Abstract
VP22 is a major tegument protein of equine herpesvirus type 1 (EHV-1). In the present study, we examined functions of VP22 in EHV-1 replication by viral protein expression analyses in cells infected with the VP22-deficient virus. The expressions of several viral proteins in the cells infected with the VP22-deficient virus were lower than those in the cells infected with the parent virus. One of the weakly expressed proteins was identified as ICP4, which is a major regulatory protein encoded by an immediate early gene of EHV-1. A real-time PCR analysis showed that the mRNA expression of ICP4 was the same in cells infected with the parent and VP22-deficient viruses. Hence, VP22 appears to promote synthesis of ICP4 post-transcriptionally.
Keywords: EHV-1, ICP4, VP22
Equine herpesvirus type 1 (EHV-1), a member of the subfamily Alphaherpesvirinae, genus Varicellovirus, causes abortion in pregnant mares, respiratory infection in young horses and neurological diseases in horses of all ages [16]. EHV-1 virion is composed of four concentric compartments: the core containing a linear double-stranded DNA, the capsid, the tegument, and the envelope. The tegument proteins, which are encoded by about 20 viral genes, comprise an amorphous region located between the capsid and the envelope [20]. In the present study, we focused on EHV-1 tegument protein VP22 (EVP22) which is composed of 304 amino acids (aa) encoded by ORF11 [1], because of its high abundance in the virion and high conservation among alphaherpesvirinae including herpes simplex virus type 1 (HSV-1), pseudorabies virus (PRV), and bovine herpesvirus type 1 (BHV-1) [10]. VP22 homologs are involved in viral replication or viral pathogenisity. For example, the VP22s of Marek’s disease virus serotype 1 (MDV-1) and varicella-zoster virus (VZV) are essential for viral replication in cell culture [2, 3]. On the other hand, BVP22 and HVP22 are not essential for viral replication in cell culture, but they were shown to increase the pathogenicity in a natural host (BVP22) [14, 15] and in an animal model (HVP22) [4, 19], respectively. In addition, we previously reported that EVP22 is not essential for viral growth but required for efficient viral growth in cultured cells [18]. In the previous study, we indicated that the growth of VP22 deletion EHV-1 were significantly lower than those of parent and revertant virus, which suggests that EVP22 is required for efficient viral growth in cultured cells [18]. The tegument protein encoded by ORF52 in murid herpesvirus 68 (subfamily Gammaherpesvirinae) genome is a structural homologue of HSV-1 VP22 (HVP22) [7]. These reports raise the possibility that VP22 proteins in alphaherpesviruses and VP22-like tegument protein in gammaherpesviruses have roles in viral replication.
It is known that the defect of HVP22 results in shutoff of protein synthesis at late stages in infection of HSV-1 [4, 17, 21, 22]. An HVP22 null mutant increased the amount of 80S ribosomes, suggesting that HVP22 is associated with the translation elongation [17]. However, it remains unclear whether VP22s of other herpesviruses including EHV-1 promote protein synthesis.
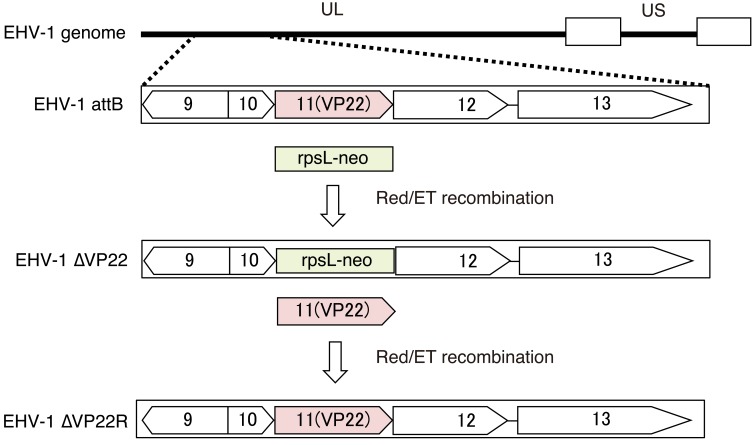
In the present study, we aimed to examine whether the defect of EVP22 affects protein synthesis using an EVP22-deficient virus (EHV-1∆VP22), its parent virus (EHV-1 attB), and EHV-1∆VP22 revertant virus (EHV-1∆VP22R), which were constructed from pAb4p BAC previously (Fig. 1) [9, 18]. To choose the viruses which excised the BAC sequence from viral genome, recombinant viruses were purified by three rounds of plaque purification using GFP fluorescence as a marker. Purified viruses were grown in primary cells originating from fetal equine kidney. The titers of stock viruses were determined by plaque assay using Madin-Darby bovine kidney (MDBK) cells.
Fig. 1.
A schematic diagram of the genome structure of parent virus and mutant viruses [18].
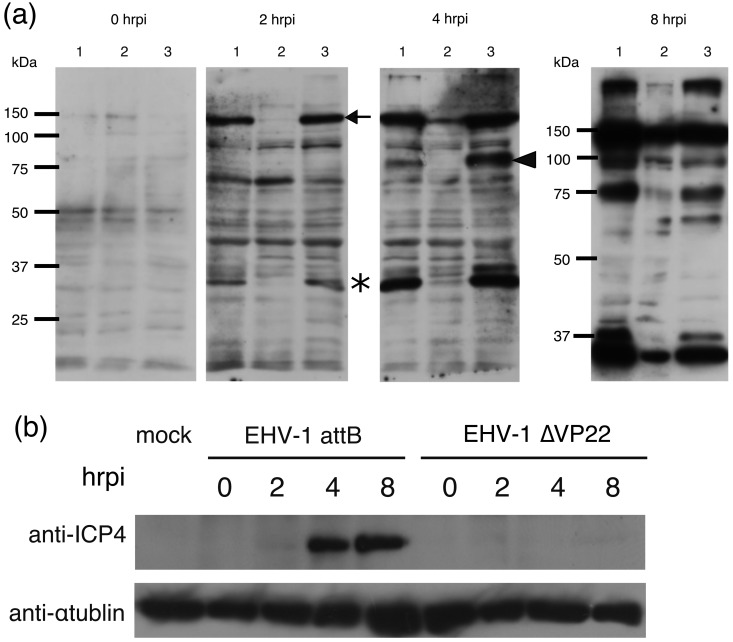
At first, we compared viral protein expression levels between EHV-1 attB-, EHV-1∆VP22- and EHV-1∆VP22R-infected cells to reveal whether EVP22 was involved in viral protein synthesis. MDBK cells grown in Minimum Essential Medium α (MEMα) (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) supplemented with 3% fetal bovine serum (FBS) were seeded in a 6-well plate and inoculated with EHV-1 attB, EHV-1∆VP22 and EHV-1∆VP22R at a multiplicity of infection (MOI) of 3 plaque-forming unit (PFU)/cell, respectively. After 1 hr adsorption, cells were washed three times with MEM and incubated at 37°C in a 5% CO2 atmosphere in 2 ml/well of MEM, and this time point was defined as 0 hrpi. At 0, 2, 4 and 8 hr post infection (hrpi), the cells were washed with phosphate-buffered saline (PBS, pH7.1–7.7) and collected in SDS sample buffer. The lysates were separated by SDS polyacrylamide gel, transferred to a PVDF membrane and reacted with a rabbit antiserum raised against EHV-1 virion at 1:1,000 (kindly provided by Dr. Kirisawa, Rakuno Gakuen University, Sapporo, Japan) [12]. The bound antibodies were detected by anti-rabbit IgG-HRP (horseradish peroxidase) (ICN/Cappel, Cincinnati, OH, U.S.A.) and visualized with enhanced ECL Western Blotting Detection Reagents (GE Healthcare, Uppsala, Sweden). No obvious difference was observed among EHV-1 attB-, EHV-1∆VP22- and EHV-1∆VP22R-infected cells at 0 hrpi (Fig. 2a). However, at 2 hrpi, two bands with molecular mass at ca. 150 kDa (arrow) and 40 kDa (asterisk) were detected in EHV-1 attB-infected cells and EHV-1∆VP22R-infected cells but not in EHV-1∆VP22-infected cells (Fig. 2a). Furthermore, at 4 hrpi, we found another band at ca. 90 kDa (arrowhead) that was expressed in EHV-1 attB-infected cells and EHV-1∆VP22R-infected cells but not in EHV-1∆VP22-infected cells (Fig. 2a). ICP4 was reported to be expressed during the early stage of EHV-1 infection [6] at a molecular mass ca. 150 kDa protein, suggesting that the 150 kDa protein was ICP4.
Fig. 2.
Protein expression comparison by Western blotting using anti-EHV-1 antibody (a). MDBK cells were infected with EHV-1 attB (lane 1), EHV-1∆VP22 (lane 2) and EHV-1∆VP22R (lane 3) at an MOI of 3 PFU/cell and analyzed by Western blotting with antibody to EHV-1 virions at 0, 2, 4 and 8 hrpi. Molecular sizes are shown on the left (a). Expression of ICP4s of EHV-1 attB, EHV-1∆VP22 and EHV-1∆VP22R were analyzed by Western blotting using anti-ICP4 antibody (b).
Herpesvirus gene expression has a classical regulatory cascade conserved across the herpesvirus family [8, 13]. There are four types of genes: 1) immediate-early (IE) genes that do not require any additional protein synthesis for their expression, 2) early (E) genes whose transcription is promoted by the IE gene products and totally independent of viral DNA synthesis, 3) leaky-late (L1) genes whose expression is promoted by some of the E gene products and augmented by viral DNA synthesis, and 4) true-late (L2) genes whose expression is also promoted by some of the E gene products and totally dependent on viral DNA synthesis [8, 13]. ORF64 encoding ICP4 (1,487 aa) lies inside two inverted repeats of the short component in the EHV-1 genome [6]. ORF64 is the only IE gene of EHV-1 [6]. ICP4 plays a crucial role as a transcription activator for early genes [11, 23, 24].
We prepared anti-ICP4 antibodies to detect ICP4 as follows. A DNA fragment corresponding to 46–351 aa of ICP4 was amplified by PCR and ligated into pGEX-6P-1 (GE Healthcare) (pGST-ICP4). Escherichia coli BL21 cells were transformed with pGST-ICP4 DNA. Protein expression was induced by adding 0.1 mM IPTG (isopropyl-β-D-1-thiogalactopyranoside, GIBCO, Life Technologies, Grand Island, NY, U.S.A.). The GST-ICP4 fusion protein was purified using glutathione-Sepharose 4B beads (GE Healthcare) from the bacterial lysate. Specific pathogen free male BALB/c mice (SLC, Hamamatsu, Japan) were immunized with the fusion protein in TiterMax Gold (TiterMax U.S.A., Inc., Narcross, GA, U.S.A.). The animal experiments were certified and conducted under the guidelines of the Animal Care and Use Committee of Gifu University (certification number 14095). The same samples used in the Fig. 1a were analyzed with the anti-ICP4 antibodies (1:5,000). ICP4 in EHV-1 attB-infected cells was detected from 4 hrpi. On the other hand, ICP4 in EHV-1 ∆VP22-infected cells was not detected during the time examined (0–8 hrpi) (Fig. 2b). These data indicated that the band, which was not detected in EHV-1∆VP22 at 2 hrpi in the Fig. 1a, is ICP4 and that expression level of ICP4 was decreased to be marginally detectable.
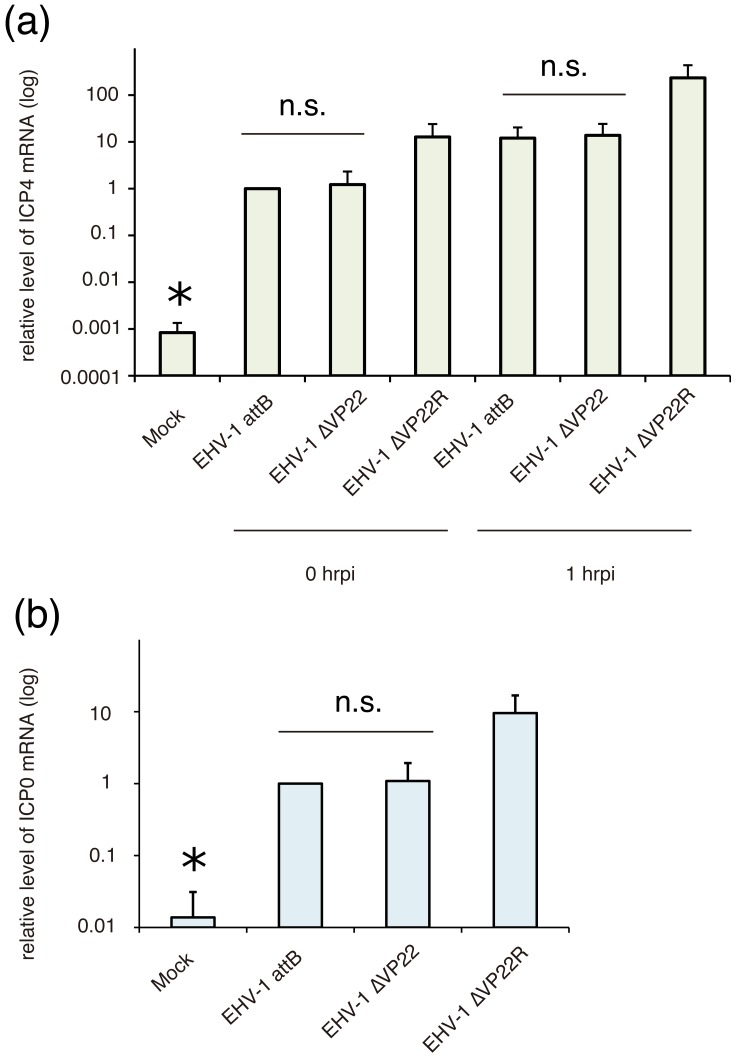
Next, to investigate the function of EVP22 on transcription and/or translation in the EHV-1 replication, we quantified the transcription levels of ICP4 mRNA. MDBK cells were inoculated with EHV-1 attB, EHV-1∆VP22 and EHV-1∆VP22R at an MOI of 3 PFU/cell. After 1 hr adsorption, cells were washed three times with MEM and incubated at 37°C in a 5% CO2 atmosphere in 1 ml/well of MEM, and this time point was defined as 0 hrpi. Total RNA and DNA of infected cells were extracted using AllPrep DNA/RNA Mini Kit (QIAGEN K.K., Hilden, Germany) at 0 and 1 hrpi. cDNA was synthesized from 0.5 µg of total RNA using ReverTra Ace (TOYOBO, Osaka, Japan). The real-time PCR assay was carried out using SYBR Premix Ex Taq II (Takara Bio Inc., Otsu, Japan) according to the manufacturer’s instructions. GAPDH (Glyceraldehyde 3-phosphate dehydrogenase) gene was used as a housekeeping gene. Primer pairs used for quantitative realtime PCR were listed in Table 1. The PCR was carried out with using a following program: 1 cycle of 95°C for 5 min, 45 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, and 72°C for 3 min. The fluorescence was measured at the end of each cycle. The copy number of ICP4 mRNA was calculated by absolute quantification normalized by the GAPDH mRNA and genome DNA. Relative level of ICP4 mRNA of mock sample was 0.0008, and we concluded this value was below the threshold. The transcription levels of ICP4 mRNA of EHV-1 attB and EHV-1∆VP22 were not significantly different at both 0 and 1 hrpi (Fig. 3a). In addition, we investigated transcription levels of an early gene, ICP0, to evaluate whether reduced expression level of ICP4 affects the transcription of early genes of EHV-1. The result of quantitative real-time PCR showed that transcription level of ICP0 mRNA was not different between EHV-1 attB and EHV-1∆VP22 (Fig. 3b). Relative level of ICP0 mRNA of mock sample was 0.014, and we concluded this value was below the threshold. The ICP4 and ICP0 mRNA transcription levels of EHV-1∆VP22R were about 10-fold higher than those of EHV-1 attB and EHV-1∆VP22. These data suggested that deletion of VP22 did not affect the transcription level of ICP4 mRNA but affected the translation of ICP4 mRNA and that ICP4 could act as the transactivator for early genes even though its protein expression level was marginally detectable. VP16, which is encoded by OFR12 gene and locate next to ORF11, has been known as transactivator of ORF64 encoding ICP4 [5]. We speculate whether decreased expression level of ICP4 might depend on decreased expression level of VP16. However, as shown in Fig. 3, mRNA levels of ICP4 were not different between EHV-1 attB and EHV-1∆VP22. Because there is no report that VP16 is involved in protein expression of ICP4, we conclude that decreased expression levels of ICP4 would depend on ORF11 deletion, not ORF12 expression level.
Table 1. PCR primers used in this study.
| Primers | Sequence (5′-3′) | Purpose |
|---|---|---|
| pGST-ICP4-F | CGGGATCCCTGTACGGCAGCCAGAACATG | pGEX-ICP4 |
| pGST-ICP4-R | CGGAATTCCTTGCGCTTCGACGCTCC | |
| rICP4-F | TCTTGCCCGCTGAGCTTAGA | Real-time PCR |
| rICP4-R | TGTGGGACGACCCCGAGATA | |
| rICP0-F | CCGGTATCTTTGGCAGTTGT | Real-time PCR |
| rICP0-R | CAATGTGCTTCTGAGACCGA | |
| GAPDH-F | CTCTTTTCCAGCAATGCCTC | Real-time PCR |
| GAPDH-R | TGTGTTCAAGTTTCCCACCA | |
Fig. 3.
mRNA levels of ICP4 (a) and ICP0 (b) in EHV-1 attB-, EHV-1∆VP22- and EHV-1∆VP22R-infected cells. MDBK cells were inoculated with EHV-1 attB, EHV-1∆VP22 and EHV-1∆VP22R at an MOI of 3. After 0 and 1 hrpi, total RNA and DNA was extracted. Quantification of mRNA was carried out by real-time quantitative RT-PCR. Expression levels of ICP4 and ICP0 mRNA were normalized with the GAPDH mRNA levels and genome DNA, and expressed relatively as the ratio against the mRNA levels in EHV-1 attB-infected cells at 0 hrpi. There are no significant (n.s.) differences between EHV-1 attB and EHV-1∆VP22. * means “below the threshold”.
Because the ICP4 and ICP0 mRNA transcription levels of EHV-1∆VP22R were different from those of EHV-1 attB, EHV-1∆VP22R might have some mutations in its genome. To confirm that there are no mutations in the EHV-1∆VP22 genome, the complete genome sequence of EHV-1∆VP22 (accession no. LC193725) was determined on an Illumina MiSeq platform according to the manufacturer’s protocols. In the EHV-1∆VP22 genome, ORF11 (encoding EVP22) was confirmed to be replaced with the rpsL-neo cassette gene sequence. One mutation was found in ORF75, in which one nucleotide (adenine) was inserted and caused a frameshift mutation. This mutation was also found in EHV-1 attB, which was used as a parent strain of the recombinant viruses in our laboratory. Therefore, we concluded that EHV-1∆VP22 had no influential mutation in the coding regions other than the mutation in VP22. Additional mutations might exist in EHV-1∆VP22R. Taken together, we concluded that EVP22 was not involved in the transcription of ICP4 mRNA at the IE stage of EHV-1 infection. However, further investigation using another revertant viruses might be required to evaluate whether EVP22 is strictly involved in transcription.
In the present study, we aimed to examine whether EVP22 is involved in promotion of viral protein expression. The present results reveal a novel role of EVP22: EVP22, like HVP22, increases viral protein expression post-transcriptionally. There is possibility that the lack of VP22 causes significant damages on virus productive cycle because of its high abundance in virions. Therefore, it is not certain whether EVP22 increases viral protein expression post-transcriptionally by direct or indirect mechanisms. Importantly, we demonstrated that EVP22 is involved in protein expression in the immediate early stage, ie, ICP4 expression, while HVP22 promote protein synthesis in the late stage of infection [4]. It is the first evidence that EVP22 promotes protein expression at the immediate early stage of infection, which is unique to EHV-1. EVP22 might be required for not only the expression of ICP4 but also the expression of other some viral proteins selectively by unknown mechanism. It might be related with the deficient growth ability of EHV-1∆VP22 as described previously [18]. Further experiments are needed to clarify the detail mechanisms how VP22 regulates translation of the EHV-1 genome.
Acknowledgments
We thank Dr. Rikio Kirisawa, Rakuno Gakuen University, Japan for kindly providing anti-EHV-1 antibody. This work was supported by JSPS KAKENHI Grant Number 24380165.
REFERENCES
- 1.Barnett B. C., Dolan A., Telford E. A., Davison A. J., McGeoch D. J.1992. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J. Gen. Virol. 73: 2167–2171. doi: 10.1099/0022-1317-73-8-2167 [DOI] [PubMed] [Google Scholar]
- 2.Che X., Reichelt M., Sommer M. H., Rajamani J., Zerboni L., Arvin A. M.2008. Functions of the ORF9-to-ORF12 gene cluster in varicella-zoster virus replication and in the pathogenesis of skin infection. J. Virol. 82: 5825–5834. doi: 10.1128/JVI.00303-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorange F., Tischer B. K., Vautherot J. F., Osterrieder N.2002. Characterization of Marek’s disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76: 1959–1970. doi: 10.1128/JVI.76.4.1959-1970.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy C., Mbong E. F., Baines J. D.2009. VP22 of herpes simplex virus 1 promotes protein synthesis at late times in infection and accumulation of a subset of viral mRNAs at early times in infection. J. Virol. 83: 1009–1017. doi: 10.1128/JVI.02245-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott G., O’Hare P.1995. Equine herpesvirus 1 gene 12, the functional homologue of herpes simplex virus VP16, transactivates via octamer sequences in the equine herpesvirus IE gene promoter. Virology 213: 258–262. doi: 10.1006/viro.1995.1568 [DOI] [PubMed] [Google Scholar]
- 6.Grundy F. J., Baumann R. P., O’Callaghan D. J.1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172: 223–236. doi: 10.1016/0042-6822(89)90124-4 [DOI] [PubMed] [Google Scholar]
- 7.Hew K., Dahlroth S. L., Pan L. X., Cornvik T., Nordlund P.2015. VP22 core domain from Herpes simplex virus 1 reveals a surprising structural conservation in both the Alpha- and Gammaherpesvirinae subfamilies. J. Gen. Virol. 96: 1436–1445. doi: 10.1099/vir.0.000078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honess R. W., Roizman B.1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasem S., Yu M. H., Yamada S., Kodaira A., Matsumura T., Tsujimura K., Madbouly H., Yamaguchi T., Ohya K., Fukushi H.2010. The ORF37 (UL24) is a neuropathogenicity determinant of equine herpesvirus 1 (EHV-1) in the mouse encephalitis model. Virology 400: 259–270. doi: 10.1016/j.virol.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 10.Kelly B. J., Fraefel C., Cunningham A. L., Diefenbach R. J.2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145: 173–186. doi: 10.1016/j.virusres.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 11.Kim S. K., Shakya A. K., O’Callaghan D. J.2016. Full trans-activation mediated by the immediate-early protein of equine herpesvirus 1 requires a consensus TATA box, but not its cognate binding sequence. Virus Res. 211: 222–232. doi: 10.1016/j.virusres.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirisawa R., Kobayashi T., Uematsu R., Ikeda A., Kuroiwa R., Urakami A., Iwai H.2003. Growth of recombinant equine herpesvirus 1 (EHV-1) replaced with passage-induced mutant gene 1 and gene 71 derived from an attenuated EHV-1 in cell cultures and in the lungs of mice. Vet. Microbiol. 95: 159–174. doi: 10.1016/S0378-1135(03)00154-8 [DOI] [PubMed] [Google Scholar]
- 13.Kozak M., Roizman B.1974. Regulation of herpesvirus macromolecular synthesis: nuclear retention of nontranslated viral RNA sequences. Proc. Natl. Acad. Sci. U.S.A. 71: 4322–4326. doi: 10.1073/pnas.71.11.4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X., Chow B., Babiuk L. A.1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15: 1057–1064. doi: 10.1016/S0264-410X(97)00008-X [DOI] [PubMed] [Google Scholar]
- 15.Liang X., Chow B., Li Y., Raggo C., Yoo D., Attah-Poku S., Babiuk L. A.1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69: 3863–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lunn D. P., Davis-Poynter N., Flaminio M. J., Horohov D. W., Osterrieder K., Pusterla N., Townsend H. G.2009. Equine herpesvirus-1 consensus statement. J. Vet. Intern. Med. 23: 450–461. doi: 10.1111/j.1939-1676.2009.0304.x [DOI] [PubMed] [Google Scholar]
- 17.Mbong E. F., Woodley L., Dunkerley E., Schrimpf J. E., Morrison L. A., Duffy C.2012. Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J. Virol. 86: 12351–12361. doi: 10.1128/JVI.01975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada A., Izume S., Ohya K., Fukushi H.2015. Equine herpesvirus type 1 tegument protein VP22 is not essential for pathogenicity in a hamster model, but is required for efficient viral growth in cultured cells. J. Vet. Med. Sci. 77: 1293–1297. doi: 10.1292/jvms.14-0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pomeranz L. E., Blaho J. A.2000. Assembly of infectious Herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74: 10041–10054. doi: 10.1128/JVI.74.21.10041-10054.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roizmann B., Desrosiers R. C., Fleckenstein B., Lopez C., Minson A. C., Studdert M. J., The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses1992. The family Herpesviridae: an update. Arch. Virol. 123: 425–449. doi: 10.1007/BF01317276 [DOI] [PubMed] [Google Scholar]
- 21.Sciortino M. T., Taddeo B., Poon A. P., Mastino A., Roizman B.2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. U.S.A. 99: 8318–8323. doi: 10.1073/pnas.122231699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sciortino M. T., Taddeo B., Giuffrè-Cuculletto M., Medici M. A., Mastino A., Roizman B.2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 81: 10924–10932. doi: 10.1128/JVI.01239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith R. H., Caughman G. B., O’Callaghan D. J.1992. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J. Virol. 66: 936–945. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith R. H., Holden V. R., O’Callaghan D. J.1995. Nuclear localization and transcriptional activation activities of truncated versions of the immediate-early gene product of equine herpesvirus 1. J. Virol. 69: 3857–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]