Abstract
The Human Cytochrome P450 (CYP) Allele Nomenclature Database, a critical resource for the pharmacogenetics and genomics communities, has transitioned to the Pharmacogene Variation (PharmVar) Consortium. In this report we provide a summary of the current database, provide an overview of the PharmVar consortium, and highlight the PharmVar database which will serve as the new home for pharmacogene nomenclature.
THE HUMAN CYTOCHROME P450 (CYP) ALLELE NOMENCLATURE DATABASE
An international group of leading experts in the emerging field of pharmacogenetics (PGx) came together in the mid 1990s to find a systematic way to catalog allelic variants of cytochrome P450 2D6 (CYP2D6).1 The proposed star (*) nomenclature system provided the field with a “common language” to name a growing number of variants being discovered. Anticipating an explosion of single nucleotide polymorphism (SNP) discovery for other genes in the CYP superfamily involved in the metabolism of xenobiotics, including many clinically used drugs, Ingelman‐Sundberg et al. proposed to apply the general nomenclature rules used for CYP2D6 to all other polymorphic CYP genes and eventually also cytochrome P450 oxidoreductase (POR).2 In 2002, the Human Cytochrome P450 (CYP) Allele Nomenclature Database (“Nomenclature website” for short) and Committee was established to systematically classify an increasing number of CYP allelic variants.3 The star (*) nomenclature was readily embraced by the pharmacogenetics field and the Nomenclature website was swiftly accepted as the central hub for CYP allele definition. The Nomenclature website has been maintained at the Karolinska Institutet under the leadership of Magnus Ingelman‐Sundberg since its inception at http://www.cypalleles.ki.se/.4 Being the leading authority for P450 nomenclature, PGx experts rely heavily on the catalog of haplotype definitions listed on the Nomenclature website for basic and translational research as well as many clinical applications including genotype test reporting. One prominent example is the Clinical Pharmacogenetic Implementation Consortium (CPIC), which facilitates the use of pharmacogenetic tests for patient care by creating freely available evidence‐based gene/drug clinical practice guidelines (https://cpicpgx.org/). The CPIC utilizes allele designations as defined by the Nomenclature website. The Pharmacogenomics Knowledge Base (PharmGKB; https://www.pharmgkb.org), a leading pharmacogenomics knowledge resource, also relies on the Nomenclature website for CYP allele definitions. Standardized allele definitions are also integral to the development of bioinformatic tools to automate the assignment of star allele genotypes from multiple genotyping platforms, including panels, array platforms, and Next‐Generation Sequencing. Examples include the Pharmacogenomic Clinical Annotation Tool (PharmCAT), which takes genomic data to make haplotype calls and get CPIC annotations (https://www.pharmgkb.org/page/pharmcat); Astrolabe, a tool developed to assign CYP2D6 genotype from Whole Genome Sequence data (currently expanded to CYPs 2C9 and 19)5; or tools that are integrated into commercial array analysis algorithms, such as the DMET and PharmacoScan Console Software packages.
Needless to say, the Nomenclature website has become an integral part of pharmacogenetics over the years as it has established itself as the authority for issuing and maintaining allele designations.
After dedicating many years of his distinguished career to developing and maintaining the CYP Allele Nomenclature resource together with Sarah Sim, who served as the Webmaster for 12 years, both Magnus Ingelman‐Sundberg and Sarah Sim have retired from the nomenclature site to focus on other projects. This resource has provided an invaluable service to the drug metabolism and PGx communities, and would be a critical loss to these communities should it cease to exist. Therefore, a group of stakeholders came together to develop a plan to maintain and sustain what was unanimously agreed on to be an essential resource. By the same token, the group also identified a number of limitations and opportunities to raise pharmacogene variation to a new level. To that end, the PharmVar Steering Committee consisting of 19 members representing academia, clinical, and commercial interests was assembled to discuss a path forward. Over the past year a series of teleconferences were organized under the leadership of Andrea Gaedigk, Steve Leeder, and Teri Klein, and in May of 2017 a Face‐to‐Face meeting was held at Children's Mercy Kansas City to discuss “must‐have” features of a future database, revisions to the nomenclature system itself, a standardized and transparent submission process and allele designation. Furthermore, mechanisms for financial support were discussed, including seeking National Institutes of Health (NIH) funding to support the development of a “Next‐Generation” pharmacogene database operated through a consortium model, the Pharmacogene Variation (PharmVar) Consortium.
THE PHARMACOGENE VARIATION (PHARMVAR) CONSORTIUM
The PharmVar Consortium will serve as the new hub for pharmacogene nomenclature. As a first step, the Human Cytochrome P450 (CYP) Allele Nomenclature Database has transitioned to Children's Mercy Kansas City from its current location at the Karolinska Institutet in Sweden, on September 26, 2017 (Figure 1). The PharmVar database will be an interactive state‐of‐the art database and serve as a centralized “Next‐Generation” Pharmacogene Variation data repository. The inaugural version, to be launched in early 2018, will be kick‐started with three high‐priority genes, CYP2C9, CYP2C19, and CYP2D6. Over the course of the first year of the project all other CYP genes that have been housed by the Nomenclature website will be transferred into the PharmVar database. Once a gene is transferred, it will no longer be updated on the Nomenclature website pages (a link through PharmVar will allow continued access to a “legacy” version). As the PharmVar database grows, other PGx genes will be added to the PharmVar database in the future. Of particular interest will be clinically actionable genes such as DPYD and SLCO1B1, for which the CPIC is publishing dosing guidelines and for which no systematic nomenclature exists. PharmVar will not be limited to genes involved in drug metabolism, and plans to eventually also house genes contributing to drug transport and response.
Figure 1.

The Human Cytochrome P450 (CYP) Allele Nomenclature Database has transitioned to the Pharmacogene Variation Consortium (PharmVar). PharmVar will closely work with the PharmGKB for data curation and dissemination.
Figure 2.
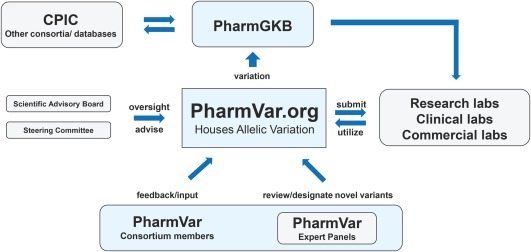
Overview of the Pharmacogene Variation Consortium.
The PharmVar Consortium, operating as a Pharmacogenetic Research Network (http://www.pgrn.org/pharmvar.html) resource, invites experts with an interest in PGx variation to become a PharmVar community member. The PharmVar database will accept submissions from research, clinical, and commercial laboratories following a standardized submission protocol and criteria (forms are available at www.PharmVar.org).
In addition to a transparent submission and review process, the PharmVar database will assign unique version numbers to haplotype definitions, genes, and database releases to enable robust tracking of nomenclature data and simplifying the task of determining the exact allele definitions used in a particular analysis. All variants will consistently be linked to rs numbers/dbSNP identifiers, if available, and the user will be able to list all alleles carrying a particular SNP of interest. The database will also include a number of other functional features, including the generation of customizable tables of genes or gene regions of interest, sequence alignments, and have the option of downloading variant sequences, among other features. Information regarding the functional consequences of allelic variation will be added in the future and synchronized with PharmGKB and CPIC annotations through direct communication with these collaborating groups. Allelic variants will be mapped to a gene's most recent RefSeqGene sequence, to its Locus Reference Genomic sequence (LRG, a stable genomic framework that never changes), and to the human Reference Genome Builds GRCh37/hg19 and GRCh38/hg38 for easy referencing across numbering systems. For many of the CYP genes listed on the Nomenclature website, the reference sequence used to define allelic variation does not match the RefSeqGene. Among those are CYP2D6, which is annotated using M33388 and not the NG_008376.3 RefSeq, or CYP2B6 and CYP3A5, which are mapped to RefSeq records that have been removed by the NCBI. In these cases, legacy numbering will be provided to facilitate cross‐referencing. Of note, some allele definitions will require revision. For instance, for CYP2D6 SNP coordinates past + 600 will shift due to a 1‐bp gain and two 1‐bp losses distinguishing M33388 from the gene's NG_008376.3 RefSeq. Accordingly, the SNP defining CYP2D6*4 will be shown as 1847G>A when mapped to NG_008376.3. Although these revisions may require updating on, e.g., test reports, standardized mapping of allelic variation is a prerequisite to facilitating genomic data analysis, automated pipelines, and standardized reporting.
In addition to being a searchable database, PharmVar will also offer a number of graphical display functions, including comparisons of user‐selected sequences and the assembly of custom tables showing genes or gene regions for SNPs of interest. In addition to content displayed on the website, PharmVar will provide a capability for bulk data downloads as well as an Application Programming Interface (API) to enable programmatic access to database content.
The PharmVar database will be a user‐friendly resource for pharmacogene variation data and directly accessible to all users (requests for allele designations will, however, require a user account). PharmVar will coordinate its efforts with the PharmGKB as well as other stakeholders such as the CPIC to provide unified and standardized pharmacogene variation designation and information regarding translation into phenotype to the PGx community. Coordinated efforts with the PharmGKB will include, for instance, the curation of variants and their incorporation into the annotations on the PharmGKB and deposit variants as part of their regular submissions to ClinVar. The PharmVar Consortium with its rich expertise and close collaborations with the PharmGKB and CPIC is uniquely positioned to contribute to the advancement of genome‐informed medicine and PGx‐guided individualized drug therapy at large.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
ACKNOWLEDGMENTS
PharmVar and PharmGKB are supported by the National Institute of General Medical Sciences Pharmacogenetics 1 R24 GM123930‐01 and R24 GM61374, respectively. Figure 1 was created by Thao P. Do of pgrn.org and used with their permission for this report.
References
- 1. Daly, A.K. et al Nomenclature for human CYP2D6 alleles. Pharmacogenetics 6, 193–201 (1996). [DOI] [PubMed] [Google Scholar]
- 2. Ingelman‐Sundberg, M. , Daly, A.K. , Oscarson, M. & Nebert, D.W. Human cytochrome P450 (CYP) genes: recommendations for the nomenclature of alleles. Pharmacogenetics 10, 91–93 (2000). [DOI] [PubMed] [Google Scholar]
- 3. Oscarson, M. & Ingelman‐Sundberg, M. CYPalleles: A web page for nomenclature of human cytochrome P450 alleles. Drug Metab. Pharmacokinet. 17, 491–495 (2002). [DOI] [PubMed] [Google Scholar]
- 4. Sim, S.C. & Ingelman‐Sundberg, M. Update on allele nomenclature for human cytochromes P450 and the Human Cytochrome P450 Allele (CYP‐Allele) Nomenclature Database. Methods Mol. Biol. 987, 251–259 (2013). [DOI] [PubMed] [Google Scholar]
- 5. Twist, G.P. et al Constellation: A tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole‐genome sequences. NPJ Genomic Med. 2, 16039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


