Abstract
Transmission of commensal intestinal bacteria between humans could promote health by establishing, maintaining and replenishing microbial diversity in the microbiota of an individual. Unlike pathogens, the routes of transmission for commensal bacteria remain unappreciated and poorly understood, despite the likely commonalities between both. Consequently, broad infection control measures that are designed to prevent pathogen transmission and infection, such as oversanitation and the overuse of antibiotics, may inadvertently affect human health by altering normal commensal transmission. In this Review, we discuss the mechanisms and factors that influence host-to-host transmission of the intestinal microbiota and examine how a better understanding of these processes will identify new approaches to nurture and restore transmission routes that are used by beneficial bacteria.
The human intestinal microbiota is one of the most densely populated microbial communities known to exist1. This community has important metabolic and protective roles in human health through metabolizing indigestible carbohydrates, producing vitamins, preventing infection by pathogenic bacteria and modulating host immune responses2. Bacteria form the majority of the microbial biomass in the human gastrointestinal microbiota and they carry out the majority of the metabolic activities2,3. Most of the bacteria in the gastrointestinal tract reside in the large intestine, with the rest found primarily in the small intestine and stomach4. The majority of these bacteria belong to two main phyla — the Bacteroidetes and Firmicutes. These phyla, together with the Proteobacteria, Actinobacteria, Synergistetes and Fusobacteria, contain almost all of the bacterial species found in the human gastrointestinal tract5–7. Most of these species are obligate anaerobes; however, the extent of aerotolerance varies among species in the Actinobacteria and Proteobacteria phyla8,9. Despite their abundance in the human gastrointestinal tract, these species represent only a small subset of all of the bacterial taxa on Earth10. Furthermore, many of these bacterial taxa are not found replicating outside of the intestinal environment, which reflects their adaptation to this specific niche11,12.
The factors that determine the optimal microbial community of an individual at any point in time are varied and include age, host genetics, diet and the local environment. Therefore, a core ‘healthy’ microbiota that is common to all individuals does not exist. Furthermore, the distinction between health-associated commensal bacteria and harmful pathogenic bacteria is not always clear, as some bacterial species can promote health or cause disease depending on the specific strain or their location in the body. For example, Bacteroides fragilis produces immunomodulatory capsular polysaccharides that stimulate the production of anti-inflammatory cytokines. If this bacterium translocates from the intestine to the peritoneum, then the capsular polysaccharides can cause inflammation, which results in the formation of an abscess13. Abscesses can be considered as beneficial to the host by limiting the spread of disease; however, if left untreated they can cause obstructions and further bacterial dissemination if ruptured14. Depending on the strain and the virulence factors that are present, Escherichia coli is either considered to be a normal commensal of the intestinal microbiota or a pathogen15. Similarly, the gastric bacterium Helicobacter pylori is associated with an increase in the incidence of peptic ulcers and stomach cancer, but a decreased incidence of oesophageal cancer16. In general, we consider a diverse microbiota that is abundant in beneficial species, such as members of the Bacteroidaceae, Ruminococcaceae and Lachnospiraceae families in the Bacteroidetes and Firmicutes phyla, and with few pathobionts, such as many members of the Proteobacteria phylum, indicative of a healthy state17.
The presence of the intestinal microbiota in the human gut is the result of extensive immigration and competition that continues throughout life. Facultative anaerobic bacteria initially colonize the gastrointestinal tract at birth and during the first three years of life. However, these bacteria are gradually replaced by obligate anaerobes as the gastrointestinal tract becomes more anaerobic and the infant transitions to a solid food diet18,19. The colonization success of these health-associated commensal bacteria is attributable to their ability to spread and to be maintained in human populations20. Thus, transmission is an essential feature of the human microbiota that relies on the strategies used by bacteria to exit from one host (donor) and stably colonize another (recipient) (FIG. 1). The ubiquitous and sometimes exclusive presence of this select group of enteric bacteria in human populations demonstrates the existence of host-adapted colonization processes and refined co-evolved transmission networks2,11,12.
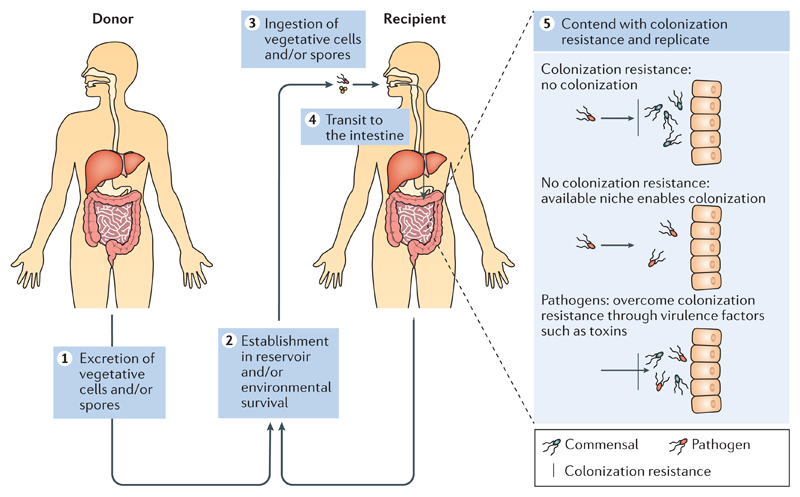
Figure 1. Transmission of pathogenic and commensal intestinal bacteria.
Intestinal pathogens and commensal bacteria use similar mechanisms to transmit between hosts. Egestion from the host in faecal matter is the first stage in transmission (step 1). To promote dispersal and subsequent ingestion by a new host, pathogens may induce diarrhoea in the donor. Once in the external environment, survival mechanisms, such as aerotolerance, viable but non-culturable dormancy and sporulation, are used by these predominately anaerobic bacteria to survive and transmit. Environmental reservoirs, such as people, food, animals and the built environment, will function as a source or sink for transmission (step 2). Once ingested by a new host (step 3), the bacterium transits to the intestines (step 4). Competition from the resident microbiota can prevent colonization (step 5, see colonization resistance); however, bacteria can colonize if a niche is unoccupied (step 5, see no colonization resistance). The restoration of bacterial species functions to maintain colonization resistance and promote the diversity of health-associated bacteria in the gut. Pathogens can overcome colonization resistance through the induction of the expression of virulence factors, such as toxins, which can lead to inflammation and perturb the resident microbiota (step 5, see pathogens). Metabolism of nutrients and replication promote persistence and support further replication and subsequent onward transmission as the recipient now becomes a donor.
Several fundamental questions that concern the transmission of the intestinal microbiota remain unanswered. How have strictly anaerobic bacteria been transmitted between humans over time to allow co-evolution to occur? In addition, how have changes in Western lifestyles during the past several decades affected the co-evolved transmission networks? At a practical level, how do we study and monitor commensal transmission so that we can preserve and nurture commensal routes of transmission to improve human health? In this Review, we discuss the routes and mechanisms that enable the intestinal microbiota to be transmitted between hosts and we describe how studies of pathogen transmission can provide a framework to study commensal transmission. We discuss the reservoirs of the intestinal microbiota and provide examples of how human activities are influencing its transmission. In addition, we outline the factors that can perturb our intestinal microbiota and the approaches that we can take to restore the microbiota to promote health. We conclude by reflecting on how modern human health regimes may be adversely affecting the transmission of health-promoting intestinal bacteria.
Routes of transmission
Most of our knowledge on the transmission mechanisms used by intestinal bacteria is derived from the study of pathogen transmission; this provides a conceptual framework to begin to understand commensal transmission21. Both commensal and pathogenic intestinal bacteria are primarily transmitted between hosts through the faecal–oral route. Commensal intestinal bacteria can also be transmitted through the vaginal–oral route at birth and through breast milk in early life. The transmission routes of commensal and pathogenic bacteria are distinguished largely by the colonization strategy that is used once inside the host. Commensal bacteria provide health benefits to the host that are a result of their colonization, whereas, depending on their virulence and infectious dose, pathogen colonization can cause disease.
Shared transmission routes of commensal and pathogenic intestinal bacteria
The first step in a typical transmission route for an intestinal pathogen is the shedding of the bacterium from the host in faecal matter, which is followed by changes in bacterial metabolism or cellular architecture to maximize survival in the external environment. The pathogen must then persist in the external environment, possibly by using reservoirs, such as animals, the built environment, water sources or food chains, to increase the likelihood of entering a new susceptible host. Once the bacterium has successfully persisted in the external aerobic environment and has been ingested by a new host, it must colonize otherwise it will rapidly transit through the gut. Colonization includes passage through the stomach, the establishment of a niche in the intestinal environment, the use of available nutrients, and replication to a level that will ensure stability and survival (FIG. 1). A newly colonized host can then become a donor for the onward transmission of that bacterium. The colonizing species will encounter competition from the resident microbiota, and this colonization resistance has important roles in preventing invasion by pathogenic bacteria and in maintaining intestinal homeostasis4. The resident microbiota can compete directly through the use of available nutrients or by the secretion of toxins that target neighbouring bacteria, as has been demonstrated for B. fragilis4,22. In addition to competition between bacterial species, the metabolism of available dietary substrates can facilitate cross-feeding between species, thus promoting cooperation and the colonization of competing species3,23,24.
It is likely that intestinal commensal bacteria use the same, or similar, strategies to those used by pathogenic bacteria to transmit between hosts. Recent evidence indicates that many of the survival mechanisms and environmental reservoirs are also common between pathogenic and commensal bacteria. Moreover, colonization factors, such as flagella and fimbriae, are also shared; these appendages are not unique to pathogens and are also a feature of commensal intestinal bacteria, including Roseburia spp. and Bifidobacterium spp.25,26. Last, sequence-based studies of pathogen transmission networks have revealed that bacteria can disseminate both locally and globally through their human hosts, which indicates that the transmission of commensal bacteria is not spatially restricted27,28.
Distinguishing the routes of transmission of commensal and pathogenic intestinal bacteria
Despite the similarities mentioned above, there are substantial differences between the mechanisms used by intestinal pathogens and commensal bacteria to transmit. Depending on the colonizing dose, host susceptibility and environment, a pathogen can exist in a low-level asymptomatic state or can induce a high-level symptomatic super-shedding state in the host29. The low-level asymptomatic state is typically associated with relatively little perturbation of the intestinal microbiota and lower levels of transmission, thus rendering the host a silent carrier of potential pathogens. Bacteria such as enteropathogenic E. coli, Vibrio cholerae and Clostridium difficile use virulence factors, such as toxins, during pathogenesis to maximize their colonization, despite causing severe inflammatory symptoms and intestinal disease. The host can restrict pathogen colonization through the secretion of antimicrobial peptides, such as neutrophil gelatinase-associated lipocalin (NGAL), which prevents microbial siderophores from binding to essential iron30. However, some pathogens can circumvent this response by producing modified siderophores, such as salmochelins, that are not bound by NGAL15. Any resulting intestinal disease typically results in substantial perturbation and instability in the commensal microbiota, which often results in diarrhoea that may promote rapid pathogen dispersal and transmission at the expense of commensal colonization and host health. Therefore, one distinction between pathogen and commensal colonization in this context is that pathogenic bacteria use a host-derived inflammation state to spread, whereas commensals do not, and therefore either decrease in number or are lost during dysbiosis31,32.
Host selection of commensal bacteria
There is emerging evidence that hosts preferentially select communities of commensal bacteria through the modulation of the intestinal environment by a combination of host genetics and immune responses. Variation in genetic profiles between individuals is known to alter many aspects of health and disease, and it is now clear that it may also influence the composition of commensal bacterial communities. Despite initial studies that concluded that human genetics does not substantially contribute to determining the bacterial species acquired 19,33, recent studies have identified the presence of bacterial species that are associated with specific genetic polymorphisms, including abundant health-associated Faecalibacterium spp.34,35. It has also been demonstrated that specific genes influence bacterial colonization. For example, expression of the fucosyltransferase 2 (FUT2) gene results in the presentation of fucosylated substrates on intestinal epithelial cells, thus enhancing the recruitment of particular species of commensal bacteria to the epithelium and protecting against the translocation of pathogenic bacteria36,37. The association between host genetics and the community composition of the microbiota remains poorly understood; however, it is now evident that host genetics may have an essential role in determining the optimal microbiota community for promoting health.
In addition to host genetics, the host immune system can distinguish between commensal and pathogenic bacteria to elicit different downstream signalling responses, through innate immune receptors, such as Toll-like receptors and nucleotide-binding oligomerization domain-like receptors (NOD-like receptors). The recognition of commensal bacteria generally promotes intestinal homeostasis, whereas the recognition of pathogens results in a pro-inflammatory response38. Studies in genetically modified mouse models have shown that the absence of caspase 3 and caspase 4, which are involved in cell apoptosis and inflammatory responses, can also substantially alter the composition of the microbiota and disease susceptibility39. Host-derived antimicrobial peptides that are produced as part of a pro-inflammatory response to pathogens have been shown to specifically recognize pathogen lipopolysaccharide (LPS) structures and do not bind to LPS on commensal bacteria, owing to an altered charge on the commensal cell surface that prevents binding40. These results demonstrate that host selection of the composition of the microbiota could be determined through host genetic background or induced in response to the presence of pathogens or commensals.
Host behaviours, such as dietary choices, may also determine the composition of the intestinal microbiota. This selection commences at birth. The presence of indigestible human milk oligosaccharides in breast milk promotes the expansion of commensal species, in particular Bifidobacterium spp., which have a wide range of glycoside hydrolases that can degrade these complex sugars before metabolizing them41,42. By importing human milk oligosaccharides into the bacterial cell before degrading them, Bifidobacterium spp. also limit nutrient availability to any pathogens that may be present in the intestinal environment41,42. The composition of the microbiota in adults can also vary substantially with diet. For example, the prevalence of Ruminococcus bromii is known to increase in people who consume diets that are high in resistant starches43,44. Taken together, it is clear that the combination of host genetics, responses to bacterial stimuli and environmental factors, such as diet, determines the current and optimal microbiota for an individual.
Survival in the environment
Once excreted from the body in faeces, intestinal bacteria must tolerate the local environment to enter and colonize a new host (FIG. 1). Environmental stresses include the toxic effects of atmospheric oxygen, ultraviolet radiation, a lack of nutrients, adverse temperatures and desiccation. Bacteria have evolved various strategies, such as a reduction or changes in metabolism, and, for some taxa, the development of a protective structure, such as a spore, to remain viable during environmental stress. These strategies are essential to ensure that commensals survive and are successfully transmitted to a new host (FIG. 2a).
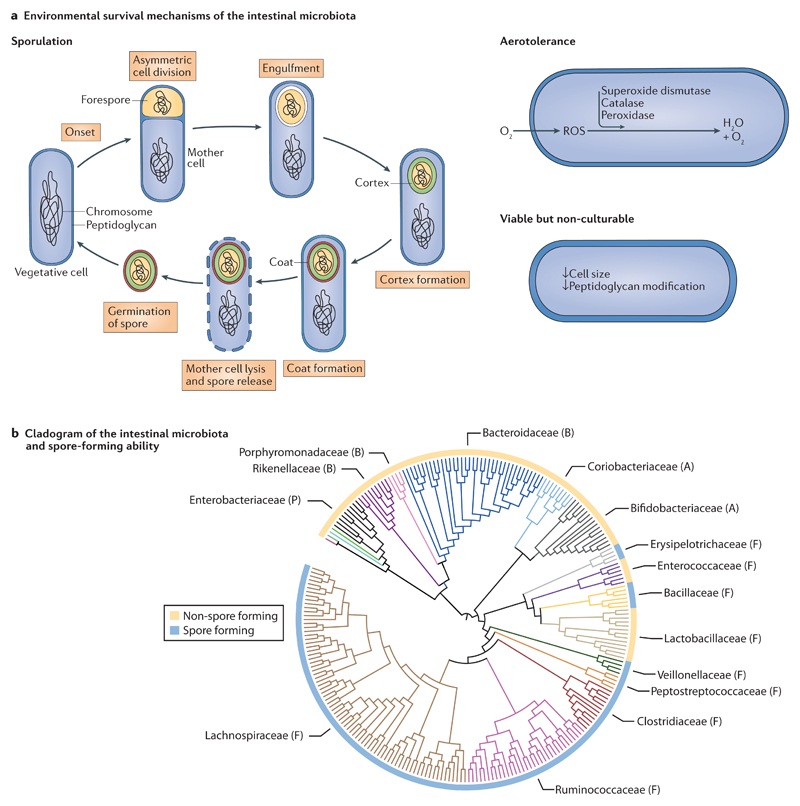
Figure 2. Environmental survival mechanisms and sporulation transmission dynamics of the intestinal microbiota.
a | Environmental survival mechanisms. Sporulation is a feature of specific members of the Firmicutes phylum and involves a series of well-defined stages that incorporate a complete structural remodelling of the bacterial cell to form a resilient spore. Aerotolerance through the degradation of harmful reactive oxygen species (ROS) that can accumulate in the bacterial cell is primarily, but not exclusively, a feature of aerobic bacteria. Presented is a simplified representation of the key elements. The viable but non-culturable state includes some morphological changes, but the extent and characteristics of this phenotype in the intestinal microbiota are unknown. b | Representative cladogram of the main human intestinal microbiota families and their associated sporulation ability. Internal clade colours represent different taxonomic families. Family names are presented, with the phylum that the family belongs to indicated by a letter in brackets: P, Proteobacteria; B, Bacteroidetes; A, Actinobacteria and F, Firmicutes. The most abundant families that have the greatest known metabolic outputs in the intestinal microbiota are the Bacteroidaceae, Ruminococcaceae and Lachnospiraceae families. The circle that surrounds the cladogram indicates known sporulation ability. Not all bacterial species in a taxon that contains spore-forming species have been demonstrated to form spores.
Sporulation
Specific members of the Firmicutes phylum produce resilient, metabolically dormant structures known as spores that are typically, but not exclusively, produced during environmental stress. Spore integrity and fecundity are maintained by the binding of DNA to small acid-soluble proteins in the spore core, which is surrounded by several durable proteinaceous layers that have low permeability and high levels of peptidoglycan45. When more favourable environmental conditions are encountered, the spore germinates, which leads to the re-formation of a vegetative cell46. Intestinal spore-forming bacteria potentially represent up to 30% of the microbial abundance in the gut and are found in several intestinal-associated bacterial families, including the Lachnospiraceae, Ruminococcaceae, Clostridiaceae and the Peptostreptococcaceae47 (FIG. 2b). Many intestinal spore-forming bacteria are known to promote homeostasis through the induction of immunomodulatory regulatory T cells48. By resisting external environmental stresses, intestinal spores facilitate host-to-host transmission through space and time (FIG. 3). Moreover, by germinating after exposure to in vivo signals, such as intestinal bile acids, spore-forming bacteria are present in a vegetative state and are able to colonize the host47. Last, the induction of sporulation may not be restricted to a stress response. For example, under normal conditions, in the absence of an environmental stress, C. difficile continuously produces spores at a low rate, which are able to persist by adhering to intestinal epithelial cells and mucin49,50. If spores of commensal intestinal bacteria could similarly persist and germinate in response to host-derived cues, this would promote colonization resistance by ensuring that the diversity of the community is maintained. Therefore, the formation of spores permits inter-host transmission, intra-host persistence and potentially contributes to the overall diversity of the intestinal microbiota.
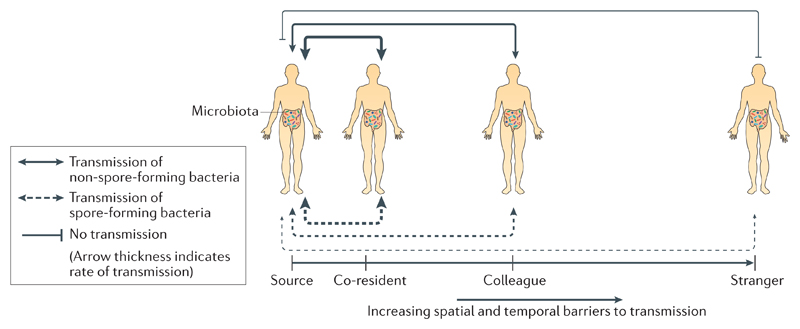
Figure 3. Inter-host transmission dynamics of spore-forming and non-spore-forming intestinal bacteria.
A hypothetical model to explain the different transmission dynamics of spore-forming and non-spore-forming bacteria. Owing to their resistance to environmental stresses and aerotolerance, spore-forming bacteria are not as spatially and temporally restricted during transmission as non-spore-forming bacteria. For individuals who are in regular contact with, and close proximity to, each other (for example, co-residents) both spore-forming bacteria and non-spore-forming bacteria can transmit with the same efficiency. However, as spatial and temporal distances increase, non-spore-forming oxygen-sensitive bacteria will become restricted in their ability to transmit until eventually transmission will not be possible. As spore-forming bacteria can remain viable for extended periods of time in external aerobic environments, they are not reliant on close contact between individuals to transmit. For example, spores that are shed by an individual can potentially be acquired by another individual weeks later.
Aerotolerance
Once shed by the host, intestinal bacteria in a vegetative state show varying levels of tolerance to atmospheric oxygen8,9,47,51–53 (Supplementary information S1 (table)). The damaging effects of oxygen in bacterial cells are due to the generation of reactive oxygen species (ROS), which damage DNA and proteins, and interfere with essential metabolic processes54. Aerobic bacteria and facultative anaerobic bacteria have evolved mechanisms to avoid and repair the damage caused by ROS, including antioxidant enzymes such as catalases, peroxidases or superoxide dismutase8,54. In a vegetative state, obligate anaerobic bacteria are typically sensitive to oxygen and may die within minutes of exposure47. Nevertheless, mechanisms to counter oxygen stress exist, even in these obligate anaerobes. Faecalibacterium prausnitzii relies on an extracellular flavin–thiol electron shuttle to grow in the presence of oxygen, which enables its survival in the oxygenated zone at the gut mucosa55. This oxygenated zone ensures that gut epithelial cells are protected from the majority of anaerobic bacteria in the lumen that could compromise the integrity of the epithelial cells56,57. The extracellular flavin–thiol electron shuttle may also promote the survival of F. prausnitzii when it is exposed to atmospheric oxygen in the presence of the antioxidants inulin, cysteine and riboflavin55,58. Other abundant intestinal bacteria, such as Roseburia spp., can only survive for a few minutes when exposed to atmospheric oxygen concentrations (Supplementary information S1 (table)). We hypothesize that Roseburia spp. either use a currently unknown survival mechanism or are extremely efficient at colonization and can readily become established in new hosts to which they are in close proximity.
Viable but non-culturable dormancy
A viable but non-culturable (VBNC) state is a form of bacterial dormancy that is not reliant on the formation of specialized resistant structures, but that typically involves a decrease in metabolic activity and the generation of a strengthened cell wall that is achieved through modifications to its peptidoglycan structure. An increase or, more typically, a decrease in cell size has also been reported59,60. These strategies all function to help bacteria withstand environmental stresses and preserve DNA integrity59–61. Similar to sporulation, the VBNC state is reversible through the removal of the inducing stress (for example, nutrient limitation or extreme temperature) or following exposure to growth stimulants, such as amino acids for E. coli or contact with intestinal cells for V. cholerae59,62. Similar to spores, VBNC bacteria can remain dormant for long periods of time. For example, Vibrio fluvialis from marine sediment was successfully cultured after 6 years of dormancy following the addition of nutrients63. The majority of VBNC bacteria that have been identified thus far are human-associated pathogens, including E. coli, Enterococcus faecium, V. cholerae, Salmonella enterica subsp. enterica serovar Typhimurium and Mycobacterium tuberculosis59,64,65. Genetic and phenotypic characterization of the VBNC state remains technically challenging, because the stimuli that are required to induce or culture bacteria from this dormant state are largely unknown or are difficult to simulate in a laboratory66. As VBNC dormancy is found in phylogenetically diverse bacterial species, it may be widespread in the intestinal microbiota and could be used as a strategy by non-spore-forming oxygen-sensitive commensal bacteria to survive in the environment until they are acquired by a new host. However, whether the VBNC state is induced in members of the commensal microbiota remains to be determined.
Reservoirs of commensal bacteria
Humans are the main reservoir of commensal intestinal bacteria, with transmission occurring readily between individuals. In addition, food, water, the environment and animals may contain microbial communities that could contribute to the human intestinal microbiota.
People
Childbirth is the first major life event in which the transmission of bacteria and colonization occur. Depending on the mode of delivery, either the birth canal of the mother, or the hospital environment and the skin of the mother provide the initial inoculum of bacteria for the infant67,68. Faecal–oral transmission could also occur during vaginal delivery, which would enable the immediate transmission of members of the intestinal microbiota to neonates at birth67. Compared with neonates that are born vaginally, it is thought that the composition of the microbiota of infants that are born by caesarean section may be more analogous to the skin microbiota than the vaginal microbiota in the early days of life68. Despite this, by six weeks, differences in the infant microbiota are determined by the body site and not the mode of delivery, which indicates that microbial convergence occurs early in life69. Evidently, there is no doubt that bacteria that can only be transmitted during vaginal delivery would be unable to colonize infants who are born by caesarean section. If no attempt is made to colonize infants who are born by caesarean section with these species, then, over generations, these species may be lost from the microbiota70. This decrease in diversity may have important health implications, as highlighted by reports that have associated immune disorders, such as asthma and allergies, in adult life with the abnormal development of the infant intestinal microbiota70,71. After childbirth, inter-host transmission of intestinal bacteria continues, as shown by people who live in the same home sharing more species in common with each other than non-residents19,72–74. The transmission rate of a bacterial species is affected by the number of hosts, their level of contact and their proximity to each other, as well as by the inherent colonization resistance of the microbiota in each individual, which is largely affected by age. A healthy adult has a broadly stable and resilient intestinal microbiota compared with an infant whose intestinal microbiota is still developing72,75.
Outside of family units, the effects of social interactions on the acquisition of the microbiota in large groups are best understood in non-human primates76,77. Similar to humans, these are social animals that live and interact with each other in defined communities, and the composition of their intestinal microbiotas are influenced by the interactions of the social group. The higher the incidence of social interaction between individuals the more similar the composition of their intestinal microbiota, with species diversity increasing accordingly76,77. The prevalence of anaerobic non-spore forming bacteria in baboons was associated with close social interactions between grooming pairs76. Although humans do not engage in social grooming, we physically interact through socially acceptable activities such as hand shaking, hugging and kissing, the frequency and intimacy of which increase as an individual interacts with a close family member or friend compared with a stranger. Thus, there is likely to be several social and cultural factors that contribute to the transmission of our intestinal microbiota.
Food
Although the microbiota of an individual is largely structured and influenced by their diet, the microorganisms that are carried in food can also contribute to the intestinal microbiota78. From early life, infants acquire up to 8 × 106 bacteria daily, including intestinal-associated bacteria, through breast milk79–81. The mechanism by which intestinal bacteria translocate from the gut to the breast is unknown; however, an entero–mammary pathway that is facilitated by phagocytes that sample the gut lumen and subsequently translocate to the breast through the bloodstream has been proposed82–84. Studies of various foods by culture-based methods have estimated that adults consume between 106 and 109 microorganisms daily85. Although most of the bacteria that are ingested do not survive transit through the stomach, those that do are not thought to colonize the gastrointestinal tract long-term86. The diversity of the microbiota that is acquired through food is dependent on diet85,87; therefore, food provides a source of both exogenous bacterial species and genes for the resident microbiota to acquire through horizontal gene transfer88.
Probiotic bacteria, typically Bifidobacterium spp. and Lactobacillus spp., can be used in an attempt to promote health and have been shown to alleviate the symptoms of several illnesses, including infectious diarrhoea and atopic eczema89–91. The long-term colonization efficiency of most probiotic bacterial species is variable 92, 93; therefore, regular ingestion of probiotics is required to make a substantial long-term contribution to health81,94. However, stable gut colonization by Bifidobacterium longum six months after ingestion has been observed, which was attributed to the presence of an unoccupied niche that was vacated by a species that has similar carbohydrate-metabolizing capabilities95. Overall, the variability in probiotic efficacy, coupled with host-specific responses to probiotics, means that the health benefits of ingesting these bacteria are not fully understood or predictable96,97.
Water
Water is a major environmental reservoir for several intestinal bacterial pathogens, such as Shigella flexneri, Shigella sonnei and V. cholerae, which can cause debilitating gastrointestinal disease98. However, little is known about the fate or the effect that commensal intestinal microorganisms that are found in water have on human health. The identification of bacteria in drinking water has primarily focused on pathogens, especially readily detectable indicator microorganisms, such as E. coli; however, the distinction between commensal and pathogenic strains of this species is not always made99. Despite an emphasis on pathogen detection, sequence-based culture-independent approaches have identified human-associated Blautia spp. in rivers100. Thus, it is clear that these species are transmitted through water; however, after the appropriate water treatment procedures, any strictly anaerobic non-spore-forming intestinal bacteria are likely to be killed, and these bacteria are therefore expected to have a low transmissibility and colonization potential. Nevertheless, the full extent of the transmission of commensal intestinal bacteria through water is currently unknown.
Animals
For most people, pets are the main source of animal-derived microbiota. The microbiotas of dogs and cats include taxa that are also found in the human microbiota; for example, genera such as Roseburia, Faecalibacterium, Bacteroides, Prevotella and Ruminoccocus101,102. Farm animals are an additional source of bacteria. Analysis of the porcine intestinal microbiota has revealed similarities in taxonomic groups and functional capabilities with the human intestinal microbiota103,104. Several human-associated pathogens, such as Salmonella enterica subsp. enterica serovar Enteritidis, Campylobacter jejuni, enteropathogenic E. coli and C. difficile 105–107, are transmitted between animals and humans; therefore, the potential for animals to transmit commensal species of bacteria is plausible. The treatment of animals that are to be used as food with antibiotics has also been linked to the acquisition of antibiotic-resistant strains of bacteria in humans108,109. This highlights the need to recognize that human health can be influenced through various diverse sources that are not directly connected to our own personal health decisions.
Built environment
Both buildings and transport systems adsorb our microbiota, which creates opportunities for microbial transmission across vast spatial areas and diverse human populations110–113. Humans are one of the main sources of indoor airborne bacteria that can spread through ventilation systems114. Outdoor air can also enter a building passively110,115. Once bacteria become airborne (for example, through flushing a toilet or using a shower), viable bacteria can disperse around a room116,117. In the built environment, the greatest density of human-associated bacteria will probably be found in bathrooms. Bacteria are abundant on surfaces that have been touched by human hands, on toilet seats or on floors118,119. Skin-associated bacteria are the most dominant species on bathroom surfaces; however, a high proportion of intestinal-associated bacteria have also been found, such as members of the Bacteroidaceae, Prevotellaceae, Ruminococcaceae and Lachnospiraceae families. The presence of these intestinal-associated bacteria, together with poor hand-washing procedures, provides a reservoir for bacteria in the built environment that have the potential to transmit to humans120.
A limitation of most of the studies on bacteria in the built environment is the lack of distinction between viable bacteria that have the potential to successfully colonize a new host and non-viable bacteria, which do not. It is estimated that only 1–10% of bacterial cells that are detected by culture-independent methods are viable121. Although culture-based methods can detect the viability of bacterial cells, the bacteria obtained will be an underrepresentation of the overall diversity in the sample121,122. Knowledge of the influence of the built environment on the transmission of the intestinal microbiota is currently incomplete, but as research in this discipline increases its influence and importance will become more apparent114.
Microbiota perturbation and restoration
Throughout life, our microbiota experiences perturbations that can alter or damage its composition and functions86,123–126. Nevertheless, on the basis of current DNA sequencing and analysis technologies, the composition of the microbiota in an individual is largely stable once established127,128. This observation raises important questions about microbiota perturbation, restoration and, ultimately, microbial transmission. How is a depleted microbiota restored following perturbation? How can we promote recolonization by specific health-associated microorganisms instead of pathogens? To help answer these questions, tools are required that enable the precise tracking of bacterial strain movements and restoration once transmission is blocked (BOX 1).
Box 1. Methods to investigate human microbiota transmission.
To understand the importance, extent and the mechanisms of microbiota transmission, researchers will need to borrow concepts and use tools from the field of host–pathogen interactions. For example, mechanistic studies on host–pathogen interactions have relied on the ability of researchers to culture and genetically modify a pathogen, and on in vitro and in vivo models to test hypotheses using ‘molecular Koch’s postulates’ (REF. 152). Furthermore, genomic epidemiology tools that can distinguish between pathogenic strains and enable pathogens to be tracked at strain-level resolution as they persist, evolve and transmit within human populations could be applied to similar studies of gut commensals28,153,154. The recent renaissance in anaerobic bacterial culturing has provided an opportunity to develop methods to study the transmission of the human microbiota through the isolation, purification and sequencing of the majority of the species of gut bacteria47,145–147,155. We are now in a position to construct a molecular biology toolbox for the genetic manipulation of unstudied commensals, to explore the molecular basis of how these bacteria promote health and are transmitted in human populations. Gnotobiotic mouse models and rodents that have a human microbiota are also valuable resources, as they provide a tractable system that can monitor microbiota dynamics, such as examining the effects of different diets on the composition of the microbiota over time134.
The availability of reference genome databases that contain accurate de novo assemblies of previously uncultured, and therefore largely uncharacterized, bacterial species is essential for understanding microbial transmission within human populations. The potential for metagenomics to resolve microbial community structures beyond the species level74,156 and across taxonomic kingdoms, means that it is a particularly useful approach for the study of microbiota transmission between individuals. Microbial profiling also enables the identification of marker bacterial species or strains that are predictive of diseases such as inflammatory disorders or malnutrition125,157. When combined with large-scale sampling by the public (citizen science), this technology enables the monitoring of health-associated and disease-associated bacterial species in populations158. Investment in tools, resources and expertise to promote metagenomics as an alternative to 16S rRNA gene amplicon sequencing for microbial community profiling is therefore worthwhile. User-friendly and/or open-source software and pipelines for analysing metagenomics data are needed and are becoming available as the use of metagenomics to study microbial communities gains support among microbiota researchers74,159.
Depending on the extent of the perturbation in the microbiota, and subsequent exposure to bacteria, the composition of the microbiota may be restored to a similar state or assume a new stable state that is composed of different bacterial species129. Therefore, a perturbation in the community will provide an opportunity for an externally derived bacterium to establish itself by reducing or eliminating competition from a resident species that occupies the same niche and requires the same nutrients95,130. Factors that affect microbiota perturbation are varied and range from antibiotic use, infection with a pathogen, a change in diet or travel18,86,123,131. Changes in the composition of the microbiota have mostly been studied at the individual level; however, there is increasing evidence that suggests that changes in Western lifestyles and diet are altering the intestinal microbiota at larger population levels. Recently, it was observed that many traditional rural hunter–gatherer societies and agrarian groups that follow non-Western social behaviours and do not commonly use antibiotics or disinfectants have a more diverse intestinal microbiota that includes bacterial species that are now absent from the intestinal microbiota of developed world populations132,133.
Any perturbation in an individual that eliminates certain bacterial species, or selects for some at the expense of other species, will prevent further onward transmission to other hosts70,134. If the perturbation happens at the population level then the effects may be compounded at a larger scale. For example, the consumption of a high-fat low-fibre diet, which is typical of Western populations, has been shown to cause the extinction of intestinal bacteria in mice if the diet is consumed over several generations134. Similarly, the use of antibiotics can negatively affect the diversity of intestinal bacteria, with repeated use preventing the restoration of the microbiota124. Although antibiotics and disinfection measures are essential for disease control and a high-calorie diet has greatly decreased undernutrititon in Western societies; in this context, these changes may result in the indiscriminate elimination of commensal species, which could affect the diversity of the microbiota and microbial transmission70,135. Indeed, a study in which the intestinal microbiotas of individuals who resided in either the United States or traditional agrarian societies in Papua New Guinea were compared attributed a lower α-diversity within, and higher β-diversity between, individuals in the United States cohort to decreased inter-host microbial transmission136.
In addition to the observed decrease in the diversity of the intestinal microbiota in Western societies, an increase in autoimmune and allergic diseases in the developed world has been observed70,137,138. Originally termed the ‘hygiene hypothesis’, there is increasing evidence in humans and animal models that exposure to microorganisms early in life promotes the maturation of the immune system and decreases the incidence of autoimmune-related diseases137,139–141. Consistent with this hypothesis, the use of antibiotics in childhood has been associated with an increased likelihood of developing paediatric inflammatory bowel disease and a predisposition to asthma and obesity in later life124,142. These examples illustrate the importance of efficient microbial transmission networks and the potential effect on human health when they fail.
Direct interventions currently provide the most immediate solution to establish or restore a diverse and beneficially functional microbiota across all age groups. Recent interventions in this area have included swabbing neonates born by caesarean section with gauze that has been pre-incubated in the vagina of the mother to mimic the natural transmission of the vaginal microbiota to the child143. In adults who are susceptible to recurrent infections with C. difficile, faecal microbiota transplantation (FMT) from a healthy donor has proven extremely effective at resolving such infections144 (BOX 2). As the number of human gut commensal species that have been isolated and archived as pure cultures continues to increase47,145–147, the development of live biotherapeutics (BOX 2) for the treatment of disorders other than C. difficile infection will become feasible. Next-generation probiotics and functional foods that make use of the numerous diverse beneficial bacteria other than the widely used Lactobacillus spp. and Bifidobacterium spp., could potentially provide health benefits to individuals and to the wider interconnected human population. The cumulative potential impact of this new field of biotherapeutics and functional foods is immense, particularly as the health effect of the intestinal microbiota extends beyond intestinal diseases to immune development and our general well-being2.
Box 2. Artificial transmission: faecal microbiota transplantation to treat disease.
Chronic pathological imbalances in the composition of the intestinal microbiota (dysbiosis) are associated with a range of clinical symptoms, diseases and poorly understood syndromes. Therefore, methods for effective microbiota replacement, restoration and manipulation are at the forefront of translational research in this field. Much of the pioneering microbiota transplantation work has focused on developing treatments for recurrent Clostridium difficile infection (CDI). Individuals are particularly susceptible to CDI after perturbation of their commensal microbiota, typically following a course of antibiotics160. In cases in which antibiotic treatment fails and CDI recurs, faecal microbiota transplantation (FMT) provides another treatment option144. During FMT, stool from a healthy individual is collected and screened for known pathogens before being transferred to a patient. Despite its unpleasant nature and lack of standardization, FMT has proven extremely efficacious for the prevention of recurrent CDI, with reported success rates of >80%, which has prompted the establishment of stool banks, clinical guidelines and regulation for FMT in Europe and the United States144,161–163.
Nevertheless, owing to the inherent variation in microbial species among healthy donors, universal standardization of FMT is challenging. Issues such as optimal donor selection and methods for stool collection, pathogen screening, processing, storage and therapeutic administration have therefore stimulated the development of more standardized alternatives162,164. Chief among these are ‘live biotherapeutics’, which are biological products that contain viable defined mixtures of microorganisms that are intended for the prophylactic and therapeutic treatment of disease165. With promising proof-of-concept results in mouse models and humans, and the increase in commercial activity to develop such medicines, the development of live biotherapeutics for the treatment of CDI remains an active area of research131,148,166.
The successes of FMT and live biotherapeutics are largely attributed to the restoration of a healthy and functional gut microbiota in individuals who have severe dysbiosis131,167. A feature of FMT in particular is the substantial and sudden restorative shift in the microbiota, which would never be achievable through natural transmission alone. Post-FMT, the microbiota profile of the recipient often has a remarkable similarity to the microbiota profile of the donor167. Moreover, strains that originate from the donor can coexist with conspecific strains of the recipient months after FMT156. Thus, the transplanted microbiota may restore health by reintroducing bacterial taxa that were absent from the recipient before FMT, supporting the expansion of the recipient’s own commensal taxa and eventually re-establishing an optimal microbial community that is suited to their genetics, diet and lifestyle.
At a broader level, changes in living practices can promote the transmission of, and colonization by, health-associated commensals at the expense of pathogen colonization. A course of broad-spectrum antibiotics to treat a gastrointestinal infection also eliminates many beneficial commensals, thus rendering the microbiota-deficient host susceptible to infections with other pathogens131,148. Indeed, narrow-spectrum antibiotics, or antimicrobials that have specific targets, such as bacteriocins or phage therapies, are desirable and under development as alternatives to treatment with broad-spectrum antibiotics149. The effects of antibacterial hygiene products and hand sanitizers on the intestinal microbiota are unclear and require further study150; however, efforts to use them more selectively may reduce the depletion of commensal bacteria. Hygiene practices currently act to decrease the total number of bacteria on a surface or individual, whereas a more targeted approach that specifically removes pathogenic microorganisms should be given greater consideration151. We anticipate that with more research we should be able to outline new approaches to safely preserve and nurture the transmission of commensal bacteria while maintaining protection from pathogens.
Conclusions and future perspectives
The factors that influence our intestinal microbiota are becoming more apparent, partly owing to technological advances in microbiology, genomics and bioinformatics, and partly owing to the realization that assembling and maintaining a healthy intestinal microbiota may depend not only on our diet, lifestyle choices and general health but also on the microbiota and the health of others. The health status of the donors that we acquire our microbiota from may affect the composition of our own intestinal microbiota. In theory, donors that have the greatest diversity of commensal bacteria in the highest numbers are most likely to replenish the depleted microbiota of potential recipients (FIG. 4). Suboptimal donors were once healthy donors, but antibiotic exposure or other disease conditions caused a decrease in the diversity of the intestinal microbiota. The microbiota of suboptimal donors may potentially include higher levels of pathogens, which may be transmitted at a higher frequency than in healthy donors.
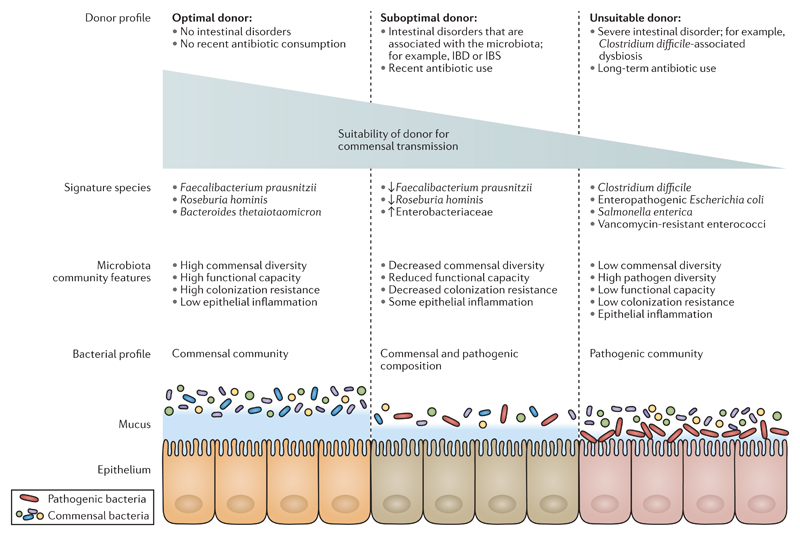
Figure 4. Transmission of commensal intestinal bacteria is influenced by donor health status.
Healthy donors who have no history of intestinal disorders or recent antibiotic treatment will typically have a diverse intestinal microbiota that exhibits high colonization resistance. Healthy donors are optimal donors of commensal microorganisms because they will regularly contribute health-associated bacteria to their environment. Conversely, donors who have lower levels of commensal diversity, decreased colonization resistance and a higher proportion of pathogenic bacteria are not considered optimal donors. These suboptimal and unsuitable donors would be more likely to shed pathogenic bacteria into the external environment that are not beneficial to human health. The signature species that categorize donors in this model are not comprehensive and are included on the basis of current research in the field. IBD, inflammatory bowel disease; IBS, irritable bowel syndrome. Figure is adapted with permission from REF.4, Wiley.
In this Review, we propose the concept of ‘spreading health through our microbiota’ as a basis to nurture, preserve and restore our microbiota to promote health. Given the potential for long-term consequences for health at all stages of life, we suggest that a greater understanding of the transmission of commensal microbiota should become a public health priority. We also advocate for lifestyle choices that promote the transmission of beneficial bacteria between individuals as a means to preserve and restore health. Traditionally, the study of bacterial transmission networks has focused on pathogens because restricting pathogen transmission is important for preventing the spread of disease. The tools and knowledge that we have acquired from these efforts can also be applied to the study of intestinal commensal transmission. The challenge moving forward will be to apply this knowledge to demonstrate and validate the hypothesis that commensal bacteria spread health.
Supplementary Material
Acknowledgements
The authors thank A. Zhu for critical reading of the manuscript during preparation. H.P.B. is funded through a Medical Research Council (MRC) grant (PF451), B.A.N. is funded through a Wellcome Trust grant (098051), S.C.F. is funded through an Australian National Health and Medical Research Council (NHMRC) grant (1091097). Work in the Lawley laboratory is supported through a MRC grant (PF451) and a Wellcome Trust grant (098051).
Footnotes
Competing interests statement
The authors declare no competing interests.
Peritoneum
A membrane that lines the abdominal cavity and provides a supportive role to internal body organs, including those of the gastrointestinal tract.
Pathobionts
Members of the commensal microbiota that may become pathogenic under certain circumstances.
Facultative anaerobic bacteria
Species that can grow and survive in aerobic and anaerobic conditions.
Infectious dose
The minimum number of bacteria required to cause an infection in a host.
Colonization resistance
The capacity of the resident microbiota to prevent the establishment of new species within the community, particularly the establishment of pathogens. Colonization resistance is a feature of a stable health-associated microbiota.
Homeostasis
A stable state. The overall maintenance of precise conditions in the microbiota that promote colonization resistance, even when subjected to external perturbations or stresses.
Colonizing dose
The minimum number of bacteria required to stably colonize a new host.
Super-shedding state
A host state, typically associated with infection by pathogens, that results in the release of numerous bacteria or spores into the external environment.
Antimicrobial peptides
A diverse range of proteins that are secreted by the host as a defence mechanism against pathogens or by microorganisms to target other microorganisms in close proximity.
Siderophores
Low-molecular-weight, iron-chelating agents that are secreted by bacteria and fungi to acquire iron from the surrounding environment.
Dysbiosis
A low-diversity microbiota with reduced colonization resistance that is typically associated with inflammation and outgrowth of facultative anaerobic Proteobacteria and pathogens.
Fucosylated
The attachment of a fucose molecule to a protein. Fucosylation of epithelial cells by the host can provide a protective role through the subsequent recruitment of commensal bacteria.
Toll-like receptors
A family of transmembrane protein receptors, characterized by the presence of a Toll and interleukin-1 receptor (TIR) domain, that recognize specific microorganism-associated molecular patterns and initiate an immune response.
Nucleotide-binding oligomerization domain-like receptors
(NOD-like receptors). A family of intracellular protein receptors that recognize microorganism-associated molecular patterns and initiate an immune response.
Desiccation
The process of liquid removal or drying out, which is usually deleterious to a bacterial cell.
Vegetative cell
The form of a bacterial cell that reproduces through binary fission.
Aerobic bacteria
Species that can only grow and survive in the presence of oxygen.
Obligate anaerobic bacteria
Species that can only grow and survive in the absence of oxygen.
Flavin–thiol electron shuttle
A process that involves the transfer of electrons to oxygen through riboflavin and thiol, which enables survival and growth in the presence of oxygen.
Social grooming
Cleaning and grooming carried out by animals, particularly primates, on other individuals in their community, which has hygienic and social roles.
Horizontal gene transfer
The transfer of genetic material between different strains or species that occurs independently of vertical transmission during replication.
Probiotics
Live microorganisms that when administered in adequate amounts confer a health benefit to the host.
Indicator microorganisms
Microorganisms that are used to assay hygiene levels of foods or water, in which the quantity of the microorganisms present is inversely related to the quality or hygiene level of the product being tested.
Undernutrition
A situation whereby an individual does not consume enough nutrients, which can have adverse effects on health.
α-diversity
The ecological diversity at a single site, as measured by the number of different species and their abundance.
β-diversity
A measure of the difference in ecological diversity between different sites.
Functional foods
Foods that contain additional elements to promote health, such as probiotics, prebiotics, vitamins or minerals.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 3.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 4.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [This landmark study characterizes the microbiota across different human body sites.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [This study provides a detailed description and phylogeny of more than 1,000 cultured species from the intestinal microbiota.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolfe RD, Hentges DJ, Campbell BJ, Barrett JT. Factors related to the oxygen tolerance of anaerobic bacteria. Appl Environ Microbiol. 1978;36:306–313. doi: 10.1128/aem.36.2.306-313.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andriantsoanirina V, Allano S, Butel MJ, Aires J. Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe. 2013;21:39–42. doi: 10.1016/j.anaerobe.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 11.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meehan CJ, Beiko RGA. Phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surana NK, Kasper DL. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol Rev. 2012;245:13–26. doi: 10.1111/j.1600-065X.2011.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow W-H, et al. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 1998;58:588–590. [PubMed] [Google Scholar]
- 17.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2010;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falkow S. Who speaks for the microbes? Emerg Infect Dis. 1998;4:495–497. doi: 10.3201/eid0403.980342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkow S. What is a pathogen? ASM News. 1997;63:359–365. [Google Scholar]
- 22.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turroni F, et al. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett. 2014;357:23–33. doi: 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 26.Neville BA, et al. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS ONE. 2013;8:e68919. doi: 10.1371/journal.pone.0068919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat Rev Microbiol. 2008;6:904–912. doi: 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 31.Stecher B, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera-Chávez F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2008;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turpin W, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 36.Pham TA, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bry L, Falk PG, Midtvedt T, Gordon JIA. Model of host–microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 38.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Brinkman BM, et al. Gut microbiota affects sensitivity to acute DSS-induced colitis independently of host genotype. Inflamm Bowel Dis. 2013;19:2560–2567. doi: 10.1097/MIB.0b013e3182a8759a. [DOI] [PubMed] [Google Scholar]
- 40.Cullen TW, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charbonneau MR, et al. A microbial perspective of human developmental biology. Nature. 2016;535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 45.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 47.Browne HP, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [This study shows that a selection of chloroform-resistant bacteria from human faeces can induce regulatory T cells in mice and have potential therapeutic properties through the alleviation of colitis in mice.] [DOI] [PubMed] [Google Scholar]
- 49.Janoir C, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun. 2013;81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mora-Uribe P, et al. Characterization of the adherence of Clostridium difficile spores: the integrity of the outermost layer affects adherence properties of spores of the epidemic strain R20291 to components of the intestinal mucosa. Front Cell Infect Microbiol. 2016;6:99. doi: 10.3389/fcimb.2016.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duncan SH, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 52.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 53.Tally FP, Stewart PR, Sutter VL, Rosenblatt JE. Oxygen tolerance of fresh clinical anaerobic bacteria. J Clin Microbiol. 1975;1:161–164. doi: 10.1128/jcm.1.2.161-164.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan MT, et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 2012;6:1578–1585. doi: 10.1038/ismej.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan MT, van Dijl JM, Harmsen HJM. Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium Faecalibacterium prausnitzii alive at ambient air. PLoS ONE. 2014;9:e96097. doi: 10.1371/journal.pone.0096097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Signoretto C, Lleo MM, Tafi MC, Canepari P. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl Environ Microbiol. 2000;66:1953–1959. doi: 10.1128/aem.66.5.1953-1959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rittershaus ES, Baek S-H, Sassetti CM. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 2013;13:643–651. doi: 10.1016/j.chom.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senoh M, et al. Conversion of viable but nonculturable Vibrio cholerae to the culturable state by co-culture with eukaryotic cells. Microbiol Immunol. 2010;54:502–507. doi: 10.1111/j.1348-0421.2010.00245.x. [DOI] [PubMed] [Google Scholar]
- 63.Amel BK, Amine B, Amina B. Survival of Vibrio fluvialis in seawater under starvation conditions. Microbiol Res. 2008;163:323–328. doi: 10.1016/j.micres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Xu HS, et al. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 65.Gupte AR, De Rezende CL, Joseph SW. Induction and resuscitation of viable but nonculturable Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol. 2003;69:6669–6675. doi: 10.1128/AEM.69.11.6669-6675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Microbiol. 2010;8:890–896. doi: 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 67.Backhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu DM, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [This perspective proposes that changes in human ecology can compromise the composition of the human microbiota with associated negative effects on human health.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2014;2:25. doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song SJ, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [In this study, a SNP-based metagenomic pipeline is developed to demonstrate mother-to-infant vertical transmission, with early-colonizing bacteria being predominately non-spore forming and late-colonizing bacteria being predominately spore-forming, which indicates temporal environmental survival by spore-forming bacteria.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faith JJ, Colombel J-F, Gordon JI. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proc Natl Acad Sci USA. 2015;112:633–640. doi: 10.1073/pnas.1418781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tung J, et al. Social networks predict gut microbiome composition in wild baboons. eLife. 2015;4:e05224. doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moeller AH, et al. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2:e1500997. doi: 10.1126/sciadv.1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heikkila MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95:471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- 80.Fernandez L, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 82.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 83.Vazquez-Torres A, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 84.Martin R, et al. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol. 2004;15:121–127. [Google Scholar]
- 85.Lang JM, Eisen JA, Zivkovic AM. The microbes we eat: abundance and taxonomy of microbes consumed in a day’s worth of meals for three diet types. PeerJ. 2014;2:e659. doi: 10.7717/peerj.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leff JW, Fierer N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE. 2013;8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brito IL, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kalliomaki M, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 90.Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. 2010;11:CD003048. doi: 10.1002/14651858.CD003048.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hill C, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 92.Malinen E, et al. PCR-ELISAII: analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Syst Appl Microbiol. 2002;25:249–258. doi: 10.1078/0723-2020-00117. [DOI] [PubMed] [Google Scholar]
- 93.Charbonneau D, Gibb RD, Quigley EM. Fecal excretion of Bifidobacterium infantis 35624 and changes in fecal microbiota after eight weeks of oral supplementation with encapsulated probiotic. Gut Microbes. 2013;4:201–211. doi: 10.4161/gmic.24196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maldonado-Gómez MX, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 96.Reid G, et al. Responders and non-responders to probiotic interventions: how can we improve the odds? Gut Microbes. 2010;1:200–204. doi: 10.4161/gmic.1.3.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sanders ME, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Natchu UC, Bhatnagar S. Diarrhoea in children: identifying the cause and burden. Lancet. 2013;382:184–186. doi: 10.1016/S0140-6736(13)60941-1. [DOI] [PubMed] [Google Scholar]
- 99.Tallon P, Magajna B, Lofranco C, Leung KT. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Poll. 2005;166:139–166. [Google Scholar]
- 100.Koskey AM, et al. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep. 2014;6:696–704. doi: 10.1111/1758-2229.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol. 2011;76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 102.Hand D, Wallis C, Colyer A, Penn CW. Pyrosequencing the canine faecal microbiota: breadth and depth of biodiversity. PLoS ONE. 2013;8:e53115. doi: 10.1371/journal.pone.0053115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamendella R, Domingo JW, Ghosh S, Martinson J, Oerther DB. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. 2011;11:103. doi: 10.1186/1471-2180-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moller-Stray J, et al. Two outbreaks of diarrhoea in nurseries in Norway after farm visits, April to May 2009. Euro Surveill. 2012;17:20321. doi: 10.2807/ese.17.47.20321-en. [DOI] [PubMed] [Google Scholar]
- 106.Toro M, et al. Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl Environ Microbiol. 2016;82:6223–6232. doi: 10.1128/AEM.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Knetsch CW, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 2014;19:20954. doi: 10.2807/1560-7917.es2014.19.45.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iovine NM, Blaser MJ. Antibiotics in animal feed and spread of resistant Campylobacter from poultry to humans. Emerg Infect Dis. 2004;10:1158–1189. doi: 10.3201/eid1006.040403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta A, et al. Antimicrobial resistance among Campylobacter strains, United States, 1997– 2001. Emerg Infect Dis. 2004;10:1102–1109. doi: 10.3201/eid1006.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dunn RR, Fierer N, Henley JB, Leff JW, Menninger HL. Home life: factors structuring the bacterial diversity found within and between homes. PLoS ONE. 2013;8:e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lax S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meadow JF, et al. Bacterial communities on classroom surfaces vary with human contact. Microbiome. 2014;2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hsu T, et al. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems. 2016;1:e00018–16. doi: 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kelley ST, Gilbert JA. Studying the microbiology of the indoor environment. Genome Biol. 2013;14:202. doi: 10.1186/gb-2013-14-2-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kembel SW, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barker J, Jones MV. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J Appl Microbiol. 2005;99:339–347. doi: 10.1111/j.1365-2672.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 117.Perkins SD, Mayfield J, Fraser V, Angenent LT. Potentially pathogenic bacteria in shower water and air of a stem cell transplant unit. Appl Environ Microbiol. 2009;75:5363–5372. doi: 10.1128/AEM.00658-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Flores GE, et al. Microbial biogeography of public restroom surfaces. PLoS ONE. 2011;6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gibbons SM, et al. Ecological succession and viability of human-associated microbiota on restroom surfaces. Appl Environ Microbiol. 2015;81:765–773. doi: 10.1128/AEM.03117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Snelling AM, Saville T, Stevens D, Beggs CB. Comparative evaluation of the hygienic efficacy of an ultra-rapid hand dryer versus conventional warm air hand dryers. J Appl Microbiol. 2010;110:19–26. doi: 10.1111/j.1365-2672.2010.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vaishampayan P, et al. New perspectives on viable microbial communities in low-biomass cleanroom environments. ISME J. 2013;7:312–324. doi: 10.1038/ismej.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsai FC, Macher JM. Concentrations of airborne culturable bacteria in 100 large US office buildings from the BASE study. Indoor Air. 2005;15:71–81. doi: 10.1111/j.1600-0668.2005.00346.x. [DOI] [PubMed] [Google Scholar]
- 123.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Korpela K, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. 2016;7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 126.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2012;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [This review uses ecological metacommunity theory to provide a framework to understand how the intestinal microbiota is assembled and to predict how it responds to disturbances.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clemente JC, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Obregon-Tito AJ, et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6:6505. doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martinez I, et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 137.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [ Supporting the hygiene hypothesis, this study describes lower rates of hay fever and eczema in large families, which is associated with reduced hygiene among siblings in larger families. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beasley R. Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet. 1998;351:1225–1232. [PubMed] [Google Scholar]
- 139.Stiemsma L, Reynolds L, Turvey S, Finlay B. The hygiene hypothesis: current perspectives and future therapies. Immunotargets Ther. 2015;4:143–157. doi: 10.2147/ITT.S61528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 141.Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 2006;7:956–960. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 143.Dominguez-Bello MG, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [ This study demonstrates the efficacy of faecal microbiota transplants to resolve C. difficile infection in a controlled clinical trial in humans. ] [DOI] [PubMed] [Google Scholar]
- 145.Lagier J-C, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Rettedal EA, Gumpert H, Sommer MOA. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5:4714. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 147.Lau JT, et al. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 2016;8:72. doi: 10.1186/s13073-016-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014;517:205–208. doi: 10.1038/nature13828. [ This study shows that a single bacterial species, Clostridium scindens, can promote resistance to infection with C. difficile in a mouse model through the synthesis of secondary bile acids that are inhibitory to this species, thus demonstrating the potential of defined bacteria-based therapies for the treatment of intestinal-associated disease. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Czaplewski L, et al. Alternatives to antibiotics — a pipeline portfolio review. Lancet Infect Dis. 2016;16:239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 150.Yee AL, Gilbert JA. Microbiome. Is triclosan harming your microbiome? Science. 2016;353:348–349. doi: 10.1126/science.aag2698. [DOI] [PubMed] [Google Scholar]
- 151.Vandegrift R, et al. Cleanliness in context: reconciling hygiene with a modern microbial perspective. bioRxiv. 2016 doi: 10.1101/095745. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Falkow S. Molecular Koch’s postulates applied to bacterial pathogenicity — a personal recollection 15 years later. Nat Rev Microbiol. 2004;2:67–72. doi: 10.1038/nrmicro799. [DOI] [PubMed] [Google Scholar]
- 153.Parkhill J, Wren BW. Bacterial epidemiology and biology — lessons from genome sequencing. Genome Biol. 2011;12:230. doi: 10.1186/gb-2011-12-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brito IL, Alm EJ. Tracking strains in the microbiome: insights from metagenomics and models. Front Microbiol. 2016;7:712. doi: 10.3389/fmicb.2016.00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014;22:267–274. doi: 10.1016/j.tim.2014.03.001. [ This review highlights the importance of a multidisciplinary approach that combines metagenomic sequencing, bioinformatics and culturing to study the human intestinal microbiota. ] [DOI] [PubMed] [Google Scholar]
- 156.Li SS, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 157.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lax S, Nagler CR, Gilbert JA. Our interface with the built environment: immunity and the indoor microbiota. Trends Immunol. 2015;36:121–123. doi: 10.1016/j.it.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Forster SC, et al. HPMCD: the database of human microbial communities from metagenomic datasets and microbial reference genomes. Nucleic Acids Res. 2015;44:D604–D609. doi: 10.1093/nar/gkv1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 161.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 162.Vyas D, Aekka A, Vyas A. Fecal transplant policy and legislation. World J Gastroenterol. 2015;21:6–11. doi: 10.3748/wjg.v21.i1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Petrof EO, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Allegretti JR, Hamilton MJ. Restoring the gut microbiome for the treatment of inflammatory bowel diseases. World J Gastroenterol. 2014;20:3468–3474. doi: 10.3748/wjg.v20.i13.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–315. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- 166.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;1:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 167.Jalanka J, et al. Long-term effects on luminal and mucosal microbiota and commonly acquired taxa in faecal microbiota transplantation for recurrent Clostridium difficile infection. BMC Med. 2016;14:155. doi: 10.1186/s12916-016-0698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.