latent luciferase with distinct substrate specificity. Several rigid luciferins emit light with both ACSLs, but styryl luciferin analogues are light-emitting substrates only for AbLL. On the other hand, an ACSL from the luminescent beetle Pyrophorus angustus lacks luciferase activity with all tested analogues, despite its higher homology to beetle luciferases. Further study of ACSLs is expected to shed light on the features necessary for bioluminescence and substrate selectivity.
TOC graphic
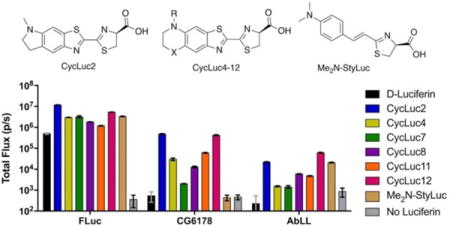
Firefly luciferase is a promiscuous enzyme. In addition to its native substrate D-luciferin, it will accept numerous synthetic luciferin analogs, resulting in light emission (Figure 1).1–5 Furthermore, luciferase is also a long-chain fatty acyl-CoA synthetase (ACSL), capable of converting fatty acids to fatty acyl-CoAs.6 Most ACSLs are promiscuous with regard to their fatty acid substrates, but are not capable of luciferase activity with D-luciferin, a compound only found naturally in bioluminescent beetles.7 Although the fruit fly fatty acyl-CoA synthetase CG6178 is 39% identical to firefly luciferase (Figure S1), it does not emit light when treated with D-luciferin, and indeed lacks the ability to adenylate D-luciferin.7
Figure 1.
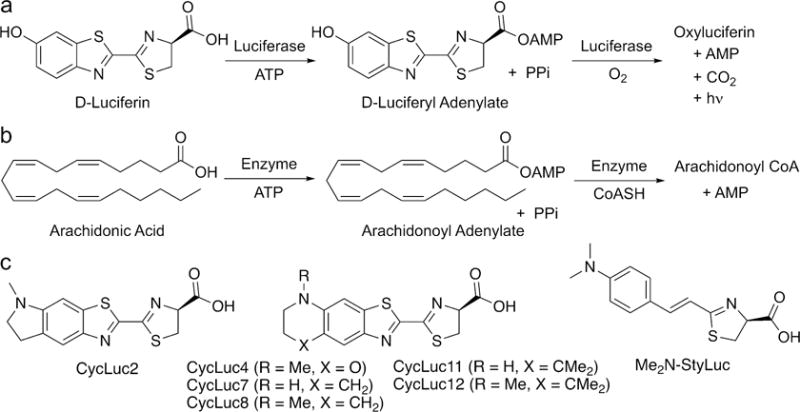
Fatty acyl-CoA synthetases and firefly luciferase catalyze similar, two-step reactions. (a) Firefly luciferase catalyzes the formation of an activated adenylate of D-luciferin followed by oxidation to an excited state oxyluciferin that is responsible for light emission. (b) Fatty acyl-CoA synthetases catalyze the formation of an activated adenylate of a free fatty acid followed by displacement of the adenylate by CoASH to form the acyl-CoA product. (c) A subset of synthetic luciferins emit light with CG6178 and/or AbLL in addition to firefly luciferase.
At first glance, the chemistry of light emission and that of fatty acid thioesterification may appear to be quite different. However, the initial step catalyzed in both reactions is essentially the same: the attack of a carboxylate on ATP to form an acyl-adenylate intermediate (Figure 1). In previous work, we hypothesized that insect ACSLs such as CG6178 could be capable of luciferase activity if given a suitable luciferin substrate (i.e., one which could be adenylated). Adenylation of a luciferin analog should activate it toward reaction with molecular oxygen,8,9 thereby allowing access to the chemistry of light emission. Supporting this hypothesis, we found that CG6178 is capable of luciferase activity when treated with the synthetic luciferin CycLuc2.8 Here we show that latent luciferase activity is not limited to CG6178 and CycLuc2. We find that the latent luciferase activity of CG6178 extends to a number of other synthetic luciferins, and that CG6178 is not the only latent luciferase among insect ACSLs.
A small panel of six luciferins was originally used to identify CG6178 as a latent luciferase.8 To probe the scope of latent luciferase activity further, we augmented this panel with ten additional rigid cyclic aminoluciferins (CycLuc3-12),3 as well as five additional acyclic aminoluciferins (Figure S2). We also included two alkenyl luciferin analogues reported by Iwano et al. that lack the canonical benzothiazole core of D-luciferin.10 Upon screening CG6178 with this expanded panel of 23 luciferin analogues, we discovered that light emission extends beyond CycLuc2 (Figure 2a and S3). Generally, rigid dialkylated luciferins are preferred (CycLuc4, 8, and 12), but CycLuc7 and CycLuc11 are also light-emitting substrates (Figure 1c). However, the bulky dialkylated CycLuc6 and CycLuc10 and alkenyl luciferin analogues did not give appreciable light emission. By contrast, all of these molecules yield bioluminescence with firefly luciferase (Figure S3).
Figure 2.
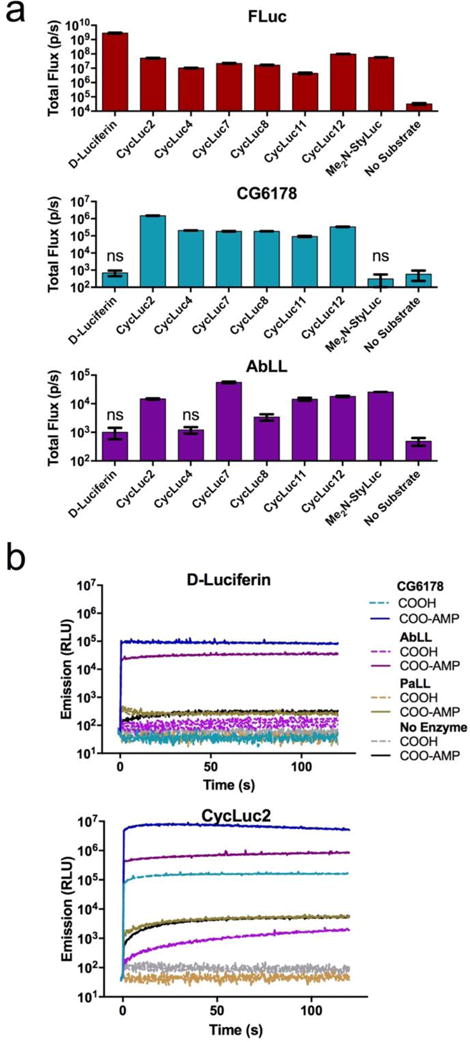
ACSL luciferase activity. (a) Identification of emissive substrates for ACSLs. Each enzyme (20 nM) was treated with the indicated luciferin analog (250 μM). Significant flux over no-substrate control was observed (p < 0.05) unless marked “ns”. (b) Burst kinetics profiles of each ACSL with D-luciferin, CycLuc2, and their pre-activated adenylates. Purified enzyme (100 nM) was rapidly injected into substrate (100 μM). Light emission was recorded every 0.5 s for 1 s pre-injection and 120 s post-injection. Background luminescent signal in the absence of enzyme is shown for reference (in gray). All assays were performed in triplicate, are represented as the mean ± SEM, and are presented on the same log scale.
Having established that latent luciferase activity is not limited to a single substrate, we next asked whether other ACSLs were capable of light emission. Oba et al. have characterized several insect ACSLs as “luciferase-like” enzymes – that is, enzymes with high homology to luciferase, but no luciferase activity with D-luciferin. An ACSL from the non-luminous Japanese click beetle Agrypnus binodulus, AbLL, possesses high sequence identity to firefly luciferase (46%, Figure S1) and to other beetle luciferases (e.g., Pyrophorus plagiophthalmus luciferase, 55%).11 The Pyrophorus angustus Luciferase-Like protein (PaLL) from the luminous Panamanian click beetle also possesses high sequence identity to firefly luciferase (46%, Figure S1) and even higher identity to its own dorsal and ventral luciferases (58% each).12 AbLL and PaLL were thus considered to be potential latent luciferases.
We expressed and purified recombinant AbLL and PaLL, and screened both against our large panel of luciferin analogues. We found that AbLL also exhibits latent luciferase activity, but with a lower maximal rate of photon flux and a substrate selectivity that is markedly different than CG6178 (Figure 2a and S3). Like CG6178, AbLL accepts CycLuc2, 7, 8, 11 and 12 as substrates. Interestingly, AbLL also accepts the styryl luciferin Me2N-StyLuc (Figure 1c), which yields no signal with CG6178. On the other hand, PaLL surprisingly lacked any luciferase activity with the entire palette of luciferins screened (Figure S3), despite its higher homology to beetle luciferases (Figure S1). Circular dichroism measurements suggested the ACSLs were all properly folded, with melting temperatures slightly higher than firefly luciferase (Figure S4).
To better understand the basis for light emission by the latent luciferases CG6178 and AbLL, we compared their burst kinetics to that of firefly luciferase. Upon rapid injection of enzyme into substrate, firefly luciferase produces a high initial rate of photon emission, followed by a decay to a lower level steady state.13 The decay is thought to be a result of product inhibition, and is more pronounced with the tighter-binding synthetic luciferins than with D-luciferin, presumably because their respective products are more potent inhibitors (Figure S5). While CG6178 does exhibit a rapid burst of light upon injection, it shows no product inhibition (Figure 2b and S5). AbLL similarly shows a lack of product inhibition, but also takes much longer to reach a steady-state level of light emission. Bioluminescence from luciferins such as CycLuc2 continued to increase even two minutes after injection (Figure 2b and S5).
For bioluminescence to occur, the (latent) luciferase must first adenylate the luciferin substrate, then activate it for reaction with oxygen.8 To dissect the importance of each of these catalytic steps to light emission, we bypassed the first step by synthesizing the intermediate adenylates of D-luciferin and CycLuc2. As previously reported, D-luciferin adenylate (Figure 1) supports bioluminescence from CG6178.8 Here we find that AbLL can also oxidize D-luciferin adenylate to give light emission (Figure 2b and S6). Similarly, CycLuc2 adenylate is a bioluminescent substrate for both CG6178 and AbLL, yielding brighter emission than CycLuc2 itself, and suggesting that adenylation is the rate-limiting step for both enzymes (Figure 2b and S6). Interestingly, while the initial burst of AbLL is greatly increased using CycLuc2 adenylate, the subsequent photon flux also continues to climb somewhat over the next several minutes. It thus appears that both the adenylation and oxidation steps are less efficient in AbLL than for either firefly luciferase or CG6178. Using either of the pre-adenylated luciferins, PaLL gives at most a very weak initial burst of light, and no substantive sustained emission over the background chemiluminescence of the adenylate in the presence of BSA, suggesting that the catalytic oxidation of luciferins is almost completely absent in this fatty acyl-CoA synthetase (Figure 2b and S6). One candidate sequence for substrate selectivity between these enzymes are the residues within and flanking Motif 2 (Figure S1), as mutations of AbLL in this region have been reported to allow weak luminescence with D-luciferin.14
We next expressed mammalian codon-optimized ACSLs in Chinese Hamster Ovary (CHO) cells, and compared their bioluminescence performance to that of codon-optimized firefly luciferase (luc2). Each was treated with a subset of the luciferin analogues, encompassing the preferred substrates for each enzyme (Figure 3). CG6178 displayed a surprisingly high level of bioluminescence in live cells compared to firefly luciferase, most notably with CycLuc2. Although photon flux from firefly luciferase was considerably higher than that of CG6178 at low concentrations of CycLuc2, the emission plateaued at ~30 μM, presumably due to product inhibition. At 250 μM CycLuc2, the photon emission from CG6178 was comparable to that of CycLuc2 with firefly luciferase (Figure 3). Peak CG6178 emission occurred at an extracellular CycLuc2 concentration of about 0.625 mM, and thereafter slightly decreased, consistent with substrate inhibition rather than product inhibition (Figure 3 and Supporting Information). On the other hand, there was no bioluminescence when CG6178-expressing cells were treated with D-luciferin, even at millimolar concentrations.
Figure 3.
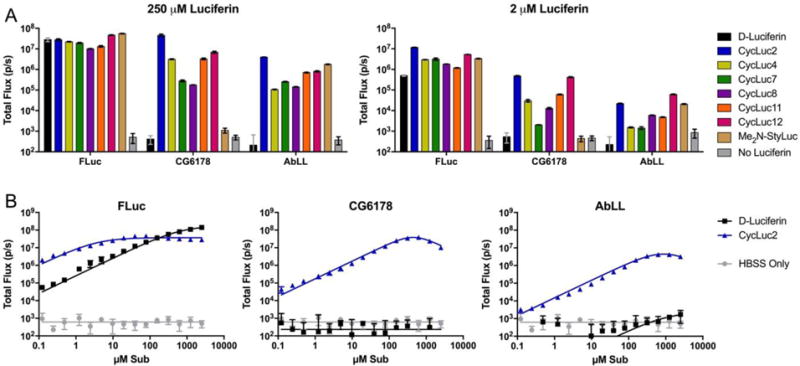
ACSL activity in live CHO cells. (a) Live CHO cells expressing the indicated ACSL treated with either high or low dose of each luciferin analogue. (b) Dose-response curves of live CHO cells expressing the indicated enzyme treated with D-luciferin or CycLuc2 up to 2.5 mM. The assays were performed in triplicate and are represented as the mean ± SEM.
For the most part, the latent luciferase AbLL behaves like a dimmer version of CG6178 (Figure 3). It is possible that this reflects a lower cellular expression level of AbLL compared to CG6178. However, their relative performance in cells largely mirrors what is seen with purified protein in vitro. CycLuc2 and CycLuc12 were preferred over CycLuc7, presumably due to their higher cell permeability. As with CG6178, no signal was observed from AbLL-expressing cells treated with D-luciferin. Furthermore, in sharp contrast to CG6178, the styryl luciferin analogue Me2N-StyLuc resulted in substantial bioluminescent signal with AbLL. We therefore sought to determine whether the latent luciferase activity of AbLL with styryl luciferin analogues could be further improved while maintaining selectivity against CG6178. Using a simplified route to this class of luciferins (Scheme S1), we synthesized two new analogues that replace the dimethylamino group of Me2N-StyLuc with azetidine or pyrrolidine, respectively (Figure 4). None of the styryl analogues emit light with CG6178, but all three are substrates for firefly luciferase and AbLL. Remarkably, bioluminescence from AbLL with pyrrolidine 5 is >10-fold brighter than with Me2N-StyLuc (Figure 4). Thus, subtle substrate modification can result in further improvements in latent luciferase emission and selectivity without the need for mutation of the enzyme itself. The design of unnatural ‘neo-substrates’ has been reported for PINK1 kinase15 and farnesyl transferase,16 but this general strategy remains an underexploited avenue to modify enzyme behavior. Potentially, luciferin analogues could be designed for a still broader range of fatty acyl-CoA synthetases,17 from a range of organisms, and include even more distantly related adenylating enzymes.18
Figure 4.
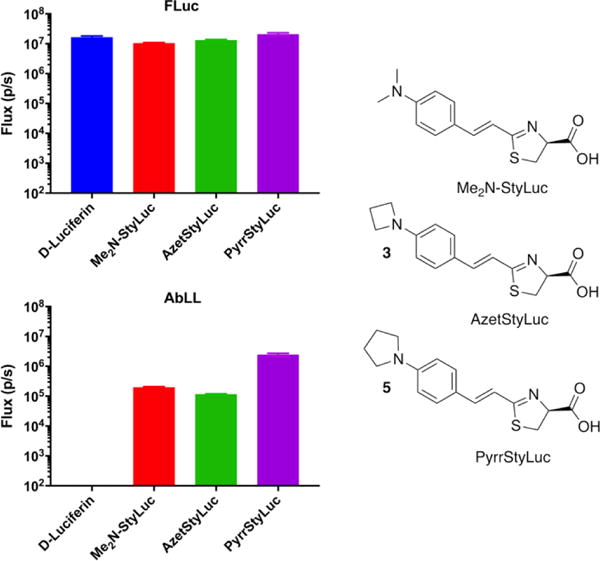
Photon flux from live CHO cells expressing FLuc or AbLL after treatment with 100 μM of D-luciferin or the indicated styryl luciferin analogue. The assays were performed in triplicate and are represented as the mean ± SEM.
Based on the surprising finding that a fruit fly enzyme is able to act as a luciferase when treated with the synthetic luciferin CycLuc2,8 we profiled the latent luciferase activity of three insect proteins with high homology to firefly luciferase (CG6178, AbLL, and PaLL). Using an expanded panel of synthetic luciferin analogues, we found that CG6178 is not unique in its ability to exhibit luciferase activity. AbLL, an ACSL from a nonluminescent click beetle, was identified as a second latent luciferase, capable of bioluminescence with styryl analogues that do not emit with CG6178. On the other hand, PaLL lacked luciferase activity with any of the tested luciferins, despite its higher homology to canonical luciferases. Further study to reveal the molecular basis for the substrate discrimination of these and other ACSLs is expected to enhance our understanding of the essential features required for luciferase activity and lead to improved and more selective bioluminescent reporters.
Methods (additional Methods in SI)
General
Chemicals were purchased from Aldrich, Matrix Scientific, Oakwood, or TCI. ATP was purchased from MP Biomedicals and Sigma. D-luciferin was purchased from Gold Bio and 6ʹ-NH2LH2 from Marker Gene Technologies. Luciferin analogues CycLuc1-12, 6ʹ-MeNHLH2, 6ʹ-Me2NLH2, 6ʹ-EtNHLH2, 6ʹ-iPrNHLH2, 6ʹ-iBuNHLH2, 6ʹ-PrOHNHLH2, Me2N-StyLuc (2b in ref.10), and AkaLumine (3b) were synthesized in-house.19,3,10,20 D-Luciferin adenylate and CycLuc2 adenylate were prepared as previously reported.8 Luminescence screening and CHO cell assays were performed using an IVIS-100. Data acquisition and analysis was performed with Living Image® software, and reported as total flux (p/s) for each ROI corresponding to each well of the 96-well plate. Burst assays were performed as described in the SI.
Purified Protein Luminescence Screening Assays
Luminescence assays were initiated by adding 30 μL of purified luciferase in enzyme buffer (20 mM Tris [pH 7.4], 0.1 mM EDTA, 1 mM TCEP, and 0.8 mg/mL BSA) to 30 μL 2× substrate in substrate buffer (20 mM Tris [pH 7.4], 0.1 mM EDTA, 8 mM MgSO4, and 4 mM ATP) in a black 96-well plate (Costar 3915). Imaging was performed one minute after enzyme addition using a Xenogen IVIS-100 at a final enzyme concentration of 20 nM and final substrate concentration of 250 μM.
CHO cell luminescence assays
Chinese hamster ovary (CHO) cells were grown in a CO2 incubator at 37°C with 5% CO2 and were cultured in F-12K Nutrient Mixture (GIBCO) supplemented with 10% fetal bovine serum and 100 U mL−1 penicillin/streptomycin. The WT firefly luciferase luc2 gene (Promega) and codon-optimized CG6178 and AbLL were cloned into the BamHI and NotI sites of pcDNA 3.1. CHO cells were plated 24 hr prior to transfection at 20,000 cells per well in 96-well black tissue culture-treated plates (Costar 3916), then transfected with Lipofectamine 2000 (0.075 μg DNA/well). Assays were performed in triplicate 24 hr after transfection. Transfected CHO cells were washed with HBSS, then incubated with 60 μL of substrate in HBSS at final concentrations ranging from 0.122 to 2,500 μM. Imaging was performed using the IVIS-100 three minutes after addition of substrate.
Synthesis of new styryl luciferins
4-(azetidin-1-yl)benzaldehyde (1)
To a solution of 4-fluorobenzaldehyde (2.00 g, 16.1 mmol) in DMSO (80.5 mL) was added anhydrous K2CO3 (6.66 g, 3.0 eq) followed by the addition of azetidine hydrochloride (2.24 g, 24.0 mmol, 1.50 eq) with stirring at 110 °C overnight. The mixture was then cooled to room temperature, diluted with water (200 mL), and extracted with ethyl acetate (3 × 100 mL). The organic layer was washed with brine (50 mL) and dried over sodium sulfate, then concentrated in vacuo. Purification by flash chromatography (20% ethyl acetate/hexanes) afforded 5 as a pale yellow solid (1.81 g, 70% yield). 1H-NMR (500 MHz, CDCl3): δ 9.76 (s, 1H), 7.74 (d, J = 8.6 Hz, 2H), 6.46 (d, J = 8.2 Hz, 2H), 4.08 (t, J = 7.4 Hz, 4H), 2.60 – 2.37 (m, 2H). LCMS: 162.1 (M+H).
(E)-3-(4-(azetidin-1-yl)phenyl)acrylonitrile (2)
To a suspension of 95% sodium hydride (72.0 mg, 2.86 mmol) in THF (9.30 mL) at 0 °C was added diethyl cyanomethylphosphonate (0.33 mL, 2.04 mmol). Stirring for 30 minutes yielded a clear solution. To this solution, 4-(azetidin-1-yl)benzaldehyde (1) (300 mg, 1.86 mmol) in THF (2.0 mL) was added dropwise. The reaction was stirred for 2 hours, then quenched with water (20 mL) and extracted with ethyl acetate (2 × 25 mL). The combined organic layers were washed with brine, dried over Na2SO4 and the solvent was removed by rotary evaporation. The resulting residue was purified by flash chromatography (10% ethyl acetate/hexanes) to give 2 (222 mg, 65%) as a green solid. 1H-NMR (500 MHz, CDCl3): δ 7.30-7.25 (m, 3H), 6.37 – 6.18 (m, 2H), 5.57 (d, J = 16.5 Hz, 1H), 3.96 (app t, J = 7.3 Hz, 4H), 2.48 – 2.21 (m, 2H). 13C NMR (125 MHz, CDCl3) δ 153.4, 150.7, 128.9, 122.2, 119.7, 110.7, 89.7, 51.8, 16.6. HRMS (M+H)+ Calculated for C12H12N2 : 185.1073 and found: 185.1060.
(S,E)-2-(4-(azetidin-1-yl)styryl)-4,5-dihydrothiazole-4-carboxylic acid (3)
D-cysteine hydrochloride (73.2 mg, 0.604 mmol) was dissolved in 15 mL of 50 mM aqueous sodium phosphate buffer, pH 8, and degassed under argon. This was added to a solution of 2 (52 mg, 0.302 mmol) in 15 mL of degassed DMF and stirred at 100 °C for 24 h. The reaction was then cooled to room temperature, diluted with sodium phosphate buffer and extracted with ethyl acetate. The aqueous phase was acidified to pH 4 with 1M HCl and extracted with ethyl acetate (2 × 50 mL). The combined organic layers were dried over Na2SO4 and the solvent was removed by rotary evaporation. The crude was purified using flash chromatography (10% MeOH/DCM) to afford 3 (23.5 mg, 27%). 1H-NMR (500 MHz, CD3OD): δ 7.47 (d, J = 8.6 Hz, 2H), 7.34 (d, J = 15.7 Hz, 1H), 6.89 (d, J = 15.8 Hz, 1H), 6.44 (d, J = 8.5 Hz, 2H), 5.11 (t, J = 8.4 Hz, 1H), 4.00 (t, J = 7.3 Hz, 4H), 3.80 – 3.65 (m, 2H), 2.53 – 2.35 (m, 2H). 13C NMR (125 MHz, CD3OD) δ 176.3, 172.5, 153.8, 147.5, 130.1, 122.7, 112.1, 110.6, 73.4, 51.4, 33.9, 16.0. HRMS (M+H)+ Calculated for C15H17N2O2S: 289.1005 and found: 289.0994.
(E)-3-(4-(pyrrolidin-1-yl)phenyl)acrylonitrile (4)
To a suspension of 95% sodium hydride (130.0 mg, 3.23 mmol) in THF (9.30 mL) at 0 °C was added diethyl cyanomethylphosphonate (0.523 mL, 3.23 mmol). Stirring for 30 minutes yielded a clear solution. To this solution, 4-(pyrrolidin-1-yl)benzaldehyde (325.6 mg, 1.86 mmol) in THF (2.0 mL) was added dropwise. The reaction was stirred for 2 hours, then quenched with water (20 mL) and extracted with ethyl acetate (2 × 25 mL). The combined organic layers were washed with brine, dried over Na2SO4 and the solvent was removed by rotary evaporation. The resulting residue was purified by flash chromatography (10% ethyl acetate/hexanes) to give 4 (224 mg, 60.7%) as a light green solid. 1H NMR (500 MHz, CDCl3): δ 7.31 (d, J = 8.8 Hz, 2H), 7.26 (d, J = 16.4 Hz, 1H), 6.53 (d, J = 8.7 Hz, 2H), 5.56 (d, J = 16.4 Hz, 1H), 3.36 (app t, J = 6.5 Hz, 4H), 2.16 – 1.91 (m, 4H).13C NMR (125 MHz, CDCl3): δ 150.8, 149.7, 129.0, 120.9, 120.0, 111.6, 88.5, 47.6, 25.5. HRMS (M+H)+ Calculated for C13H14N2: 199.1230 and found: 199.1213.
(S,E)-2-(4-(pyrrolidin-1-yl)styryl)-4,5-dihydrothiazole-4-carboxylic acid (5)
D-cysteine (43.0 mg, 0.353 mmol) and nitrile 4 (46.5 mg, 0.235 mmol) were added to ethanol (2 mL). This solution was degassed under argon for 5 minutes and then heated to 70 °C for 24h. The reaction was then cooled to room temperature, evaporated, and diluted with water (5 mL). Extraction with ethyl acetate (2 × 5 mL) was performed to remove the unreacted nitrile, then the aqueous layer was acidified to pH 4 with 1M HCl to precipitate the product. HPLC purification (10-90% acetonitrile/water) yielded 5 as a dark brown solid (20.6 mg, 0.068 mmol) in 29% yield. 1H NMR (500 MHz, CD3OD) δ 7.29 (d, J = 8.7 Hz, 2H), 6.95 (d, J = 16.0 Hz, 1H), 6.75 (d, J = 16.0 Hz, 1H), 6.46 (d, J = 8.8 Hz, 2H), 4.85 (t, J = 9.1 Hz, 1H), 3.47 (dd, J = 10.7, 9.1 Hz, 1H), 3.37 (dd, J = 10.7, 9.3 Hz, 1H), 3.28 – 3.15 (m, 4H), 1.97 – 1.89 (m, 4H). 13C NMR (125 MHz, CD3OD) δ 177.0, 170.0, 149.2, 142.6, 128.8, 122.3, 115.9, 111.3, 80.4, 47.1, 35.3, 25.0. HRMS (M+H)+ Calculated for C16H18N2O2S: 303.1162 and found: 303.1145.
Supplementary Material
Acknowledgments
This work was supported by the US National Institutes of Health (R01EB013270 and R21EB020243).
Footnotes
Supporting Information
Supporting methods, figures, scheme, and NMR spectra (PDF). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Adams ST, Miller SC. Beyond D-luciferin: expanding the scope of bioluminescence imaging in vivo. Curr Opin Chem Biol. 2014;21:112–120. doi: 10.1016/j.cbpa.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jathoul AP, Grounds H, Anderson JC, Pule MA. A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging. Angew Chem Int Ed Engl. 2014;53:13059–13063. doi: 10.1002/anie.201405955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mofford DM, Reddy GR, Miller SC. Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over D-luciferin. J Am Chem Soc. 2014;136:13277–13282. doi: 10.1021/ja505795s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinhardt RC, O’Neill JM, Rathbun CM, McCutcheon DC, Paley MA, Prescher JA. Design and Synthesis of an Alkynyl Luciferin Analogue for Bioluminescence Imaging. Chem Eur J. 2016;22:3671–3675. doi: 10.1002/chem.201503944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takakura H, Kojima R, Kamiya M, Kobayashi E, Komatsu T, Ueno T, Terai T, Hanaoka K, Nagano T, Urano Y. New class of bioluminogenic probe based on bioluminescent enzyme-induced electron transfer: BioLeT. J Am Chem Soc. 2015;137:4010–4013. doi: 10.1021/ja511014w. [DOI] [PubMed] [Google Scholar]
- 6.Oba Y, Ojika M, Inouye S. Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 2003;540:251–254. doi: 10.1016/s0014-5793(03)00272-2. [DOI] [PubMed] [Google Scholar]
- 7.Oba Y, Ojika M, Inouye S. Characterization of CG6178 gene product with high sequence similarity to firefly luciferase in Drosophila melanogaster. Gene. 2004;329:137–145. doi: 10.1016/j.gene.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Mofford DM, Reddy GR, Miller SC. Latent luciferase activity in the fruit fly revealed by a synthetic luciferin. Proc Natl Acad Sci U S A. 2014;111:4443–4448. doi: 10.1073/pnas.1319300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branchini BR, Behney CE, Southworth TL, Fontaine DM, Gulick AM, Vinyard DJ, Brudvig GW. Experimental Support for a Single Electron-Transfer Oxidation Mechanism in Firefly Bioluminescence. J Am Chem Soc. 2015;137:7592–7595. doi: 10.1021/jacs.5b03820. [DOI] [PubMed] [Google Scholar]
- 10.Iwano S, Obata R, Miura C, Kiyama M, Hama K, Nakamura M, Amano Y, Kojima S, Hirano T, Maki S, Niwa H. Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron. 2013;69:3847–3856. [Google Scholar]
- 11.Oba Y, Iida K, Ojika M, Inouye S. Orthologous gene of beetle luciferase in non-luminous click beetle, Agrypnus binodulus (Elateridae), encodes a fatty acyl-CoA synthetase. Gene. 2008;407:169–175. doi: 10.1016/j.gene.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Oba Y, Kumazaki M, Inouye S. Characterization of luciferases and its paralogue in the Panamanian luminous click beetle Pyrophorus angustus: A click beetle luciferase lacks the fatty acyl-CoA synthetic activity. Gene. 2010;452:1–6. doi: 10.1016/j.gene.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Fraga H. Firefly luminescence: a historical perspective and recent developments. Photochem Photobiol Sci. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 14.Oba Y, Iida K, Inouye S. Functional conversion of fatty acyl-CoA synthetase to firefly luciferase by site-directed mutagenesis: A key substitution responsible for luminescence activity. FEBS Lett. 2009;583:2004–2008. doi: 10.1016/j.febslet.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM. A neo-substrate that amplifies catalytic activity of parkinson’s-disease-related kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novotny CJ, Hamilton GL, McCormick F, Shokat KM. Farnesyltransferase-Mediated Delivery of a Covalent Inhibitor Overcomes Alternative Prenylation to Mislocalize K-Ras. ACS Chem Biol. 2017;12:1956–1962. doi: 10.1021/acschembio.7b00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma DK, Adams ST, Jr, Liebmann KL, Miller SC. Rapid Access to a Broad Range of 6′-Substituted Firefly Luciferin Analogues Reveals Surprising Emitters and Inhibitors. Org Lett. 2017 doi: 10.1021/acs.orglett.7b02806. DOI: 10.1021/acs.orglett.7b02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsarkova AS, Kaskova ZM, Yampolsky IV. A Tale Of Two Luciferins: Fungal and Earthworm New Bioluminescent Systems. Acc Chem Res. 2016;49:2372–2380. doi: 10.1021/acs.accounts.6b00322. [DOI] [PubMed] [Google Scholar]
- 19.Reddy GR, Thompson WC, Miller SC. Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J Am Chem Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. N-Alkylated 6′-aminoluciferins are bioluminescent substrates for Ultra-Glo and QuantiLum luciferase: new potential scaffolds for bioluminescent assays. Biochemistry. 2008;47:10383–10393. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


