Abstract
We have employed as ‘gold standards’ two in-house, well-characterised and validated polyclonal antibodies, C9-L and C9-S, which detect the longer and shorter forms of C9orf72, and have compared seven other commercially available antibodies with these in order to evaluate the utility of the latter as credible tools for the demonstration of C9orf72. C9-L and C9-S antibodies immunostained cytoplasmic ‘speckles’, and the nuclear membrane, respectively, in cerebellar Purkinje cells of the cerebellum in patients with behavioural variant frontotemporal dementia (bvFTD) with amyotrophic lateral sclerosis (ALS), and in patients with ALS alone. Similar staining was seen in Purkinje cells in healthy control tissues and in other neurodegenerative disorders, and in pyramidal cells of CA4 and dentate gyrus of hippocampus. However, in the spinal cord there was little cytoplasmic staining with C9-L antibody. C9-S antibody immunostained the nuclear membrane of anterior horn cells in healthy neurons. In patients with bvFTD + ALS, or ALS alone, this C9-S nuclear staining was redistributed to the plasma membrane. In those patients with bvFTD + ALS or ALS bearing an expansion in C9orf72, none of the commercially available antibodies detected TDP-43 inclusions in anterior horn cells, nor were dipeptide repeat proteins demonstrated. Five of the commercial antibodies provided immunohistochemical staining patterns similar in morphological appearance to the in-house C9-L antibody, but distinct from C9-S antibody. However, only three showed sufficient specificity and intensity of staining for C9orf72 at acceptably low concentrations, to make them of practical value and sufficiently reliable for the detection of at least the longer form of C9orf72 protein.
Keywords: C9orf72, frontotemporal lobar degeneration, amyotrophic lateral sclerosis
Introduction
An expanded hexanucleotide (GGGGCC) repeat in a non-coding region of the C9orf72 gene is the most common cause of familial frontemporal lobar degeneration (FTLD), with or without amyotrophic lateral sclerosis (ALS), and of ALS itself, but can also occur in around 7% of apparently sporadic cases (1,2). Normally, less than 20 such repeats are present, but when expanded an excess of 36 repeats is seen, ranging upwards to as many as several thousand repeats. How the expansion in C9orf72 causes disease remains unclear. Three mechanisms have been proposed. None is mutually exclusive, and all could play some part in disease pathogenesis. A loss of function effect (haploinsufficiency) consequent upon a reduced output of C9orf72 protein has been suggested (1,3,4), with the extent of the loss being dependent upon the degree of DNA methylation (5,6). Alternatively, the formation of both sense and antisense nuclear RNA foci has been demonstrated, both in human disease (1,7–9) and in fly models (9). These might sequester RNA transcripts (1,9), or other endogenous RNA binding proteins (7,8), thereby interfering with the transcriptome. Finally, a non-ATG mediated (RAN) sense and antisense translation of the expansion itself leads to accumulation of the dipeptide repeat proteins, poly-GA, poly-GR, poly-GP, poly-PA and poly-PR (10–16), any or all of which might confer neurotoxicity. Nevertheless, how these potential effects translate into the TDP-43 proteinopathy that characterises both conditions remains to be established.
Sequence and structural analysis identifies C9orf72 as a member of the DENN (Differentially Expressed in Normal and Neoplastic cells) family of proteins (17). These are GTP-GDP exchange factors for Rab GTPases, key regulators of membrane trafficking (18,19). Alternative splicing of C9orf72 transcripts yields long and short forms of the translated protein (1). Co-immunoprecipitation studies suggest that both long and short forms of C9orf72 interact with importin-β1 and Ran-GTPase, indicating a role in nucleocytoplasmic transport (4). Loss of C9orf72 protein results in decreased Rab activation and changes in endosomal trafficking, pointing to a role in autophagy regulation (20). Such a conclusion is supported by a physical interaction between C9orf72, especially the longer form, and SMCR8/WDR41, and with the FIP200/Ulk1/ATG13/ATG101 complex, a key regulator of autophagy initiation (21–23). Hence, the expansion in C9orf72 could join other FTLD associated mutations in genes such as VCP (24), SQSTM1 (25), UBQLN2 (26), OPTN (27), TBK1 (28) and CHMP2B (29), whose protein products are well known to participate in the autophagosomal degradation of effete proteins.
Nevertheless, our understanding of disease mechanisms in FTLD and ALS associated with expansions in C9orf72 has been hampered by a lack of validated antibodies to C9orf72 protein. Recently, Xiao et al. reported two validated polyclonal C9orf72 antibodies, raised against long and short forms of C9orf72, termed C9-L and C9-S, respectively (4). In the present study, we have employed these antibodies as ‘gold standards’ of C9orf72 protein immunostaining, and have compared findings from a series of commercially available C9orf72 antibodies with these in order to evaluate the utility of these commercial antibodies for the credible demonstration of C9orf72 proteins within cells and tissues. We have also extended earlier work in C9orf72 expansion carriers and non-carriers, based on cerebellum and spinal cord (4), by investigating the hippocampus dentate gyrus granule cells and CA4 pyramidal cells, brain regions which are prone to disease in C9orf72 expansion bearers (11,13).
Patients and methods
Patients
The study group consisted of 26 patients (Supplementary Table 1). Five had behavioural variant frontotemporal dementia (bvFTD) combined with ALS (bvFTD + ALS), all with FTLD-TDP type B histology (30), three of whom bore an expansion in C9orf72. Eight patients had ALS alone, three bearing an expansion in C9orf72, four had Huntington’s disease, four had Alzheimer’s disease and five5 were healthy controls not known to have suffered from any motor or cognitive difficulties in life (Table 1). All tissues were obtained from the Manchester Brain Bank through appropriate consenting procedures for the collection and use of human brain tissues. Patients with bvFTD + ALS fulfilled Lund-Manchester clinical diagnostic criteria for FTLD (31,32). Patients with ALS fulfilled El Escorial criteria (33). Possession of an expansion in C9orf72 was evidenced by Southern blot and/or repeat primed PCR (11,13).
Table 1.
Mean (± SD) post mortem delay time and age at death in the five pathological groups studied. FTD = frontotemporal dementia; MND = motor neuron Disease.
| Group | M/F | Post mortem delay (h) | Age at death (y) |
|---|---|---|---|
| FTD + MND (n = 5) | 2/3 | 71.4 ± 38.3 | 64.8 ± 5.4 |
| MND (n = 8) | 5/3 | 54.8 ± 25.9 | 55.5 ± 10.2 |
| Huntington’s disease (n = 4) | 1/3 | 46.8 ± 7.9 | 60.8 ± 7.0 |
| Alzheimer’s disease (n = 4) | 1/3 | 93.8 ± 58.7 | 79.8 ± 5.2 |
| Healthy Controls (n = 5) | 5/0 | 80.0 ± 51.1 | 79.4 ± 14.4 |
Histological methods
Nine anti-C9orf72 antibodies were evaluated (Table 2). C9-L and C9-S antibodies were employed as ‘gold standards’ (4) for the characterisation of the other seven antibodies. Four antibodies were commercially obtained from Proteintech (Manchester). One was a polyclonal antibody (22637-1-AP) supplied as three different batches (18450, 20422 and 23058), the numbers of which refer to different rounds of purification produced from same lot of antibody, but affinity purified at different times, or from different rabbits. Nevertheless, the immunogen was the same for all. The other Proteintech antibody was a monoclonal C9orf72 antibody (66140-1-Ig). Further antibodies were obtained from Santa Cruz (sc-138763), GeneTex (GTX119776) and Abgent (AP12982b). These were employed for immunohistochemistry as described previously (11,13), following titration to determine optimal immunostaining (see Table 2).
Table 2.
Antibodies employed in the study.
| Name/Code | Type | Species | Source | Dilution |
|---|---|---|---|---|
| C9-L | polyclonal | rabbit | Xiao et al. 2015 | 1:3000 |
| C9-S | polyclonal | rabbit | Xiao et al. 2015 | 1:3000 |
| 22637-1-AP (18450) | polyclonal | rabbit | Proteintech Group | 1:400 |
| 22637-1-AP (20422) | polyclonal | rabbit | Proteintech Group | 1:2000 |
| 22637-1-AP (23058) | polyclonal | rabbit | Proteintech Group | 1:200 |
| 66140-1-Ig | monoclonal | mouse | Proteintech Group | 1:2000 |
| GTX119776 | polyclonal | rabbit | GeneTex | 1:1000 |
| sc-138763 | polyclonal | rabbit | Santa Cruz | 1:200 |
| AP12928b | polyclonal | rabbit | Abgent | 1:100 |
Paraffin sections were cut at 6 µm from formalin fixed blocks of cerebellum from all cases, and from temporal cortex with hippocampus and lumbar spinal cord from a subset of cases where immunostaining for C9orf72 within the cerebellum had been found to be robust. Antigen unmasking was performed by pressure cooking in citrate buffer (pH 6.0, 10 mM) for 30 min, reaching 120 degrees Celsius and >15 kPa pressure. A subset of sections was immunostained using TE9 buffer for antigen unmasking (4) but this did not result in any observable differences in staining pattern or intensity from those treated with citrate buffer. The presence and degree of staining of C9orf72 immunostained nerve cells was assessed microscopically according to:
0 = no stained neurons in any field.
0.5 = rare/single weakly stained neurons in the entire section.
1 = few, usually weakly stained, neurons in some but not all fields.
2 = a moderate number of mostly well stained neurons in each field.
3 = most cells strongly staining in each field.
4 = all cells strongly staining in every field.
Such assessments were performed on Purkinje cells of the cerebellum, and on granule cells of the dentate gyrus and CA4 pyramidal cells of the hippocampus.
Statistical analysis
Rating data were entered into an Excel spreadsheet and analysed using Statistical Package for Social Sciences (SPSS) software (version 17.0). Patients were stratified according to genetic and pathological subtype for statistical analysis of the effects of mutation or underlying pathology on staining. Comparisons of semi-quantitative scores for the intensity of neuronal C9orf72 immunostaining in Purkinje neurons of the cerebellum were performed using a Kruskal–Wallis test with post-hoc Mann–Whitney test. Comparisons of semi-quantitative scores for the intensity of neuronal C9orf72 immunostaining in Purkinje neurons across the different C9orf72 antibodies were also performed using a Kruskal–Wallis test. Correlation between neuronal staining scores and patient age, or post mortem delay, for each antibody, were made using Spearman’s rank correlation statistic. In all instances, significance levels were set at p < 0.05.
Results
Immunostaining within the cerebellum with C9-L antibody was seen as small cytoplasmic granules within the perikaryon and proximal dendrites of Purkinje cells, conferring a ‘speckled’ appearance to the cells; there was no nuclear membrane staining (Figure 1(a)). By contrast, C9-S antibody immunostained the nuclear membrane, but not cytoplasm, of Purkinje cells (Figure 1(b)). Although there was a weak ‘background’ parenchymal staining within the molecular and granule cell layers with C9-L antibody, there was no staining of cytoplasm or nuclear membrane of granule or basket cells. Notably, dipeptide repeat proteins within cerebellar granule cells were unstained with either C9-L or C9-S antibodies in those patients bearing an expansion in C9orf72 in whom previous work, employing both p62 immunostaining and immunostaining for poly-GA, poly-GP and poly-GR proteins (11,13), had shown these to be abundant.
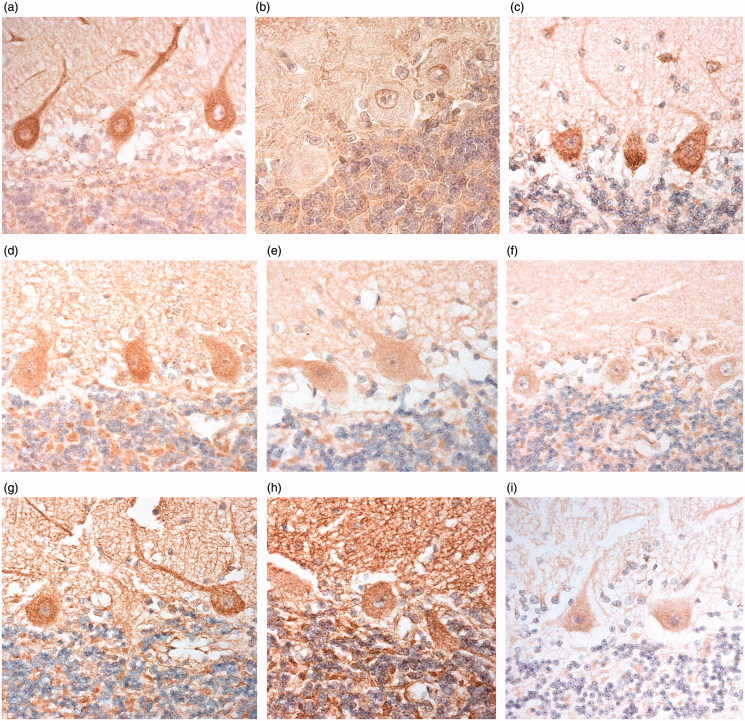
Figure 1.
Immunostaining of Purkinje cells of the cerebellum for C9orf72 protein using C9-L antibody (a), C9-S antibody (b), Proteintech monoclonal antibody (c), Proteintech polyclonal antibody, 20422 (d), GeneTex antibody (e), Proteintech polyclonal antibodies, 18450 (f) and 23058 (g), Santa Cruz antibody (h) and Abgent antibody (i). Immunoperoxidase–haematoxylin, ×400 microscope magnification.
Nonetheless, the intensity of Purkinje cell staining was highly variable between cases ranging from little or no immunostaining of ‘speckles’ (grades 0, 0.5 and 1), through moderate (grade 2), to those with strong immunostaining of many speckles (grades 3 and 4). This variation in Purkinje cell staining did not appear to relate to any particular diagnostic group (χ2 = 7.7; p = 0.102) or the presence/absence of an expansion in C9orf72 (p = 0.248). Likewise, neither patient age at death (r s = 0.167) nor post mortem delay (PMD) (r s = 0.176) appeared to influence the degree of staining.
Subsequently, a subset of 10 cases was chosen in order not to ‘waste’ antibody on most other cases where little or no immunostaining for C9-L had been obtained, and to provide meaningful comparative data when using the other C9orf72 antibodies. These cases represented all pathological subgroups and included five strongly, and five only weakly immunostained cases when using C9-L antibody on cerebellum. These cases were also used for the assessment of C9orf72 immunostaining patterns in hippocampus and spinal cord.
A similar pattern, and intensity, of staining of Purkinje cells to that observed with C9-L antibody was seen with Proteintech monoclonal antibody (Figure 1(c)), Proteintech polyclonal antibody, 20422 (Figure 1(d)), and GeneTex antibody (Figure 1(e), Table 3). A similar, but weaker, staining of Purkinje cells was seen with Proteintech polyclonal antibodies, 18450 and 23058 (Figures 1(f,g), Table 3). Santa Cruz antibody immunostained the whole cerebellar section and did not clearly distinguish the speckled staining in Purkinje cell somata from other cerebellar tissue components (Figure 1(h)). There was very little or no immunostaining with Abgent antibody, though when present this did appear similar to the other antibodies in terms of Purkinje cell staining (Figure 1(i)). ‘Background’ parenchymal staining in the molecular and granule cell layers also varied in intensity being generally very weak or absent in C9-L immunostaining, but ranging from weak to intense with all other antibodies (Table 3). Variable glial cell immunostaining was also seen in the white matter, ranging from absent or weak to moderate or strong (Table 3). As with C9-L immunostaining, there were no correlations between intensity of staining and age at death (p = 0.799–0.927) or PMD (p = 0.596–0.973) for any of the four Proteintech antibodies, or the GeneTex antibody.
Table 3.
Relative staining of cerebellar structures by each antibody.
| Antibody | Purkinje cells | Molecular layer | Granule cell layer | Glial cells |
|---|---|---|---|---|
| C9-L | +++ | 0/+ | 0/+ | 0 |
| C9-S | +++ | + | + | 0 |
| 22637-1-AP (18450) | + | + | + | + |
| 22637-1-AP (20422) | +++ | ++ | ++ | + |
| 22637-1-AP (23058) | + | + | + | + |
| 66140-1-Ig | +++ | 0/+ | 0/+ | + |
| GTX119776 | +++ | ++ | ++ | ++ |
| sc-138763 | +/++ | +++ | +++ | +++ |
| AP12928b | 0/+ | 0/+ | 0/+ | 0 |
Comparison of scores for intensity of Purkinje cell immunostaining made on the subset of 10 cases showed a significant overall difference in scores between C9-L, Proteintech 18450, 20422 and 23058 antibodies, Proteintech monoclonal antibody and GeneTex antibody (χ2 = 16.6; p = 0.005). Scores for immunostaining with C9-L antibody were significantly higher than those for Proteintech 18450 (p = 0.044) and 23058 (p = 0.027) antibodies (which did not differ from each other), but not significantly different from Proteintech 20422 (p = 0.876) and monoclonal (p = 0.938) antibodies, or GeneTex antibody (p = 0.082) (which again did not differ significantly from each other).
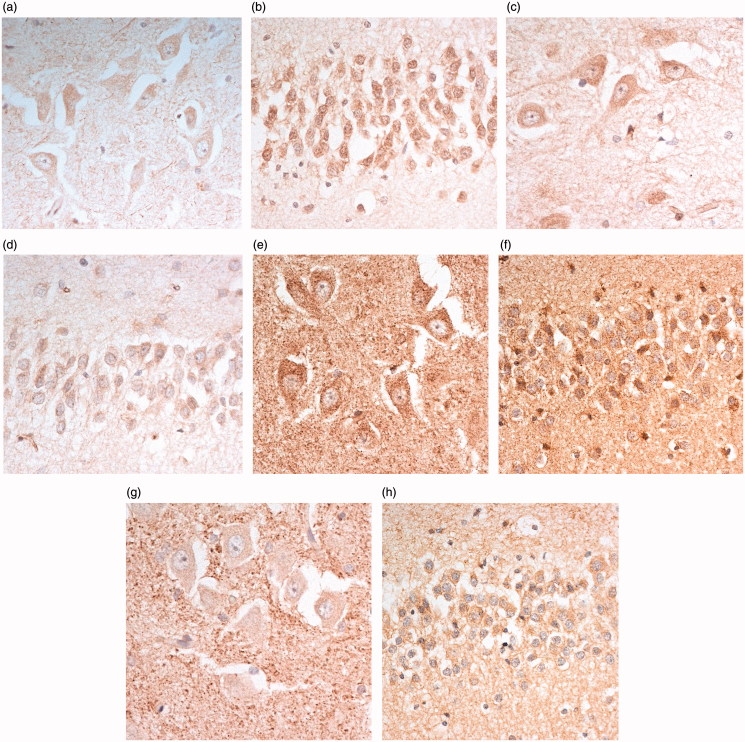
In the hippocampus, C9-L immunostaining was seen as small granules within the cytoplasm and proximal dendrites of pyramidal cells of area CA4 (Figure 2(a)), and neurons of the dentate gyrus (Figure 2(b)). Again, there was no nuclear membrane staining, or immunostaining of dipeptide repeat proteins in such cells. Similar findings were made using the Proteintech monoclonal antibody (Figure 2(c,d)) and the polyclonal 20422 antibody. The other two Proteintech polyclonal antibodies (18450 and 23058) barely immunostained such cells, even at high antibody concentrations (not shown). The GeneTex antibody strongly immunostained the cytoplasm of CA4 neurons (Figure 2(e)) and dentate gyrus granule cells (Figure 2(f)), and also strongly immunostained parenchymal structures resembling (synaptic) clusters around CA4 neurons (Figure 2(e)), a finding not seen with the C9-L or Proteintech antibodies. These same structures were also prominently immunostained with Santa Cruz antibody (Figure 2(g)), though here the cytoplasm of CA4 neurons (Figure 2(g)) and dentate gyrus granule cells (Figure 2(h)) was barely stained. The Abgent antibody showed little or no immunostaining of any region of the hippocampus (not shown).
Figure 2.
Immunostaining of pyramidal cells of CA4 region (a,c,e,g), and dentate gyrus granule cells (b,d,f,h), of the hippocampus for C9orf72 protein using C9-L antibody (a and b, respectively), Proteintech monoclonal antibody (c and d respectively), GeneTex antibody (e and f, respectively) and Santa Cruz antibody (g and h, respectively). Note immunostaining of ‘synaptic clusters’ in CA4 region using GeneTex and Santa Cruz (e and g, respectively) antibodies. Immunoperoxidase–haematoxylin, ×400 microscope magnification.
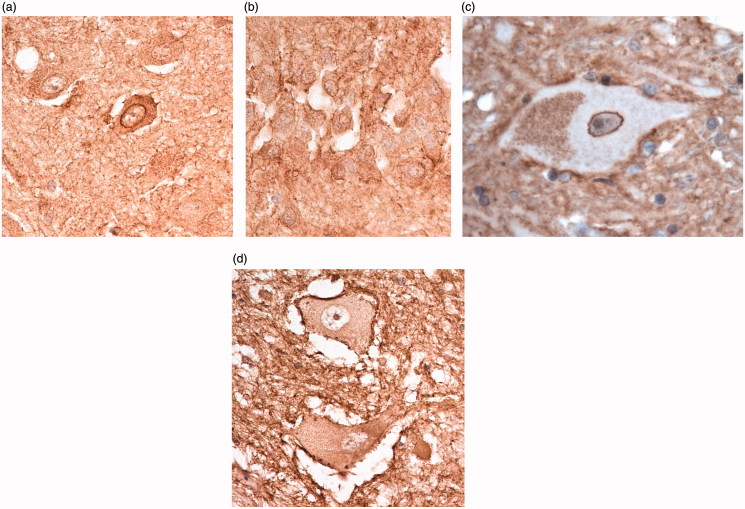
In the spinal cord, C9-L immunostaining was mostly seen as a few, weakly immunostained, small granules within the cytoplasm and proximal dendrites of some anterior horn cells (Figure 3(a)), although occasionally others were strongly immunostained. Again, there was no nuclear membrane staining in these cells, or immunostaining of structures resembling the TDP-43 inclusions present in such cells in the patients with bvFTD + ALS, or ALS alone, with or without an expansion in C9orf72. Similar findings were made using Proteintech monoclonal antibody (Figure 3(b)) and the polyclonal 20422 (Figure 3(c)) antibody, and when using GeneTex (Figure 3(d)) and Santa Cruz (Figure 3(e)) antibodies. Cells were unstained using Abgent antibody (Figure 3(f)).
Figure 3.
Immunostaining of anterior horn cells of the spinal cord for C9orf72 protein using C9-L antibody (a), Proteintech monoclonal antibody (b), Proteintech polyclonal antibody, 20422 (c), GeneTex antibody (d), Santa Cruz antibody (e) and Abgent antibody (f). Immunoperoxidase–haematoxylin, ×400 microscope magnification.
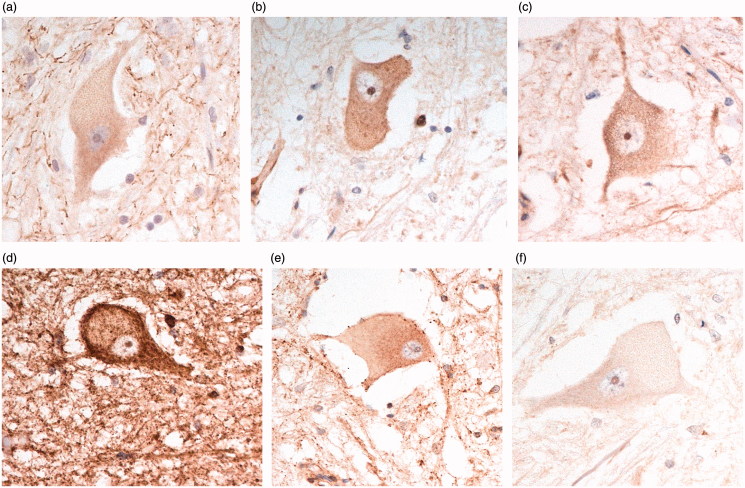
By contrast, C9-S antibody immunostained the nuclear membrane, but not cytoplasm, of Purkinje cells (see Figure 1(b)). Those cases that showed strong cytoplasmic labelling with C9-L antibody also showed strong nuclear labelling with C9-S antibody, and vice versa. A similar pattern of nuclear membrane staining was seen in pyramidal cells of CA4 (Figure 4(a)) and granule cells of the dentate gyrus (Figure 4(b)) in all pathological groups, and in FTLD patients with and without an expansion in C9orf72. In the spinal cord, C9-S immunostaining detected the nuclear membrane in normal cells (Figure 4(c)), but in patients with bvFTD + ALS, or ALS alone, with or without an expansion in C9orf72, there was translocation of this immunostaining to the plasma membrane (Figure 4(d)). This pattern of (nuclear) staining was not seen in any of the brain regions investigated with any of the commercial antibodies.
Figure 4.
Immunostaining for C9orf72 protein using C9-S antibody showing nuclear membrane staining in pyramidal neurones of CA4 region (a), dentate gyrus granule cells (b) and normal anterior horn cells (c). In bvFTD + MND, there is loss of nuclear membrane staining which is replaced by immunostaining of the external plasma membrane (d). Immunoperoxidase–haematoxylin, ×400 microscope magnification.
Discussion
Previously, C9-L antibody demonstrated large ‘speckles’ in the cytoplasm of Purkinje cells of the cerebellum and diffuse cytoplasmic labelling in spinal motor neurons. Conversely, C9-S labelled the nuclear membrane of healthy neurons, with apparent relocation to the plasma membrane in diseased motor neurons in bvALS + FTD or ALS alone (4). In the present study, we have made similar observations in Purkinje cells of patients with bvFTD + ALS and in ALS alone, as well as in other neurodegenerative disorders and healthy controls. However, it is unclear what these speckled structures in Purkinje cells represent. Although C9orf72 may have a role in autophagy regulation, this morphological pattern of Purkinje cell immunostaining has not been found using a range of markers of endosomes, lysosomes and autophagosomes (4). Further work, involving for example immunogold labelling studies, will need to be carried out in order to determine the identity of the subcellular structures which underpin the speckled Purkinje cell cytoplasmic staining pattern observed with C9-L antibody, and to investigate how the C9-S epitopes might traffic from nuclear to cytoplasmic membrane in some FTD/ALS patients. Nonetheless, it is notable that the intensity of cytoplasmic staining in Purkinje cells is far greater than that seen in hippocampal and spinal neurons, consistent with studies showing higher expression levels of C9orf72 protein in cerebellum compared to other brain regions (1,3), implying that such cells have greater dependency on C9orf72 protein for maintenance of their health and vitality irrespective of what the precise roles of the long and short form of the protein might be.
Of the seven commercially available C9orf72 antibodies tested, a similar pattern of Purkinje cell immunostaining was noted in five, being similar in intensity in Proteintech polyclonal antibody 22637-1-AP (batch 20422) and Proteintech monoclonal antibody 66140-1-Ig, and also in the GeneTex antibody GTX119776, to that seen with C9-L antibody; a similar, but much weaker pattern of immunostaining was seen using Proteintech 22637-1-AP batches 18450 and 23058 polyclonal antibodies. The Santa Cruz and Sigma antibodies failed to detect this pattern of Purkinje cell immunostaining. Variations in staining pattern and intensity may to some extent reflect different epitopes or protein conformations recognised by each antibody. Hence, we would conclude that commercially available Proteintech polyclonal antibody 22637-1-AP (batch 20422), Proteintech monoclonal antibody 66140-1-Ig, and GeneTex antibody GTX119776, can be confidently employed for the detection of C9orf72 protein in immunohistochemical studies. The observations that none of the five commercially available antibodies also immunostained the nuclear membrane of Purkinje cells implies that all of these only detect the long form of C9orf72 protein, a conclusion consistent with information provided by the manufacturers whereby only a single band at around 50–55 KDa was detected by each commercial antibody, akin to that detected by C9-L (at 54.8KDa), and none detected a band equivalent to C9-S at 24.8 KDa (4).
Earlier studies using certain commercial antibodies, including the Santa Cruz antibody employed here (sc-138763) and an antibody from Sigma (HPA023873), revealed a pattern of immunostaining for C9orf72 within area CA4 of hippocampus which resembled ‘synaptic clusters’ (34–36). In the present study, this same pattern of staining was again seen with the Santa Cruz antibody, and also to a lesser extent with GeneTex antibody, but not with any of the Proteintech antibodies, or with C9-L or C9-S (see Figure 2), suggesting this immunostaining may not relate to C9orf72 protein and questions the specificity of the Santa Cruz and GeneTex antibodies.
The high level of variability of C9orf72 immunostaining between cases is difficult to explain and did not relate to age of the individual, pathological group or post mortem delay period. Potential explanations might lie with brain pH at time of death, since this is a more reliable index of tissue quality than post mortem delay (37). Preservation of C9orf72 protein within tissues after death, like many other brain proteins, may be highly susceptible to degradation in a low (i.e. <6.0) pH environment. Alternatively, the efficiency of immunostaining may relate to the length of time tissues may have been stored in formalin fixation before blocking and processing into wax sections. In the present study, unfortunately, it was not possible to assess these potential confounders as brain pH values were not available for most cases, and it was not possible to retrospectively estimate how long tissues might have been stored in formalin prior to processing.
There is strong evidence for a loss of function effect associated with the expansion consequent upon reduced gene expression (1,3,4), this being dependent upon the degree of DNA methylation (5,6). Nonetheless, we found no significant differences in the level of immunostaining for C9orf72 in cases bearing expansions in C9orf72 compared to those without expansion. However, in formalin fixed paraffin embedded tissues, immunohistochemistry does not necessarily reflect gene expression levels, and as such cannot be taken as a direct measure of protein output. The lack of immunostaining for any structures resembling dipeptide repeat proteins in neurons in cerebellum granule cells, or cells of the dentate gyrus or CA4 regions of the hippocampus, in C9orf72 expansion bearers indicates that C9orf72 protein is not a component of these pathological features. A loss of C9-S, importin-β1 and Ran-GTPase nuclear membrane labelling in anterior horn cells containing TDP-43 inclusions in MND cases with expansions in the C9orf72 gene (4) suggests changes in C9-S protein function may determine, or result from, TDP-43 mislocalisation.
In summary, we have evaluated the staining properties of seven commercially available antibodies against C9orf72 protein compared to two well-characterised in-house antibodies (4). We find three of these commercial antibodies provided immunohistochemical staining patterns similar to the in-house C9-L antibody, but distinct from C9-S antibody. These appear reliable for the detection of at least the longer form of C9orf72 protein in normal and diseased tissues.
Supplementary Material
Acknowledgements
We acknowledge the support of the Manchester Brain Bank by Alzheimer’s Research UK and Alzheimer’s Society through their funding of the Brains for Dementia Research (BDR) Programme. Manchester Brain Bank also receives Service Support costs from the Medical Research Council. This work was supported by Medical Research Council of UK, grant number G0701041 (SPB), The James Hunter ALS Initiative and ALS Canada/Brain Canada Hudson Translational Team Grant (JR).
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Supplementary material available online
References
- 1. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. . Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. . A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, et al. . Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779.e5–.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, et al. . Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol. 2015;78:568–83. [DOI] [PubMed] [Google Scholar]
- 5. Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, et al. . C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, Fratta P, et al. . The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol. 2015;129:715–28. [DOI] [PubMed] [Google Scholar]
- 7. Cooper-Knock J, Walsh MJ, Higginbottom A, Highley RJ, Dickman MJ, Edbauer D, et al. . Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooper-Knock J, Higginbottom A, Stopford MJ, Highley RJ, Ince PG, Wharton SB, et al. . Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol. 2015;130:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, et al. . C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. . Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidson Y, Barker H, Robinson AC, Troakes C, Smith B, Al SS, et al. . Brain distribution of dipeptide repeat proteins in Frontotemporal Lobar Degeneration and Motor Neurone Disease associated with expansions in C9ORF72 . Acta Neuropathol Commun. 2014;2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, et al. . Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–79. [DOI] [PubMed] [Google Scholar]
- 13. Mann DMA, Rollinson S, Robinson A, Callister J, Snowden JS, Gendron T, et al. . Dipeptide repeat proteins are present in the p62 positive inclusions in patients with Frontotemporal Lobar Degeneration and Motor Neurone Disease associated with expansions in C9ORF72. Acta Neuropathol Commun. 2013;1:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, et al. . Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–94. [DOI] [PubMed] [Google Scholar]
- 15. Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. . The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. [DOI] [PubMed] [Google Scholar]
- 16. Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. . RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang D, Iyer LM, He F, Aravind L.. Discovery of Novel DENN Proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Gene. 2012;3:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marat AL, Dokainish H, McPherson PS.. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizuno-Yamasaki E, Rivera-Molina F, Novick P.. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, et al. . C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, et al. . The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao S, MacNair L, McLean J, McGoldrick P, McKeever P, Soleimani S, et al. . C9orf72 isoforms in Amyotrophic Lateral Sclerosis and Frontotemporal Lobar Degeneration. Brain Res. 2016;1647:43–9. [DOI] [PubMed] [Google Scholar]
- 23. Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. . The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, et al. . Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–81. [DOI] [PubMed] [Google Scholar]
- 25. Miller L, Rollinson S, Callister J, Young K, Harris J, Gerhard A, et al. . p62/SQSTM1 analysis in frontotemporal lobar degeneration. Neurobiol Aging. 2015;36:1603e5–9. [DOI] [PubMed] [Google Scholar]
- 26. Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. . Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. . Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. [DOI] [PubMed] [Google Scholar]
- 28. Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. . Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–6. [DOI] [PubMed] [Google Scholar]
- 29. Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, et al. . Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8. [DOI] [PubMed] [Google Scholar]
- 30. Mackenzie IRA, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, et al. . A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. . Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. [DOI] [PubMed] [Google Scholar]
- 32. Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. . Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134:2456–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124 (suppl):96–107. [DOI] [PubMed] [Google Scholar]
- 34. Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, et al. . Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135:751–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Satoh J, Tabunoki H, Ishida T, Saito Y, Arima K.. Dystrophic neurites express C9orf72 in Alzheimer's disease brains. Alzheimers Res Ther. 2012;4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, et al. . Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135:693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson AC, Palmer L, Love S, Hamard M, Esiri M, Ansorge O, et al. . Extended post-mortem delay times should not be viewed as a deterrent to scientific investigation of human brain tissue: a study from the Brains for Dementia Research Neuropathology Study Group. Acta Neuropathol. 2016;132:753–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.