Abstract
It is well established that any properly conducted biophysical studies of proteins must take appropriate account of solvent. For water-soluble proteins it has been an article of faith that water is largely responsible for stabilizing the fold, a notion that has recently come under increasing scrutiny. Further, there are some instances when proteins are studied experimentally in the absence of solvent, as in matrix-assisted laser desorption/ionization or electrospray mass spectrometry, for example, or in organic solvents for protein engineering purposes. Apart from these considerations, there is considerable speculation as to whether there is life on planets other than Earth, where conditions including the presence of water (both in liquid or vapor form and indeed ice), temperature and pressure may be vastly different from those prevailing on Earth. Mars, for example, has only 0.6% of Earth's mean atmospheric pressure which presents profound problems to protein structures, as this paper and a large corpus of experimental work demonstrate. Similar objections will most likely apply in the case of most exoplanets and other bodies such as comets whose chemistry and climate are still largely unknown.
This poses the question, how do proteins survive in these different environments? In order to cast some light on these issues we have conducted a series of molecular dynamics simulations on protein dehydration under a variety of conditions. We find that, while proteins undergoing dehydration can retain their integrity for a short duration they ultimately become disordered, and we further show that the disordering can be retarded if superficial water is kept in place on the surface. These findings are compared with other published results on protein solvation in an astrobiological and astrochemical setting. Inter alia, our results suggest that there are limits as to what to expect in terms of the existence of possible extraterrestrial forms as well to what can be achieved in experimental investigations on living systems despatched from Earth. This finding may appear to undermine currently held hopes that life will be found on nearby planets, but it is important to be aware that the presence of ice and water are by themselves not sufficient; there has to be an atmosphere which includes water vapor at a sufficiently high partial pressure for proteins to be active. A possible scenario in which there has been a history of adequate water vapor pressure which allowed organisms to prepare for a future dessicated state by forming suitable protective capsules cannot of course be ruled out.
Introduction
The protein world is extremely diverse as to function, and correspondingly (consequently, one needs to say) to structure. One can subdivide the protein world into three main classes: fibrous proteins such as keratin, fibroin, collagen, characterised by ever-extended repeated structural motifs, globular proteins that are water-soluble and which reside inside cells or in the surrounding aqueous fluid, and proteins which are embedded in lipid bilayer membranes. For all three classes, solvent (water for the first two of these, lipid for the third) is an essential requirement for maintaining stability as well as the flexibility needed for them to exert their functions. Numerous attempts to understand such dynamic behavior in proteins have been undertaken over many years, supported by ever increasing accuracy in the force fields used to study protein dynamics. But a stringent requirement, established many years ago, is for appropriate account to be taken of the solvent and its interaction with the protein. For the case of aqueous systems, approximations to discrete water models have had to be discarded (Nutt and Smith, 2007) and better models introduced (Nutt and Smith, 2007; Van Aalten et al., 1996; Levitt et al., 1997). While these inevitably increased the computational overheads, these have with time, and the continued applicability of Moore's law, become ever less of a problem.
Given this as a backdrop to future work on molecular dynamic simulations of proteins, why would anyone ever want to conduct simulations in the absence of solvent?
An important family of experimental tools for studying proteins and protein-protein interactions consists of matrix-assisted laser desorption/ionization (MALDI), time-of-flight mass spectrometry (TOF) and electrospray (Smith et al., 1990; Robinson et al., 2007; Wyttenbach and Bowers, 2007; Breuker and McLafferty, 2008; Benesch and Robinson, 2009; Liu et al., 2009; Liu and Schey, 2008; Barrera et al., 2008). Many proteins, and many protein-protein complexes, retain their structural integrity in vacuo, at least for a sufficiently long time, for many of their essential structural features to be retained and be capable of study in intimate detail (Patriksson et al., 2007; Meyer et al., 2009). All the while, water is being stripped off these entities, and one may wonder how many of the old assumptions about the importance of water in protein structure are still valid. Of course, water plays a critical role, but the structures that water has played a role in sculpting retain their integrity for some time after the water has been stripped off; short-term memory is preserved.
There are other reasons for wanting to study the properties of proteins in the absence of water or in the reduced presence of water (Smith et al., 1990; Robinson et al., 2007; Wyttenbach and Bowers, 2007; Breuker and McLafferty, 2008; Benesch and Robinson, 2009; Liu et al., 2009; Liu and Schey, 2008; Barrera et al., 2008; Patriksson et al., 2007; Meyer et al., 2009; Soares et al., 2003; Wedberg et al., 2012). These studies mostly highlight aspects of protein structural integrity but say little about the effects on activity although for certain protein engineering applications it has been shown (Klibanov, 2010) that it may be considered advantageous to conduct enzyme reactions in organic solvents.
For this work, our guiding principles were to provide insights into protein hydration and dehydration in the light of an abundance of excellent published work (Smith et al., 1990; Robinson et al., 2007; Wyttenbach and Bowers, 2007; Breuker and McLafferty, 2008; Benesch and Robinson, 2009; Liu et al., 2009; Liu and Schey, 2008; Barrera et al., 2008) but in particular by the large-scale systematic experimental study of protein hydration (Chen et al., 2008). In that seminal work, distinction was made between three categories of water molecules: individual water molecules hydrogen-bonded to donor/acceptor atoms on the surface or in crevices, bound waters immediately surrounding these, and remaining crystal waters (meaning water molecules located in the crystal structure) surrounding the former which have the normal attributes of bulk water. We adopted this solvation model and corresponding terminology in our work. We set about simulating this kind of environment for a globular protein with MW 17.6 kDa.
We are aware that we are far from being the first to carry out vacuum simulations of proteins. Indeed, the earliest molecular dynamics (MD) studies on proteins were all carried out in the absence of solvent, as alluded to above. This was due to restrictions in available computer power, and attempts were made already then to approximate for solvent effects. MD simulations in vacuo with the express purpose of studying solvation were later carried out, so that our work is not unique in that respect. For example unsolvated peptides large enough to possess secondary structure have been studied (Jarrold, 2007). This author found that some helices were much more stable in vacuum than in aqueous solution and that charge is critical for stabilizing α-helices and destabilizing β-sheets. While these are valuable observations they do not impact on either the validity or the novelty of our work. Although that work reported the formation of incipient tertiary structures, it was carried out on short peptides with a focus on secondary structure, not on large globular proteins with a focus on (de)solvation and aqueous shielding as in our case. Other published work (Seibert et al., 2009) similarly records studies on polypeptides rather than proteins. The latter authors conducted interesting melting and reannealing studies in these systems with good agreement with experiment, although we have difficulty in understanding the phrase “the native state of the peptide”, since nobody has to our knowledge ever been able to define, in a way that all can agree on, what the native state of a protein, much less that of a peptide, actually is. NMR will show which state(s) are energetically accessible at a given temperature and pressure. There is nothing about that that is necessarily “native”. Furthermore, in that work (Seibert et al., 2009) and in the previously cited study (Jarrold, 2007), the focus was on polypeptides, while we attend to the case of a large globular protein.
Much more relevant to the matter in hand is work (Arteca and Tapia, 2002, 2003) that noted that protein unfolding and refolding transitions are possible even in the absence of water. These authors studied two small proteins of similar length: the α/β protein (lysozyme MW 14.3 kDa) and a four-α-helix bundle protein (cytochrome c' MW 12.0 kDa) and found that their unfolding behavior was very different. They report that the former protein undergoes a limited reduction in secondary structure, relaxing back to what are referred to as “quasi-native states”, while cytochrome c' unfolds more slowly by first losing tertiary contacts. There is some apparent contradiction in these reports whereby the unfolding behavior was stated (Arteca and Tapia, 2002) to be different in the two cases while the second paper (Arteca and Tapia, 2003) states: “suggesting that similar unfolding pathways may be accessible to many protein sequences”. We did not set out to resolve this issue but instead worked on a somewhat larger protein (MW 17.6 kDa) that has been studied by mass spectrometry (Liu and Schey, 2008). We complement our studies by considering issues of protein structural integrity and the question, referred to above, of whether bound water has a protecting effect on the protein.
These studies are important not only in relation to what happens under various experimental conditions, or in vivo (“in vivo” here being defined as being in living systems on planet Earth). There is an ongoing search for life elsewhere in the cosmos than here on Earth, particularly on planetary neighbor Mars (we cannot quite include the current attention given the spectacular landing on comet 67P because the search there is for organic feedstock chemicals and not life itself or even proteins). Establishing what conditions would be required in order to support proteins, the most crucial of biochemicals that life depends on, must surely be one of the central aims of research in astrobiology and astrochemistry. Protein biosynthesis, correct folding and function all require water both in liquid and vapor form. This issue has been discussed in previous work (Ball 2013) which we return to below. The issue of the presence or otherwise of water on e.g. Mars has been considered in much earlier times (Herschel 1784) and one of the co-founders of the theory of evolution (Wallace 1907) declared that Mars would be devoid of life. This issue has been discussed in depth in a recent review (Rummel et al. 2014). The main conclusion concerning the potential for life on Mars stated in that review was that it was ”..… determined by locations where both of the parameters (without margins added) of temperature (above 255°K) and water activity (relative humidity > 60%) are attained. There are places/times on Mars where both of these parameters are attained within a single sol, but it is unknown whether terrestrial organisms can use resources in this discontinuous fashion.”. Arid conditions are not supportive of life. Even if certain hardened living systems might survive extreme environments they would most probably have adapted to those environments after an evolutionary history that from the start was more conducive to life. Proteins and other biomacromolecules need some water for structural integrity and functioning. Whether or not proteins exist(ed) elsewhere in the cosmos, there is a long history of protein evolution on this planet which has been studied most notably in the laboratory resurrections of ancestral proteins (Risso et al 2014).
Methods
The Yasara program (Krieger et al 2002) was used to prepare the bovine β-lactoglobulin (1beb) structure for simulation studies. The monomer coordinates were extracted from the crystal dimer. All disulfide bridges were reduced (broken) using a standard Yasara command in order to allow protein unrestricted movement and reduce shearing effects. In support of this procedure, we claim that disulfide bridge rupture is very likely one of the first things to happen in MALDI and very likely also in the extreme extraterrestrial environments. The AMBER03 force field (Case et al., 2010) was used as implemented within the Yasara program. Automatic correction of wrong isomers and cis-peptide bonds were implemented. The protein was encased in a 50Å cube filled with TIP4P water (Jorgensen and Madura 1985) at a density of 0.997g/cm3. Subsequent simulations were conducted with the same settings. Coordinates were saved at 0.5ps intervals. The NVP ensemble was used throughout, but P was set at either 0 or 1013 Hpa depending on the experiment. Prior to each experiment, the structure was relaxed and equilibrated by running a minimization at 500°C and initially at 1013 HPa. This choice of starting temperature was not arbitrary but instead required some thought. There is no way to choose a priori a representative temperature out in space. Comets can experience temperatures from -220° out in the Oort cloud to many hundreds of degrees as they graze the sun. Average surface temperatures on planets of course vary widely: Mars: -143°C to +35°C (mean -63°C), Earth: -50°C to +50°C (mean ~ +10°C), Venus: ~ +500°C. Temperatures in MALDI experiments are of course much higher than any of these so it is hard to decide what would be a suitable starting temperature for our experiments. In practice (meaning that several temperature levels were investigated prior to the final decision) the starting temperature 500°C was chosen. It is the lowest temperature that provides measurable water loss. Different hydration schemes were studied, with the aim of investigating what, if any, was the effect of a possible shielding effect of the bound waters and surrounding bulk water. As defined under Results the “hydration water” set consisted of the protein deprived of those crystal waters that were not in intimate contact (defined as those where the distance between any of the H/O atoms in HOH is not within 3.5Å of the surface). For both sets the vacuum simulations were conducted either in vacuo or in water (meaning that additional water was added to fill the MD simulation box). All simulations were conducted in a cell which was augmented by the Yasara program by 50 Å to all three cell dimensions, and the systems were equilibrated and minimized. For all simulations, the protein was fixed at the origin since it was found to drift otherwise. Automatic correction of wrong isomers and cis peptide bonds was implemented. Simulations were conducted using the same parameters as in the initial setup and minimisation. Coordinates were saved every 0.5ps. Simulations were discontinued when water was driven off completely after ~ 25 ps for the vacuum cases (upper panel in Figure 1) and to all intents and purposes permanently (lower panel)). During this time 200 structures (as referred to in the abscissae of the figures) were saved. Typically, Tables were created for bound and hydration water removal and plotted. Fixation of water atoms in the control experiment (Table 1) was done by making appropriate setting in the Yasara implementation of AMBER03.
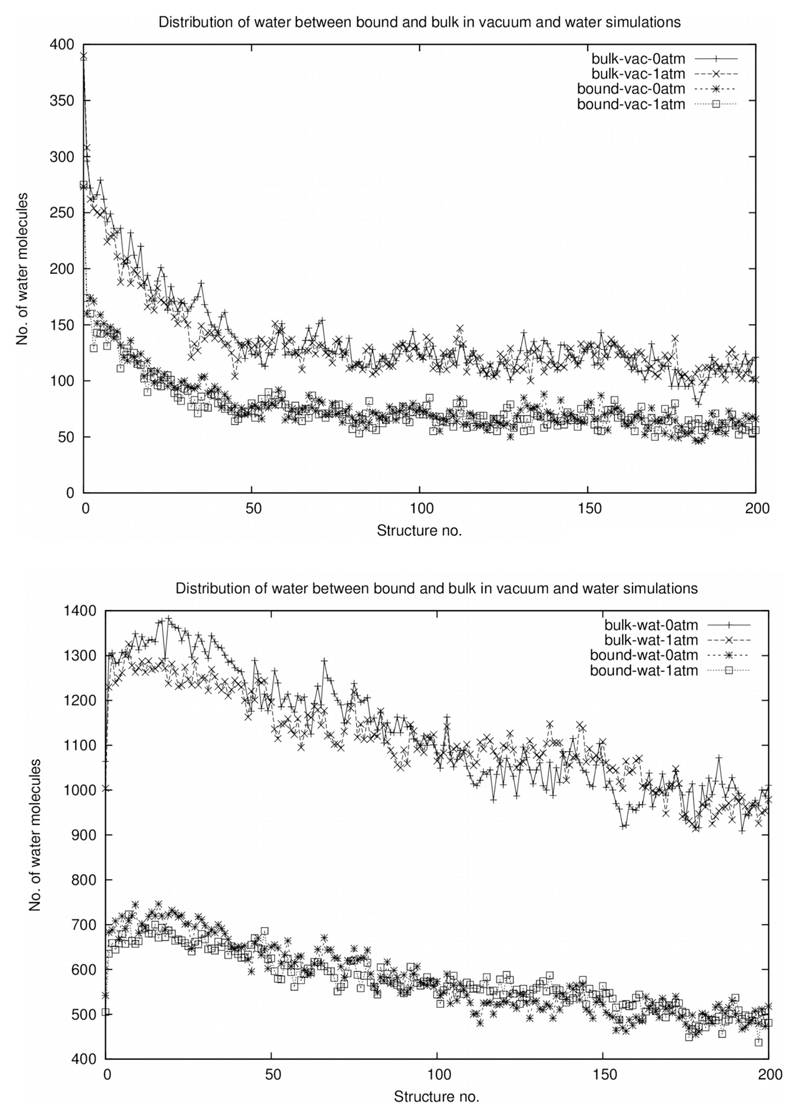
Figure 1.
100 ps simulations for the vacuum (upper panel) and water (lower panel) cases.
Table 1.
| Simulation type | ΔRMSD/0.5 ns |
|---|---|
| Bulk water under 0 atm in vacuum, waters fixed | 0.389 |
| Hydration water under 0 atm in vacuum, waters fixed | 0.032 |
| Bulk water under 0 atm in vacuum | 4.841 |
| Bulk water under 0 atm in water | 7.548 |
| Hydration water under 0 atm in vacuum | 4.263 |
| Hydration water under 0 atm in water | 3.861 |
Results
For the purposes of this study the choice of protein could be almost arbitrary. The protein bovine beta-lactoglobulin (PDB I.d. 1beb, MW 17.6 kDa) was selected as it has been studied in detail by MALDI (Liu and Schey, 2008). Following Chen et al. (2008) we use the same three categories of water molecules: water hydrogen bonded to individual groups on the protein surface, and the surrounding layer associated with that layer and a further external layer, which allows free diffusion. Within the bound water region (the first two layers of hydration referred to by Chen et al. (2008)), which we refer to collectively as “hydration water” in our simulations, diffusion of water and solutes is restricted. The simulations where the remaining crystal waters are present are called “bulk water” simulations. Further, simulations described as “in vacuum” do not have any additional water beyond those corresponding to the “hydration water” and “bulk water” cases, while those described as “in water” are carried out in a box where any space not occupied by protein and its crystal waters (which comprises members of all 3 of the Chen et al. sets) is filled with water. Four dual sets of simulations were carried out as below (see also Methods), for each case with the simulation pressure set to 1 and 0 atm respectively. The results are shown in the following figures:
Bulk water under 0 atm in vacuum (Figure 1 upper panel)
Bulk water under 1 atm in vacuum (Figure 1 upper panel)
Bulk water under 0 atm in water (Figure 1 lower panel)
Bulk water under 1 atm in water (Figure 1 lower panel)
Hydration water under 0 atm in vacuum (Figure 1 upper panel)
Hydration water under 1 atm in vacuum (Figure 1 upper panel)
Hydration water under 0 atm in water (Figure 1 lower panel)
Hydration water under 1 atm in water (Figure 1 lower panel)
For the case of bulk water in vacuum, the dehydration curves exhibit approximately hyperbolic behavior at both 0 atm and 1 atm, following a roughly similar time-course. For bulk water in water at 0 atm and 1 atm a roughly similar time-course is followed at both pressures, but the 1 atm case shows some retardation as would be expected. Hydration water in vacuum cases at 0 atm and 1 atm both show, like the corresponding bulk water cases, a hyperbolic-type behavior, time-courses are very similar. For hydration water in water at 0 atm and 1 atm we see the retardation effect at 1 atm as in the bound water case.
The original data can be inspected and downloaded from:
tables: http://adelard.org.uk/experiment/vac/hydro/dat/tables/
plots: http://adelard.org.uk/experiment/vac/hydro/dat/plots/
Note that the simulations were run for a length of time adequate to ensure that waters were stripped off exhaustively. There was no need (in this work) to run lengthier simulations.
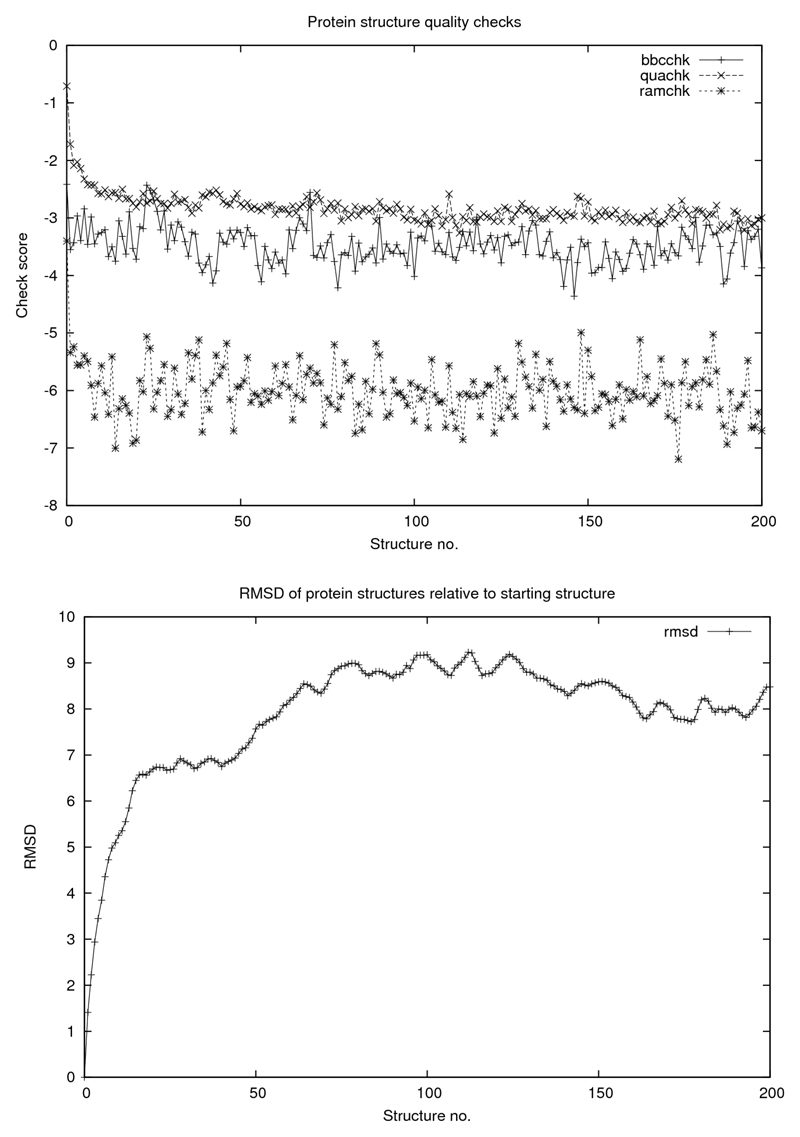
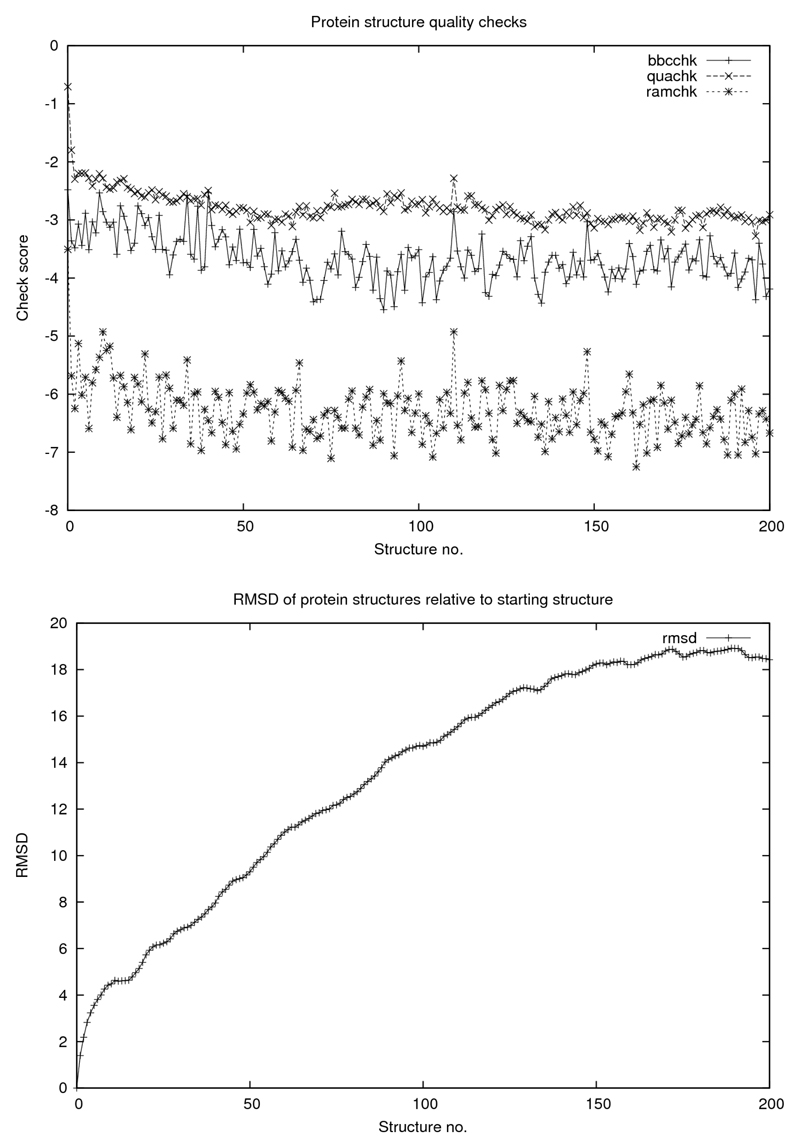
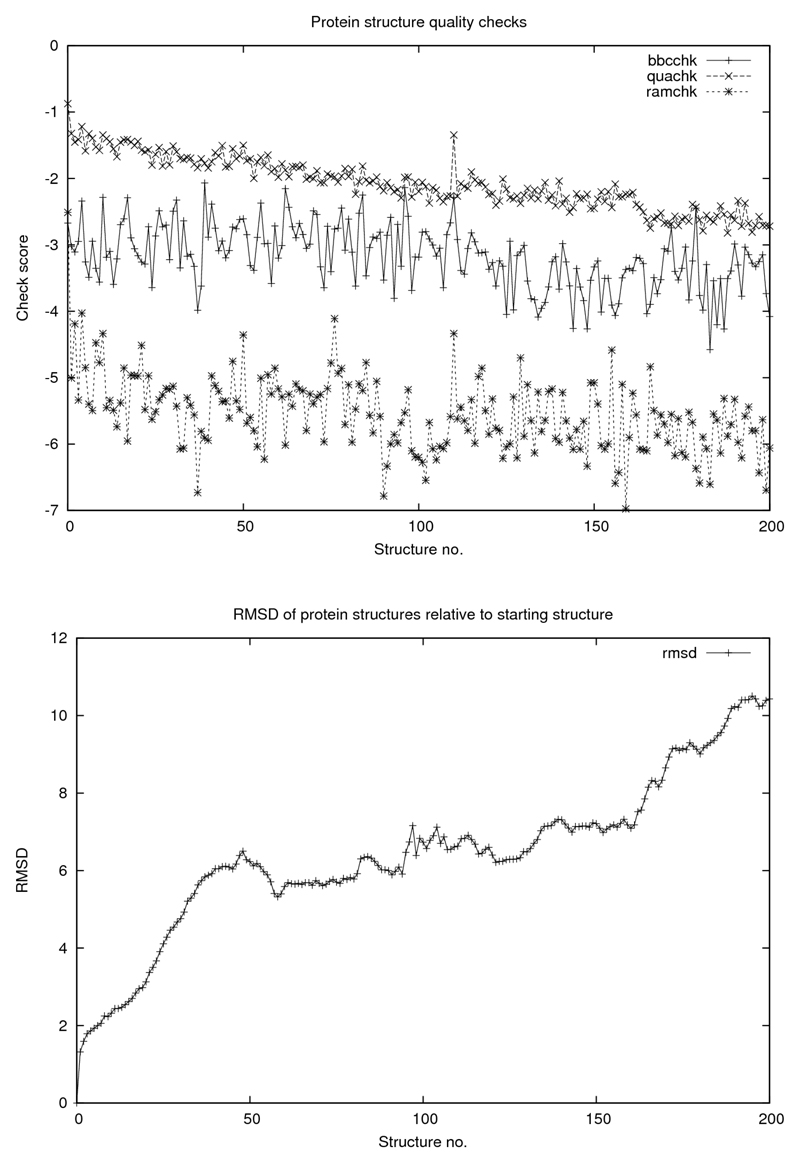
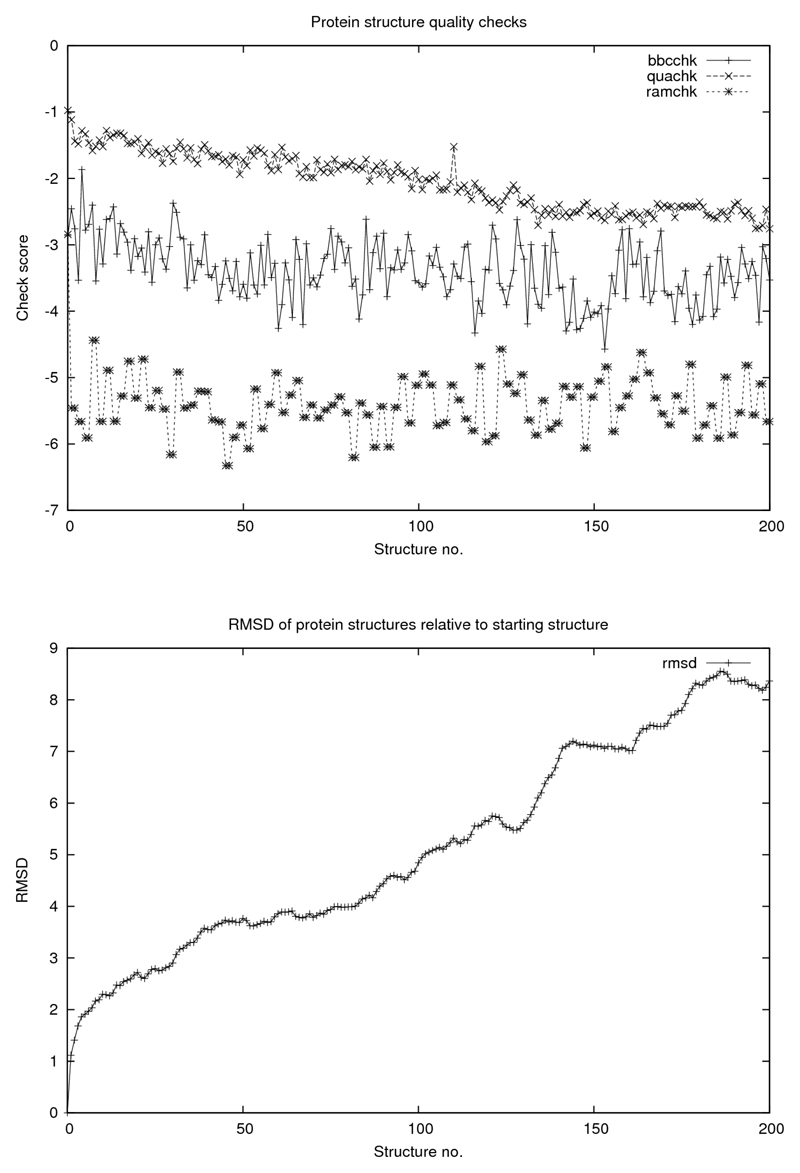
In addition to studying how the protein itself behaves under these conditions, key structural parameters for the saved structures along the simulation pathway were calculated. Firstly, for each of the protein structures generated in the four simulations, the root-mean-square displacement (RMSD) of the coordinates from the starting structure were determined using Yasara (Krieger et al., 2002). Next, certain quality control features of the protein, calculated with a view to detecting changes in the protein structure as it gets dehydrated, were determined using WHAT IF (Vriend, 1990). These were checks of backbone geometry (bbchk), bond lengths and angles (quachk) and a check of bounds in secondary structure as defined by the Ramachandran diagram (ramchk). Plots of these results are shown in Figures 2, 3, 4 and 5.
Figure 2.
WHAT IF quality checks: bbchk, quachk and ramchk as explained in text. For all figures abscissa is number of structures saved at 0.5 ps intervals corresponding to 100 ps simulations.
Figure 3.
WHAT IF quality checks: bbchk, quachk and ramchk as explained in text. For all figures abscissa is number of structures saved at 0.5 ps intervals corresponding to 100 ps simulations.
Figure 4.
WHAT IF quality checks: bbchk, quachk and ramchk as explained in text. For all figures abscissa is number of structures saved at 0.5 ps intervals corresponding to 100 ps simulations.
Figure 5.
WHAT IF quality checks: bbchk, quachk and ramchk as explained in text. For all figures abscissa is number of structures saved at 0.5 ps intervals corresponding to 100 ps simulations.
As a control experiment, designed to test to what extent the presence of bound waters aford some kind of protection to the protein structure a set of simulations was conducted in which the bound water was fixed to the protein surface. Simulations were conducted for 2ns under AMBER 03 FF in vacuum and simulation water at 500°C at ambient pressure saving structures at 100 ps intervals. The effect of water is expressed (Table 1) as change in root-mean square displacement over 0.5 nsec. (ΔRMSD / 0.5 nsec). Simulations were run only for the 0 atm case, four of them as before plus two runs with fixed waters. As shown in Table 1, there is a large effect due to the presence of water. When water is free to diffuse away or evaporate, there is a considerably increased deterioration in the integrity of the structure, as indicated in the RMSD data. This is clear evidence for a protecting effect of water on proteins. Water may not be the decisive factor in promoting a given fold, but it clearly has a stabilizing role.
Discussion and Conclusions
We have conducted a series of simulations as described above and considered the dehydration as the simulation proceeds. It has long been a credo in the protein folding and structure debate that proteins derive their stability from the entropic consequences of desolvation, in particular from hydrophobic surfaces (Kauzmann, 1959). This model is too simplistic, hydration of polar residues is at least as important (Ben-Naim, 2012; Bywater, 2013). In particular, attention must be paid to the issue of how tightly the water is bound to the protein surface and within its interstices (Chen et al., 2008). In our study, the waters behave in a similar fashion whether tightly bound or not (Figure 1A to D), but differ in their behaviour between the vacuum case where this a monotonic decline in the amount of waters adhering to the protein, while in the bound-water environment, there is an uptake of water for the first few picoseconds followed by a decline. There is slightly more water retained in vacuum compared to pressure at 1 atm (Figure 1A to D).
Concerning the protein itself and its “quality” parameters, there does not seem to be any catastrophic “collapse” in structure (of the kind described (Seibert et al., 2005) for polypeptides, for example) apart from a slight decline in core packing as the simulation proceeds although the RMSD increases in a rather dramatic way, somewhat differently for the four cases. Common to all cases is a detectable correlation between the decline in solvation and in structural integrity as judged from the RMSD. Finally, we demonstrate that the above mentioned dramatic deterioration in the protein structural integrity is counteracted by the presence of water when it is constrained to remain attached to the protein surface.
Although of course, the proteins unfold and then fragment during the mass spectrometry experiments, the relative integrity of the proteins for the initial phases of our simulations, even as water is extracted, prompts the following question: Since “hydrophobic interactions” (Kauzmann, 1959) are not keeping the protein structure intact in the gas phase as the solvent is being removed, what is stabilizing the structures? The same question may be asked in the cases where water is replaced by organic solvents (Soares et al., 2003; Chen et al., 2008; Wedberg et al., 2012; Klibanov, 2010). Van der Waals interactions still operate, and dipolar interactions will increase in strength as water is removed. These forces have an underlying quantum mechanical mechanism. It has earlier been observed (Patriksson et al., 2007) that intramolecular hydrogen bonds actually increase in number as dehydration proceeds. Although our results indicate a steady deterioration of the integrity of protein structure, it is salutary to read (Ball 2013) about “a tendency to assume that this is always a precondition of protein function has militated against a thorough investigation of what proteins can and cannot do in truly anhydrous conditions”. But the same author precedes that with: “current evidence points to a strong and sensitive general dependence of protein function on both the structure and dynamics of the hydration environment, which in turn are intimately connected to the hydrogen-bonded network of the water molecules and their interactions with donors and acceptors at the protein surface”. Our results are very much in line with this statement and they thereby represent a contribution to this “current evidence”.
Most of the solar-system planets other than Earth and known exoplanets have vastly different atmospheric conditions compared to Earth, and most have little or no water (as liquid or ice, but also vapor: low atmospheric pressure will drive off water from proteins). We therefore think it would be useful to include our results along with other current evidence in any discussions of “life on other planets”. There are clearly limits as to what to expect in terms of the existence of possible extraterrestrial forms as well to what can be achieved in terms of survivability of living systems despatched from Earth to other locations in the cosmos. Mars, in particular seems to be a very unwelcoming place (“the complete absence of organic material is generally believed to be characteristic of the entire planetary surface”) (Gurnett 2009). It is hardly likely that proteins sent from e.g. Earth would survive in such conditions, and the first steps towards their demise would be dehydration. Similar remarks may be made about comets, taking 67P as an example (Goesman et al. 2015) where a suite of 16 inorganic compounds, including many nitrogen-bearing species but no sulfur-bearing species, and four organic compounds—methyl isocyanate, acetone, propionaldehyde, and acetamide was found, but certainly no proteins. Here again, the atmosphere is too dry, or rather, simply absent. Although some of the mentioned organic compounds may have the properties of solvents, there is no evidence that there are oceans or even lakes of such liquids. Of course, protection to proteins can be afforded even in dry conditions by suitable packaging as in microspores and tiny organisms such as tardigrades, but once the proteins are removed from these containers they will degrade rapidly. We conclude that most of outer space is simply too arid for proteins to survive and thus our studies of protein dehydration point to what would happen if unprotected proteins were delivered to such locations. The only survival strategy available would be that mentioned in the Abstract, to form suitable encapsulating structures in the hope that wetter conditions might return.
Acknowledgements
We thank Gert Vriend for valuable discussions and for supplying and supporting the WHAT IF. He and Elmar Krieger are thanked for supplying the Yasara program. We also thank David Case for kindly granting an academic license for the AMBER 11 program. We wish further to thank the reviewers and editors for criticisms and helpful suggestions.
Footnotes
Author information
Both authors are freelance researchers without funding or sponsorship of any kind, and there are no conflicts of interest.
References
- Arteca G, Tapia O. J Phys Chem. 2002;B106:1081–1089. [Google Scholar]
- Arteca G, Tapia O. Mol Phys. 2003;101:2743–2753. [Google Scholar]
- Ball P. The importance of water. In: Smith IWM, Cockell CS, Leach S, editors. Astrochemistry and Astrobiology. Springer; Heidelberg New York Dordrecht London: 2013. [Google Scholar]
- Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- Benesch JLP, Robinson CV. Nature. 2009;462:576–577. doi: 10.1038/462576a. [DOI] [PubMed] [Google Scholar]
- Ben-Naim A. J Biomol Struct Dyn. 2012;30:113–124. doi: 10.1080/07391102.2012.674286. [DOI] [PubMed] [Google Scholar]
- Breuker K, McLafferty FW. Proc Nat Acad Sci USA. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater RP. J Biomol Struct Dyn. 2013;31:967–969. doi: 10.1080/07391102.2012.748531. [DOI] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz K, Roberts B, et al. AMBER. Vol. 11 University of California; San Francisco: 2010. [Google Scholar]
- Chen XF, Weber I, Harrison RW. J Phys Chem B. 2008;112:12073–12080. doi: 10.1021/jp802795a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goesmann F, Rosenbauer H, Bredehöft JH, Cabane M, Ehrenfreund P, Gautier T, Giri C, Krüger H, Le Roy L, MacDermott AJ, McKenna-Lawlor S, et al. Science. 2015;349(6247) doi: 10.1126/science.aab0689. [DOI] [PubMed] [Google Scholar]
- Gurnett DA. Trans Am Clin Climatol Assoc. 2009;120:299–325. [PMC free article] [PubMed] [Google Scholar]
- Herschel W. Phil Trans Roy Soc. 1784;1784:233. [Google Scholar]
- Jarrold MF. Phys Chem Chem Phys. 2007;9:1659–1671. doi: 10.1039/b612615d. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Madura JD. Mol Phys. 1985;56:1381–1392. [Google Scholar]
- Kauzmann W. Adv Prot Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Klibanov AM. Nature. 2010;409:241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Levitt M, Hirschberg R, Sharon KE, Daggett V. J Phys Chem B. 1997;101:5051–5056. [Google Scholar]
- Liu Z, Schey KL. J Am Soc Mass Spectrom. 2008;19:231–238. doi: 10.1016/j.jasms.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bagal D, Kitova EN, Schnier PD, Klassen JS. J Am Chem Soc. 2009;131:15980–15981. doi: 10.1021/ja9060454. [DOI] [PubMed] [Google Scholar]
- Meyer T, de la Cruz X, Orozco M. Structure. 2009;17:88–95. doi: 10.1016/j.str.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Nutt DR, Smith JC. J Chem Theory Comput. 2007;3:1550–1560. doi: 10.1021/ct700053u. [DOI] [PubMed] [Google Scholar]
- Patriksson A, Marklund E, Van der Spoel D. Biochemistry. 2007;46:933–945. doi: 10.1021/bi061182y. [DOI] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Gaucher EA, Sanchez-Ruiz JM. Proteins. 2014;82:887–896. doi: 10.1002/prot.24575. [DOI] [PubMed] [Google Scholar]
- Robinson CV, Sali A, Baumeister W. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- Rummel JD, Beaty DW, Jones MA, Bakermans C, Barlow NG, Boston PJ, Chevrier VF, Clark BC, de Vera JP, Gough RV, Hallsworth JE, et al. Astrobiology. 2014;14:887–968. doi: 10.1089/ast.2014.1227. [DOI] [PubMed] [Google Scholar]
- Soares CM, Teixeira VH, Baptista AM. Biophys J. 2003;84:1628–1641. doi: 10.1016/S0006-3495(03)74972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert MM, Patriksson A, Hess B, Van der Spoel D. J Mol Biol. 2005;354:173–183. doi: 10.1016/j.jmb.2005.09.030. [DOI] [PubMed] [Google Scholar]
- Smith RD, Loo JA, Edmonds CG, Barinaga CJ, Udseth HR. Anal Chem. 1990;62:882–899. doi: 10.1021/ac00208a002. [DOI] [PubMed] [Google Scholar]
- Van Aalten DMF, Amadei A, Bywater RP, Findlay JBC, Berendsen HJC, Sander C, Stouten PFW. Biophys J. 1996;70:684–692. doi: 10.1016/S0006-3495(96)79608-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wallace AR. Is Mars Habitable? A critical examination of Professor Lowell's book "Mars and Its Canals" with an alternative explanation. 1907 Available for download from http://people.wku.edu/charles.smith/wallace/S730.htm.
- Wedberg R, Abildskov J, Peters GH. J Phys Chem B. 2012;116:2575–2585. doi: 10.1021/jp211054u. [DOI] [PubMed] [Google Scholar]
- Wyttenbach T, Bowers MT. Ann Rev Phys Chem. 2007;58:511–533. doi: 10.1146/annurev.physchem.58.032806.104515. [DOI] [PubMed] [Google Scholar]